Non-Coding RNAs and Nucleosome Remodeling Complexes: An Intricate Regulatory Relationship
Abstract
1. Introduction
2. Nucleosome Remodelers
3. Non-Coding RNAs
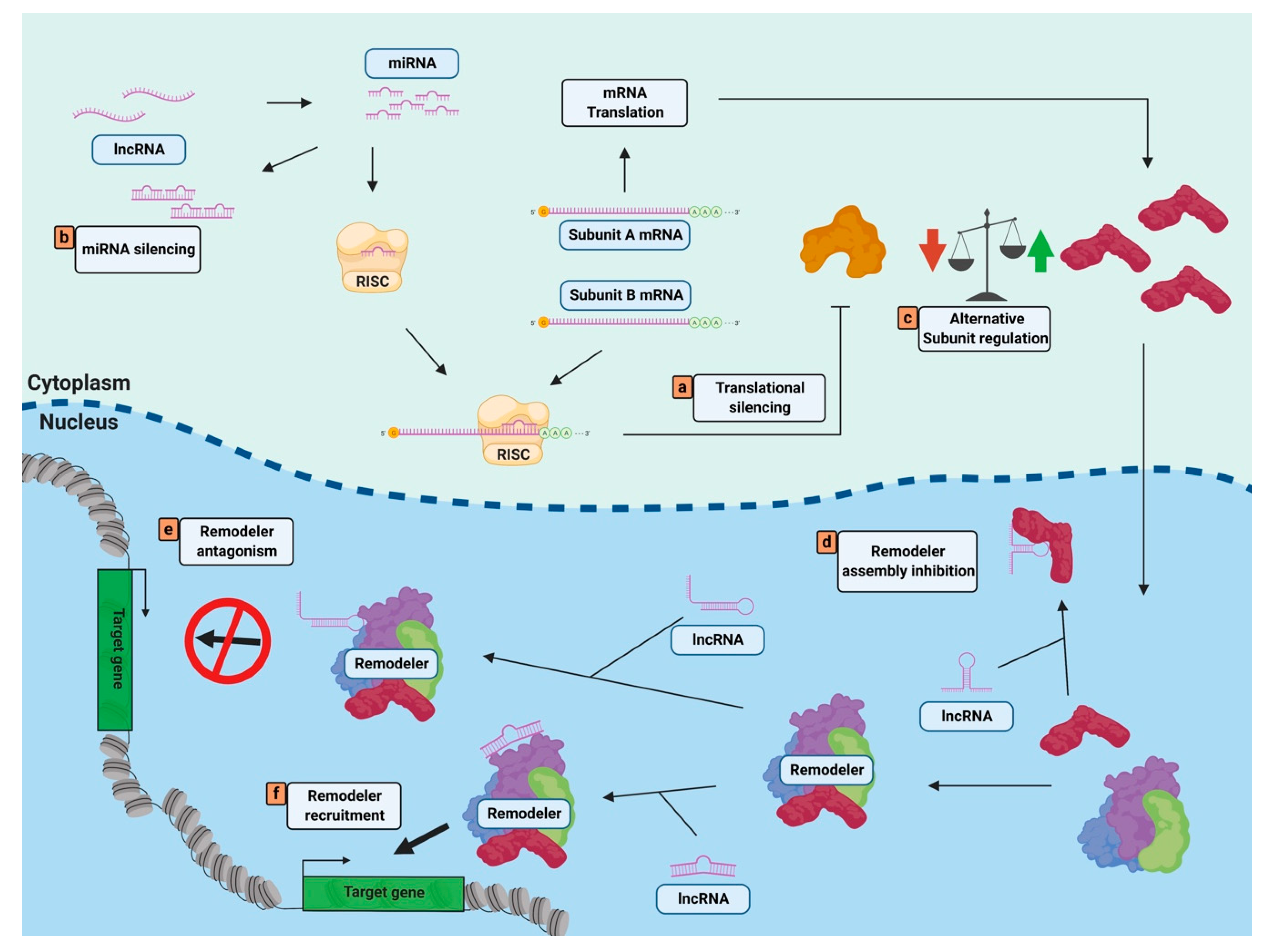
4. Non-Coding RNAs Regulate Nucleosome Remodelers through Complex Targeting or Composition
4.1. ncRNAs Can Regulate Nucleosome Remodeler Targeting
4.2. ncRNAs Can Alter Nucleosome Remodeler Complex Composition
5. Members from Each Nucleosome Remodeling Family Regulate ncRNAs
5.1. SWI/SNF Family Remodelers Regulate ncRNA Expression at Gene Regulatory Sites in Several Eukaryotic Systems
5.2. INO80 Family Nucleosome Remodelers Likely Influence the Expression of Many ncRNA Species
5.3. ISWI Nucleosome Remodelers Are Critical Regulators of Intragenic Cryptic ncRNAs.
5.4. CHD Nucleosome Remodelers Show Significant Potential as ncRNA Regulators
6. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kornberg, R.D. Chromatin Structure: A Repeating Unit of Histones and DNA. Science 1974, 184, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Cairns, B.R. The Biology of Chromatin Remodeling Complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef] [PubMed]
- Breiling, A.; Lyko, F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin 2015, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, S.; Workman, J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Hargreaves, D.C.; Crabtree, G.R. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 2011, 21, 396–420. [Google Scholar] [CrossRef]
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLoS Genet. 2013, 9, e1003569. [Google Scholar] [CrossRef]
- Dunham, I.; Kundaje, A.; Aldred, S.F.; Collins, P.J.; Davis, C.A.; Doyle, F.; Epstein, C.B.; Frietze, S.; Harrow, J.; Kaul, R.; et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar]
- Dozmorov, M.G.; Giles, C.B.; Koelsch, K.A.; Wren, J.D. Systematic classification of non-coding RNAs by epigenomic similarity. BMC Bioinform. 2013, 14, S2. [Google Scholar] [CrossRef]
- Wang, C.; Jia, L.; Wang, Y.; Du, Z.; Zhou, L.; Wen, X.; Li, H.; Zhang, S.; Chen, H.; Chen, N.; et al. Genome-wide interaction target profiling reveals a novel Peblr20-eRNA activation pathway to control stem cell pluripotency. Theranostics 2020, 10, 353–370. [Google Scholar] [CrossRef]
- Lakhotia, S.C. Non-coding RNAs have key roles in cell regulation. Proc. Indian Natl. Sci. Acad. 2016, 82, 1171–1182. [Google Scholar] [CrossRef]
- Alessio, E.; Bonadio, R.S.; Buson, L.; Chemello, F.; Cagnin, S. A single cell but many different transcripts: A journey into the world of long non-coding RNAs. Int. J. Mol. Sci. 2020, 21, 302. [Google Scholar] [CrossRef] [PubMed]
- Subhash, S.; Mishra, K.; Akhade, V.S.; Kanduri, M.; Mondal, T.; Kanduri, C. H3K4me2 and WDR5 enriched chromatin interacting long non-coding RNAs maintain transcriptionally competent chromatin at divergent transcriptional units. Nucleic Acids Res. 2018, 46, 9384–9400. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Chang, C.P. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015, 12, 1094–1098. [Google Scholar] [CrossRef]
- Flynn, R.A.; Do, B.T.; Rubin, A.J.; Calo, E.; Lee, B.; Kuchelmeister, H.; Rale, M.; Chu, C.; Kool, E.T.; Wysocka, J.; et al. 7SK-BAF axis controls pervasive transcription at enhancers. Nat. Struct. Mol. Biol. 2016, 23, 231–238. [Google Scholar] [CrossRef]
- Barisic, D.; Stadler, M.B.; Iurlaro, M.; Schübeler, D. Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors. Nature 2019, 569, 136–140. [Google Scholar] [CrossRef]
- Flaus, A.; Martin, D.M.A.; Barton, G.J.; Owen-Hughes, T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006, 34, 2887–2905. [Google Scholar] [CrossRef]
