Abstract
Reversible data hiding (RDH) is a recently emerged research domain in the field of information security domain with broad applications in medical images and meta-data handling in the cloud. The amount of data required to handle the healthcare sector has exponentially increased due to the increase in the population. Medical images and various reports such as discharge summaries and diagnosis reports are the most common data in the healthcare sector. The RDH schemes are widely explored to embed the medical reports in the medical image instead of sending them as separate files. The receiver can extract the clinical reports and recover the original medical image for further diagnosis. This manuscript proposes an approach that uses a new lossless compression-based RDH scheme that creates vacant room for data hiding. The proposed scheme uses run-length encoding and a modified Elias gamma encoding scheme on higher-order bit planes for lossless compression. The conventional Elias gamma encoding process is modified in the proposed method to embed some additional data bits during the encoding process itself. The revised approach ensures a high embedding rate and lossless recovery of medical images at the receiver side. The experimental study is conducted on both natural images and medical images. The average embedding rate from the proposed scheme for the medical images is 0.75 bits per pixel. The scheme achieved a 0 bit error rate during image recovery and data extraction. The experimental study shows that the newly introduced scheme performs better when compared with the existing RDH schemes.
1. Introduction
Recent technological advancements in computer science and communication support various sectors such as banking, business, education, and healthcare [1,2]. The healthcare sector is one of the prominent domains that require significant attention from researchers for improvement to ensure a healthy society, leading to increasing the country’s productivity. For the last two decades, a lot of research has been happening in the healthcare domain, which mainly focuses on designing and implementing computer-aided diagnosis systems, developing hardware devices to monitor healthcare parameters, and implementing various strategies for the secure transmission of healthcare data. This research focuses on designing and implementing an efficient and safe data transmission scheme suited for the healthcare sector.
Security is a broad term used to denote the set of procedures to ensure authorized access and confidentiality of the data. A massive amount of data is transmitted in the healthcare sector every day. The healthcare data includes medical images, patient reports, discharge summaries, and diagnosis reports. A significant amount of attention is required to ensure the security of this data. Cryptography is one of the most popular approaches in practice to provide confidentiality. In a cryptography scheme, the data are converted into an unreadable form with the help of an encryption scheme by taking an encryption key as the input. An authorized receiver who has the decryption key can decrypt the contents before using them. An unauthorized person can access the encrypted information during transmission using some sophisticated techniques, but they may not view the content since it is in incomprehensible form.
Several encryption schemes are in practice, and they can be broadly classified into two forms, symmetric encryption or asymmetric encryption, based on the keys used for encryption and decryption [3,4]. A symmetric encryption system uses the same key during encryption and decryption. However, an asymmetric encryption scheme uses different keys for encryption and decryption. Most of the well-known encryption schemes, such as advanced encryption standard (AES), data encryption standard (DES), Tripple DES, Blowfish, Twofish, etc., are symmetric encryption techniques. The well-known asymmetric encryption techniques in practice are encryption schemes such as Diffie–Hellman, ElGamal, and Rivest–Shamir–Adleman (RSA) [5,6].