- Neigeborn, L.; Carlson, M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 1984, 108, 845–858. [Google Scholar]
- Peterson, C.L.; Herskowitz, I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 1992, 68, 573–583. [Google Scholar] [CrossRef]
- Hassan, A.H.; Prochasson, P.; Neely, K.E.; Galasinski, S.C.; Chandy, M.; Carrozza, M.J.; Workman, J.L. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 2002, 111, 369–379. [Google Scholar] [CrossRef]
- Flanagan, J.F.; Mi, L.Z.; Chruszcz, M.; Cymborowski, M.; Clines, K.L.; Kim, Y.; Minor, W.; Rastinejad, F.; Khorasanizadeh, S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 2005, 438, 1181–1185. [Google Scholar] [CrossRef] [PubMed]
- Trotter, K.W.; Fan, H.-Y.; Ivey, M.L.; Kingston, R.E.; Archer, T.K. The HSA Domain of BRG1 Mediates Critical Interactions Required for Glucocorticoid Receptor-Dependent Transcriptional Activation In Vivo. Mol. Cell. Biol. 2008, 28, 1413–1426. [Google Scholar] [CrossRef] [PubMed]
- Szerlong, H.; Hinata, K.; Viswanathan, R.; Erdjument-Bromage, H.; Tempst, P.; Cairns, B.R. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat. Struct. Mol. Biol. 2008, 15, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, X.; Zhang, Z.; Cai, G. Structure and functional interactions of INO80 actin/Arp module. J. Mol. Cell Biol. 2019, 11, 345–355. [Google Scholar] [CrossRef] [PubMed]
- McKnight, J.N.; Jenkins, K.R.; Nodelman, I.M.; Escobar, T.; Bowman, G.D. Extranucleosomal DNA Binding Directs Nucleosome Sliding by Chd1. Mol. Cell. Biol. 2011, 31, 4746–4759. [Google Scholar] [CrossRef] [PubMed]
- Mueller-Planitz, F.; Klinker, H.; Ludwigsen, J.; Becker, P.B. The ATPase domain of ISWI is an autonomous nucleosome remodeling machine. Nat. Struct. Mol. Biol. 2013, 20, 82–89. [Google Scholar] [CrossRef]
- Castello, A.; Fischer, B.; Frese, C.K.; Horos, R.; Alleaume, A.M.; Foehr, S.; Curk, T.; Krijgsveld, J.; Hentze, M.W. Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol. Cell 2016, 63, 696–710. [Google Scholar] [CrossRef]
- Trendel, J.; Schwarzl, T.; Horos, R.; Prakash, A.; Bateman, A.; Hentze, M.W.; Krijgsveld, J. The Human RNA-Binding Proteome and Its Dynamics during Translational Arrest. Cell 2019, 176, 391–403.e19. [Google Scholar] [CrossRef]
- Bao, X.; Guo, X.; Yin, M.; Tariq, M.; Lai, Y.; Kanwal, S.; Zhou, J.; Li, N.; Lv, Y.; Pulido-Quetglas, C.; et al. Capturing the interactome of newly transcribed RNA. Nat. Methods 2018, 15, 213–220. [Google Scholar] [CrossRef]
- Maris, C.; Dominguez, C.; Allain, F.H.T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef]
- Lizarrondo, J.; Dock-Bregeon, A.C.; Martino, L.; Conte, M.R. Structural dynamics in the La-module of La-related proteins. RNA Biol. 2020, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Holley, R.W.; Apgar, J.; Everett, G.A.; Madison, J.T.; Marquisee, M.; Merrill, S.H.; Penswick, J.R.; Zamir, A. Structure of a Ribonucleic Acid. Science 1965, 147, 1462–1465. [Google Scholar] [CrossRef] [PubMed]
- van Bakel, H.; Nislow, C.; Blencowe, B.J.; Hughes, T.R. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010, 8, e1000371. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2006, 309, 1559–1563. [Google Scholar]
- Rnas, N.; Riken, T.; Gene, M.; Project, E.; When, U.; For, U.; Information, S. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-lenght cDNAs. Nature 2002, 420, 563–573. [Google Scholar]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, Biogenesis, and Their Evolving Role in Animal Development and Disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef]
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Volders, P.J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. Lncipedia 5: Towards a reference set of human long non-coding rnas. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef]
- Cheng, J.-T.; Wang, L.; Wang, H.; Tang, F.-R.; Cai, W.-Q.; Sethi, G.; Xin, H.-W.; Ma, Z. Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition. Cells 2019, 8, 1178. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, Z.; Jiang, J.; Xu, C.; Kang, J.; Xiao, L.; Wu, M.; Xiong, J.; Guo, X.; Liu, H. Endogenous miRNA Sponge lincRNA-RoR Regulates Oct4, Nanog, and Sox2 in Human Embryonic Stem Cell Self-Renewal. Dev. Cell 2013, 25, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Plath, K.; Fang, J.; Mlynarczyk-Evans, S.K.; Cao, R.; Worringer, K.A.; Wang, H.; De la Cruz, C.C.; Otte, A.P.; Panning, B.; Zhang, Y. Role of histone H3 lysine 27 methylation in X inactivation. Science 2003, 300, 131–135. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Morales, D.R.; Thomas, K.; Presser, A.; Bernstein, B.E.; Van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef]
- Banerji, J.; Rusconi, S.; Schaffner, W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell 1981, 27, 299–308. [Google Scholar] [CrossRef]
- Core, L.J.; Martins, A.L.; Danko, C.G.; Waters, C.T.; Siepel, A.; Lis, J.T. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 2014, 46, 1311–1320. [Google Scholar] [CrossRef]
- Xiong, L.; Kang, R.; Ding, R.; Kang, W.; Zhang, Y.; Liu, W.; Huang, Q.; Meng, J.; Guo, Z. Genome-wide identification and characterization of enhancers across 10 human tissues. Int. J. Biol. Sci. 2018, 14, 1321–1332. [Google Scholar] [CrossRef]
- Davidson, L.; Francis, L.; Cordiner, R.A.; Eaton, J.D.; Estell, C.; Macias, S.; Cáceres, J.F.; West, S. Rapid Depletion of DIS3, EXOSC10, or XRN2 Reveals the Immediate Impact of Exoribonucleolysis on Nuclear RNA Metabolism and Transcriptional Control. Cell Rep. 2019, 26, 2779–2791.e5. [Google Scholar] [CrossRef]
- Pefanis, E.; Wang, J.; Rothschild, G.; Lim, J.; Kazadi, D.; Sun, J.; Federation, A.; Chao, J.; Elliott, O.; Liu, Z.P.; et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 2015, 161, 774–789. [Google Scholar] [CrossRef] [PubMed]
- Tsai, P.F.; Dell’Orso, S.; Rodriguez, J.; Vivanco, K.O.; Ko, K.D.; Jiang, K.; Juan, A.H.; Sarshad, A.A.; Vian, L.; Tran, M.; et al. A Muscle-Specific Enhancer RNA Mediates Cohesin Recruitment and Regulates Transcription In trans. Mol. Cell 2018, 71, 129–141.e8. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, L.; Ren, S.; Wang, L.; Blackburn, P.R.; McNulty, M.S.; Gao, X.; Qiao, M.; Vessella, R.L.; Kohli, M.; et al. Activation of P-TEFb by Androgen Receptor-Regulated Enhancer RNAs in Castration-Resistant Prostate Cancer. Cell Rep. 2016, 15, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.; Chen, Y.; Song, H.; Li, D.; Mei, H.; Yang, F.; Fang, E.; Wang, X.; Huang, K.; Zheng, L.; et al. HPSE enhancer RNA promotes cancer progression through driving chromatin looping and regulating hnRNPU/p300/EGR1/HPSE axis. Oncogene 2018, 37, 2728–2745. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Notani, D.; Ma, Q.; Tanasa, B.; Nunez, E.; Chen, A.Y.; Merkurjev, D.; Zhang, J.; Ohgi, K.; Song, X.; et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013, 498, 516–520. [Google Scholar] [CrossRef]
- Rahnamoun, H.; Lee, J.; Sun, Z.; Lu, H.; Ramsey, K.M.; Komives, E.A.; Lauberth, S.M. RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat. Struct. Mol. Biol. 2018, 25, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Melo, C.A.; Drost, J.; Wijchers, P.J.; van de Werken, H.; de Wit, E.; Vrielink, J.A.F.O.; Elkon, R.; Melo, S.A.; Léveillé, N.; Kalluri, R.; et al. ERNAs Are Required for p53-Dependent Enhancer Activity and Gene Transcription. Mol. Cell 2013, 49, 524–535. [Google Scholar] [CrossRef]
- Schaukowitch, K.; Joo, J.Y.; Liu, X.; Watts, J.K.; Martinez, C.; Kim, T.K. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 2014, 56, 29–42. [Google Scholar] [CrossRef]
- Alfert, A.; Moreno, N.; Kerl, K. The BAF complex in development and disease. Epigenetics Chromatin 2019, 12, 1–15. [Google Scholar] [CrossRef]
- Xu, Y.; Yan, W.; Chen, X. SNF5, a core component of the SWI/SNF complex, is necessary for p53 expression and cell survival, in part through eIF4E. Oncogene 2010, 29, 4090–4100. [Google Scholar] [CrossRef] [PubMed]
- Prensner, J.R.; Iyer, M.K.; Balbin, O.A.; Dhanasekaran, S.M.; Cao, Q.; Brenner, J.C.; Laxman, B.; Asangani, I.A.; Grasso, C.S.; Kominsky, H.D.; et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011, 29, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Prensner, J.R.; Iyer, M.K.; Sahu, A.; Asangani, I.A.; Cao, Q.; Patel, L.; Vergara, I.A.; Davicioni, E.; Erho, N.; Ghadessi, M.; et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013, 45, 1392–1403. [Google Scholar] [CrossRef] [PubMed]
- Muchardt, C.; Yaniv, M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993, 12, 4279–4290. [Google Scholar] [CrossRef] [PubMed]
- Khavari, P.A.; Peterson, C.L.; Tamkun, J.W.; Mendel, D.B.; Crabtree, G.R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 1993, 366, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Raab, J.R.; Runge, J.S.; Spear, C.C.; Magnuson, T. Co-regulation of transcription by BRG1 and BRM, two mutually exclusive SWI/SNF ATPase subunits. Epigenetics Chromatin 2017, 10, 1–15. [Google Scholar] [CrossRef]
- Jégu, T.; Blum, R.; Cochrane, J.C.; Yang, L.; Wang, C.Y.; Gilles, M.E.; Colognori, D.; Szanto, A.; Marr, S.K.; Kingston, R.E.; et al. Xist RNA antagonizes the SWI/SNF chromatin remodeler BRG1 on the inactive X chromosome. Nat. Struct. Mol. Biol. 2019, 26, 96–109. [Google Scholar] [CrossRef]
- Wang, X.; Gong, Y.; Jin, B.; Wu, C.; Yang, J.; Wang, L.; Zhang, Z.; Mao, Z. Long non-coding RNA urothelial carcinoma associated 1 induces cell replication by inhibiting BRG1 in 5637 cells. Oncol. Rep. 2014, 32, 1281–1290. [Google Scholar] [CrossRef]
- Wang, Y.; He, L.; Du, Y.; Zhu, P.; Huang, G.; Luo, J.; Yan, X.; Ye, B.; Li, C.; Xia, P.; et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 2015, 16, 413–425. [Google Scholar] [CrossRef]
- Hu, G.; Gong, A.-Y.; Wang, Y.; Ma, S.; Chen, X.; Chen, J.; Su, C.-J.; Shibata, A.; Strauss-Soukup, J.K.; Drescher, K.M.; et al. LincRNA-Cox2 Promotes Late Inflammatory Gene Transcription in Macrophages through Modulating SWI/SNF-Mediated Chromatin Remodeling. J. Immunol. 2016, 196, 2799–2808. [Google Scholar] [CrossRef]
- Liu, X.; Lu, Y.; Zhu, J.; Liu, M.; Xie, M.; Ye, M.; Li, M.; Wang, S.; Ming, Z.; Tong, Q.; et al. A Long Noncoding RNA, Antisense IL-7, Promotes Inflammatory Gene Transcription through Facilitating Histone Acetylation and Switch/Sucrose Nonfermentable Chromatin Remodeling. J. Immunol. 2019, 203, 1548–1559. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Wang, H.; Hu, X.; Cao, X. lncRNA MALAT1 binds chromatin remodeling subunit BRG1 to epigenetically promote inflammation-related hepatocellular carcinoma progression. Oncoimmunology 2019, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ruhl, D.D.; Jin, J.; Cai, Y.; Swanson, S.; Florens, L.; Washburn, M.P.; Conaway, R.C.; Conaway, J.W.; Chrivia, J.C. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry 2006, 45, 5671–5677. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.; Liu, B.; Yang, L.; Zhu, X.; Zhang, D.; Wu, W.; Zhu, P.; Wang, Y.; Wang, S.; Xia, P.; et al. LncKdm2b controls self-renewal of embryonic stem cells via activating expression of transcription factor Zbtb3. EMBO J. 2018, 37, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Wong, J.; Moreno, G.T.; Young, M.K.; Côté, J.; Wang, W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 1998, 2, 851–861. [Google Scholar] [CrossRef]
- Hoffmeister, H.; Fuchs, A.; Erdel, F.; Pinz, S.; Gröbner-Ferreira, R.; Bruckmann, A.; Deutzmann, R.; Schwartz, U.; Maldonado, R.; Huber, C.; et al. CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Nucleic Acids Res. 2017, 45, 10534–10554. [Google Scholar] [CrossRef]
- Zhao, Z.; Dammert, M.A.; Grummt, I.; Bierhoff, H. LncRNA-Induced Nucleosome Repositioning Reinforces Transcriptional Repression of rRNA Genes upon Hypotonic Stress. Cell Rep. 2016, 14, 1876–1882. [Google Scholar] [CrossRef]
- Wong, L.H.; McGhie, J.D.; Sim, M.; Anderson, M.A.; Ahn, S.; Hannan, R.D.; George, A.J.; Morgan, K.A.; Mann, J.R.; Choo, K.H.A. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010, 20, 351–360. [Google Scholar] [CrossRef]
- Ren, W.; Medeiros, N.; Warneford-Thomson, R.; Wulfridge, P.; Yan, Q.; Bian, J.; Sidoli, S.; Garcia, B.A.; Skordalakes, E.; Joyce, E.; et al. Disruption of ATRX-RNA interactions uncovers roles in ATRX localization and PRC2 function. Nat. Commun. 2020, 11, 2219. [Google Scholar] [CrossRef]
- Gurney, T.; Eliceiri, G.L. Intracellular distribution of low molecular weight RNA species in hela cells. J. Cell Biol. 1980, 87, 398–403. [Google Scholar] [CrossRef]
- Ji, X.; Zhou, Y.; Pandit, S.; Huang, J.; Li, H.; Lin, C.Y.; Xiao, R.; Burge, C.B.; Fu, X.D. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 2013, 153, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Smola, M.J.; Christy, T.W.; Inoue, K.; Nicholson, C.O.; Friedersdorf, M.; Keene, J.D.; Lee, D.M.; Calabrese, J.M.; Weeks, K.M. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. USA 2016, 113, 10322–10327. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Moss, W.N.; Rutenberg-Schoenberg, M.; Simon, M.D. Probing Xist RNA Structure in Cells Using Targeted Structure-Seq. PLoS Genet. 2015, 11, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Cajigas, I.; Leib, D.E.; Cochrane, J.; Luo, H.; Swyter, K.R.; Chen, S.; Clark, B.S.; Thompson, J.; Yates, J.R.; Kingston, R.E.; et al. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development 2015, 142, 2641–2652. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.D.; Bycroft, M.; Zinzalla, G. Structure of the BRK domain of the SWI/SNF chromatin remodeling complex subunit BRG1 reveals a potential role in protein–protein interactions. Protein Sci. 2020, 29, 1047–1053. [Google Scholar] [CrossRef]
- Han, P.; Li, W.; Lin, C.H.; Yang, J.; Shang, C.; Nurnberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.Y.; Lin, C.J.; et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106. [Google Scholar] [CrossRef]
- He, C.; Sidoli, S.; Warneford-Thomson, R.; Tatomer, D.C.; Wilusz, J.E.; Garcia, B.A.; Bonasio, R. High-Resolution Mapping of RNA-Binding Regions in the Nuclear Proteome of Embryonic Stem Cells. Mol. Cell 2016, 64, 416–430. [Google Scholar] [CrossRef]
- Hong, C.F.; Lin, S.Y.; Chou, Y.T.; Wu, C.W. MicroRNA-7 compromises p53 protein-dependent apoptosis by controlling the expression of the chromatin remodeling factor SMARCD1. J. Biol. Chem. 2016, 291, 1877–1889. [Google Scholar] [CrossRef]
- Shen, J.; Xiao, Z.; Wu, W.K.K.; Wang, M.H.; To, K.F.; Chen, Y.; Yang, W.; Li, M.S.M.; Shin, V.Y.; Tong, J.H.; et al. Epigenetic silencing of miR-490-3p reactivates the chromatin remodeler SMARCD1 to promote helicobacter pylori-induced gastric carcinogenesis. Cancer Res. 2015, 75, 754–765. [Google Scholar] [CrossRef]
- Tommasi, S.; Pinto, R.; Danza, K.; Pilato, B.; Palumbo, O.; Micale, L.; De Summa, S. miR-151-5p, targeting chromatin remodeler SMARCA5, as a marker for the BRCAness phenotype. Oncotarget 2016, 7, 80363–80372. [Google Scholar] [CrossRef]
- Green, C.D.; Huang, Y.; Dou, X.; Yang, L.; Liu, Y.; Han, J.D.J. Impact of Dietary Interventions on Noncoding RNA Networks and mRNAs Encoding Chromatin-Related Factors. Cell Rep. 2017, 18, 2957–2968. [Google Scholar] [CrossRef]
- Cai, C.; Ashktorab, H.; Pang, X.; Zhao, Y.; Sha, W.; Liu, Y.; Gu, X. MicroRNA-211 expression promotes colorectal cancer cell growth in vitro and in Vivo by targeting tumor suppressor CHD5. PLoS ONE 2012, 7, e29750. [Google Scholar] [CrossRef]
- Sheng, W.; Chen, Y.; Gong, Y.; Dong, T.; Zhang, B.; Gao, W. MiR-148a inhibits self-renewal of thyroid cancer stem cells via repressing INO80 expression. Oncol. Rep. 2016, 36, 3387–3396. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chang, C.C.; Liu, T.Y.T.C.; Lee, Y.T.; Chen, Y.C.Y.L.; Yeh, K.T.; Lee, C.C.; Chen, Y.C.Y.L.; Lin, P.C.; Chang, Y.S.; Chan, W.L.; et al. Genome-wide analysis of lncRNAs in 3’-untranslated regions: CR933609 acts as a decoy to protect the INO80D gene. Int. J. Oncol. 2018, 53, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Zhu, B.; Ueda, A.; Song, X.; Horike, S.I.; Yokota, T.; Akagi, T. Baf53a is involved in survival of mouse ES cells, which can be compensated by Baf53b. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yoo, A.S.; Staahl, B.T.; Chen, L.; Crabtree, G.R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 2009, 460, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Sharp, P.A. Emerging roles for natural microRNA sponges. Curr. Biol. 2010, 20, R858–R861. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gadue, P.; Chen, K.; Jiao, Y.; Tuteja, G.; Schug, J.; Li, W.; Kaestner, K.H. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 2012, 151, 1608–1616. [Google Scholar] [CrossRef]
- Alatwi, H.E.; Downs, J.A. Removal of H2A.Z by INO 80 promotes homologous recombination. EMBO Rep. 2015, 16, 986–994. [Google Scholar] [CrossRef]
- Konev, A.Y.; Tribus, M.; Park, S.Y.; Podhraski, V.; Lim, C.Y.; Emelyanov, A.V.; Vershilova, E.; Pirrotta, V.; Kadonaga, J.T.; Lusser, A.; et al. CHD1 Motor Protein Is Required for Deposition of Histone Variant H3.3 into Chromatin in Vivo. Science 2007, 317, 1087–1090. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Sandaltzopoulos, R.; Wang, H.M.; Hamiche, A.; Ranallo, R.; Lee, K.M.; Fu, D.; Wu, C. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 2001, 8, 531–543. [Google Scholar] [CrossRef]
- Hainer, S.J.; Gu, W.; Carone, B.R.; Landry, B.D.; Rando, O.J.; Mello, C.C.; Fazzio, T.G. Suppression of pervasive noncoding transcription in embryonic stem cells by esBAF. Genes Dev. 2015, 29, 362–378. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.; Jothi, R.; Ronan, J.L.; Cui, K.; Zhao, K.; Crabtree, G.R. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. USA 2009, 106, 5187–5191. [Google Scholar] [CrossRef]
- Ho, L.; Miller, E.L.; Ronan, J.L.; Ho, W.Q.; Jothi, R.; Crabtree, G.R. EsBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 2011, 13, 903–913. [Google Scholar] [CrossRef]
- Archacki, R.; Yatusevich, R.; Buszewicz, D.; Krzyczmonik, K.; Patryn, J.; Iwanicka-Nowicka, R.; Biecek, P.; Wilczynski, B.; Koblowska, M.; Jerzmanowski, A.; et al. Arabidopsis SWI/SNF chromatin remodeling complex binds both promoters and terminators to regulate gene expression. Nucleic Acids Res. 2017, 45, 3116–3129. [Google Scholar] [CrossRef]
- Alexander, J.M.; Hota, S.K.; He, D.; Thomas, S.; Ho, L.; Pennacchio, L.A.; Bruneau, B.G. Brg1 modulates enhancer activation in mesoderm lineage commitment. Development 2015, 142, 1418–1430. [Google Scholar] [CrossRef]
- Mallappa, C.; Nasipak, B.T.; Etheridge, L.; Androphy, E.J.; Jones, S.N.; Sagerström, C.G.; Ohkawa, Y.; Imbalzano, A.N. Myogenic MicroRNA Expression Requires ATP-Dependent Chromatin Remodeling Enzyme Function. Mol. Cell. Biol. 2010, 30, 3176–3186. [Google Scholar] [CrossRef] [PubMed]
- Rawal, Y.; Chereji, R.V.; Qiu, H.; Ananthakrishnan, S.; Govind, C.K.; Clark, D.J.; Hinnebusch, A.G. SWI/SNF and RSC cooperate to reposition and evict promoter nucleosomes at highly expressed genes in yeast. Genes Dev. 2018, 32, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.C.K.; Patel, H.; Chia, M.; Moretto, F.; Frith, D.; Snijders, A.P.; van Werven, F.J. Repression of Divergent Noncoding Transcription by a Sequence-Specific Transcription Factor. Mol. Cell 2018, 72, 942–954.e7. [Google Scholar] [CrossRef]
- Marquardt, S.; Escalante-Chong, R.; Pho, N.; Wang, J.; Churchman, L.S.; Springer, M.; Buratowski, S. A chromatin-based mechanism for limiting divergent noncoding transcription. Cell 2014, 157, 1712–1723. [Google Scholar] [CrossRef] [PubMed]
- Alcid, E.A.; Tsukiyama, T. ATP-dependent chromatin remodeling shapes the long noncoding RNA landscape. Genes Dev. 2014, 28, 2348–2360. [Google Scholar] [CrossRef] [PubMed]
- Giaimo, B.D.; Ferrante, F.; Herchenröther, A.; Hake, S.B.; Borggrefe, T. The histone variant H2A.Z in gene regulation. Epigenetics Chromatin 2019, 12, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Mizuguchi, G.; Shen, X.; Landry, J.; Wu, W.H.; Sen, S.; Wu, C. ATP-Driven Exchange of Histone H2AZ Variant Catalyzed by SWR1 Chromatin Remodeling Complex. Science 2004, 303, 343–348. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Su, T.; Yen, L.; Jacquet, K.; Huang, C.; Côté, J.; Kurdistani, S.K.; Carey, M.F. EP400 Deposits H3.3 into Promoters and Enhancers during Gene Activation. Mol. Cell 2016, 61, 27–38. [Google Scholar] [CrossRef]
- Rudnizky, S.; Bavly, A.; Malik, O.; Pnueli, L.; Melamed, P.; Kaplan, A. H2A.Z controls the stability and mobility of nucleosomes to regulate expression of the LH genes. Nat. Commun. 2016, 7, 12958. [Google Scholar] [CrossRef]
- Choi, K.; Kim, J.; Müller, S.Y.; Oh, M.; Underwood, C.; Henderson, I.; Lee, I. Regulation of microRNA-mediated developmental changes by the SWR1 chromatin remodeling complex. Plant Physiol. 2016, 171, 1128–1143. [Google Scholar]
- Wang, F.; Ranjan, A.; Wei, D.; Wu, C. Comment on “A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme”. Science 2016, 353, 358a. [Google Scholar] [CrossRef]
- Watanabe, S.; Radman-Livaja, M.; Rando, O.J.; Peterson, C.L. A Histone Acetylation Switch Regulates H2A.Z Deposition by the SWR-C Remodeling Enzyme. Science 2013, 340, 195–199. [Google Scholar] [CrossRef]
- Papamichos-Chronakis, M.; Watanabe, S.; Rando, O.J.; Peterson, C.L. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 2011, 144, 200–213. [Google Scholar] [CrossRef]
- Tramantano, M.; Sun, L.; Au, C.; Labuz, D.; Liu, Z.; Chou, M.; Shen, C.; Luk, E. Constitutive turnover of histone H2A.Z at yeast promoters requires the preinitiation complex. Elife 2016, 5, 1–30. [Google Scholar] [CrossRef]
- Xue, Y.; Pradhan, S.K.; Sun, F.; Chronis, C.; Tran, N.; Su, T.; Van, C.; Vashisht, A.; Wohlschlegel, J.; Peterson, C.L.; et al. Mot1, Ino80C, and NC2 Function Coordinately to Regulate Pervasive Transcription in Yeast and Mammals. Mol. Cell 2017, 67, 594–607.e4. [Google Scholar] [CrossRef]
- Xue, Y.; Van, C.; Pradhan, S.K.; Su, T.; Gehrke, J.; Kuryan, B.G.; Kitada, T.; Vashisht, A.; Tran, N.; Wohlschlege, J.; et al. The ino80 complex prevents invasion of euchromatin into silent chromatin. Genes Dev. 2015, 29, 350–355. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gkikopoulos, T.; Schofield, P.; Singh, V.; Pinskaya, M.; Mellor, J.; Smolle, M.; Workman, J.L.; Barton, G.J.; Owen-Hughes, T. A Role for Snf2-Related Nucleosome-Spacing Enzymes in Genome-Wide Nucleosome Organization. Science 2011, 333, 1758–1760. [Google Scholar] [CrossRef] [PubMed]
- Smolle, M.; Venkatesh, S.; Gogol, M.M.; Li, H.; Zhang, Y.; Florens, L.; Washburn, M.P.; Workman, J.L. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 2012, 19, 884–892. [Google Scholar] [CrossRef]
- Whitehouse, I.; Rando, O.J.; Delrow, J.; Tsukiyama, T. Chromatin remodelling at promoters suppresses antisense transcription. Nature 2007, 450, 1031–1035. [Google Scholar] [CrossRef] [PubMed]
- Shim, Y.S.; Choi, Y.; Kang, K.; Cho, K.; Oh, S.; Lee, J.; Grewal, S.I.S.; Lee, D. Hrp3 controls nucleosome positioning to suppress non-coding transcription in eu-and heterochromatin. EMBO J. 2012, 31, 4375–4387. [Google Scholar] [CrossRef] [PubMed]
- Hennig, B.P.; Bendrin, K.; Zhou, Y.; Fischer, T. Chd1 chromatin remodelers maintain nucleosome organization and repress cryptic transcription. EMBO Rep. 2012, 13, 997–1003. [Google Scholar] [CrossRef]
- Srinivasan, S.; Dorighi, K.M.; Tamkun, J.W. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 2008, 4, e1000217. [Google Scholar] [CrossRef]
- Lusser, A.; Urwin, D.L.; Kadonaga, J.T. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 2005, 12, 160–166. [Google Scholar] [CrossRef]
- Ito, T.; Bulger, M.; Pazin, M.J.; Kobayashi, R.; Kadonaga, J.T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 1997, 90, 145–155. [Google Scholar] [CrossRef]
- LeRoy, G.; Orphanides, G.; Lane, W.S. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 1998, 282, 1900–1904. [Google Scholar] [CrossRef] [PubMed]
- Wiechens, N.; Singh, V.; Gkikopoulos, T.; Schofield, P.; Rocha, S.; Owen-Hughes, T. The Chromatin Remodelling Enzymes SNF2H and SNF2L Position Nucleosomes adjacent to CTCF and Other Transcription Factors. PLoS Genet. 2016, 12, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.A.; Baek, S.; Sung, M.H.; John, S.; Wiench, M.; Johnson, T.A.; Schiltz, R.L.; Hager, G.L. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat. Struct. Mol. Biol. 2014, 21, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Murawska, M.; Brehm, A. CHD chromatin remodelers and the transcription cycle. Transcription 2011, 2, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Walfridsson, J.; Khorosjutina, O.; Matikainen, P.; Gustafsson, C.M.; Ekwall, K. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J. 2007, 26, 2868–2879. [Google Scholar] [CrossRef]
- Schnetz, M.P.; Bartels, C.F.; Shastri, K.; Balasubramanian, D.; Zentner, G.E.; Balaji, R.; Zhang, X.; Song, L.; Wang, Z.; Laframboise, T.; et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009, 19, 590–601. [Google Scholar] [CrossRef]
- Menon, T.; Yates, J.A.; Bochar, D.A. Regulation of androgen-responsive transcription by the chromatin remodeling factor CHD8. Mol. Endocrinol. 2010, 24, 1165–1174. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patty, B.J.; Hainer, S.J. Non-Coding RNAs and Nucleosome Remodeling Complexes: An Intricate Regulatory Relationship. Biology 2020, 9, 213. https://doi.org/10.3390/biology9080213
Patty BJ, Hainer SJ. Non-Coding RNAs and Nucleosome Remodeling Complexes: An Intricate Regulatory Relationship. Biology. 2020; 9(8):213. https://doi.org/10.3390/biology9080213
Chicago/Turabian StylePatty, Benjamin J., and Sarah J. Hainer. 2020. "Non-Coding RNAs and Nucleosome Remodeling Complexes: An Intricate Regulatory Relationship" Biology 9, no. 8: 213. https://doi.org/10.3390/biology9080213
APA StylePatty, B. J., & Hainer, S. J. (2020). Non-Coding RNAs and Nucleosome Remodeling Complexes: An Intricate Regulatory Relationship. Biology, 9(8), 213. https://doi.org/10.3390/biology9080213