The confidentiality of the digital data can be ensured using image encryption techniques. It has been proven that the recent encryption techniques such as AES, DES, RSA, etc. are hard to break by malicious people. One of the key concerns in a cryptosystem is that one can easily recognize whether digital content is in its original form or encrypted form. For example, in the case of image encryption, the encrypted image will have noise-like or texture-like patterns if it is encrypted. Considering this, recently, a new scheme called visually meaning image encryption (VMIE) has been introduced [7,8]. In a VMIE scheme, the encrypted image looks like a natural image, and the attackers cannot distinguish a genuine image from an encrypted one. This provides some security since attackers will analyze encrypted images more often with the assumption that it consists of some crucial data.
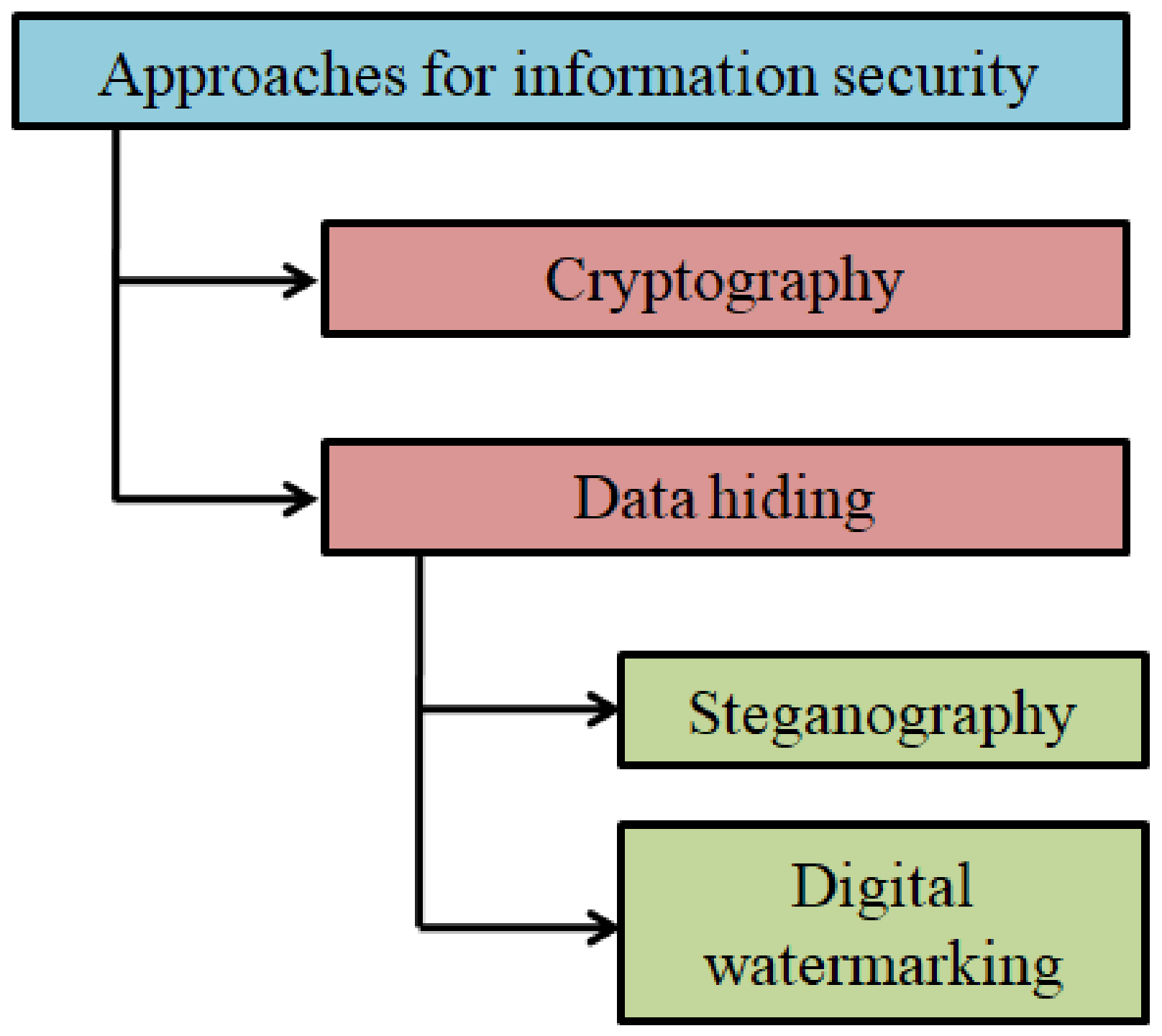
Data hiding is another key area widely studied and explored in information security [9,10,11]. In data hiding, the confidentiality of the data is maintained by embedding it in a cover medium. The cover medium can be digital content such as image, video, text, or audio [12,13]. Data hiding schemes are popularly used for steganography and watermarking. Steganography is the process of concealing a secret message in a cover medium. In contrast, instead of a secret message, a digital watermark will be embedded to facilitate data authentication or copyright protection in a watermarking scheme [14,15].
The overview of various information security approaches is schematically shown in Figure 1.
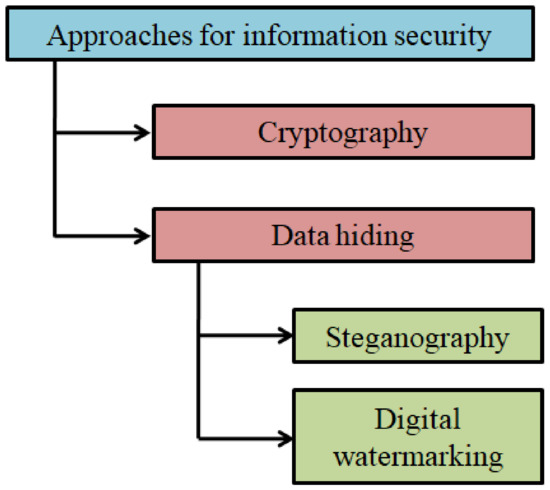
Figure 1.
Approaches to information security: an overview.
Reversible data hiding (RDH) is an emerging area in the field of data hiding. In a conventional data hiding approach, the cover medium will be permanently altered during the data hiding process. RDH is a way of hiding data in some cover medium by changing the actual content. Still, later, the original content can be losslessly recovered during the extraction of confidential information. The reversible data hiding schemes are widely used in cloud computing and medical image transmission [16,17]. A few applications are listed below:
- Common people widely use cloud services due to their convenience and availability. The cloud service provider (CSP) cannot make any permanent changes that they have received from the user. Therefore, RDH schemes can be utilized by the CSP for embedding metadata or watermark on the users’ data. The CSP can restore the image whenever the user requests specific data [18,19].
- Watermarking schemes are popular in medical image transmission to ensure authenticity on the receiver side. Conventional watermarking is not suited for watermarking medical images since it alters the medical image permanently. Alterations in the medical image due to watermarking may adversely affect the diagnosis, especially when using some computer-aided diagnosis tools. The RDH schemes can be used to embed the watermark in the medical images, which will help to ensure authenticity at the receiver side, and the original image can be restored for further diagnosis [20,21,22].
- The transmission of medical images and clinical reports from one hospital to another is prevalent due to the increase in the population and advancements in telemedicine. Handling medical images and related clinical descriptions is challenging in the hospital environment since one-to-one correspondence should be maintained. RDH schemes can be utilized in this area to transmit clinical reports and the respective medical images. An efficient RDH scheme can be used to embed the clinical information in the medical image before storing or sharing it. Further, during diagnosis, the clinical reports can be extracted, and the original image can be recovered [23,24].
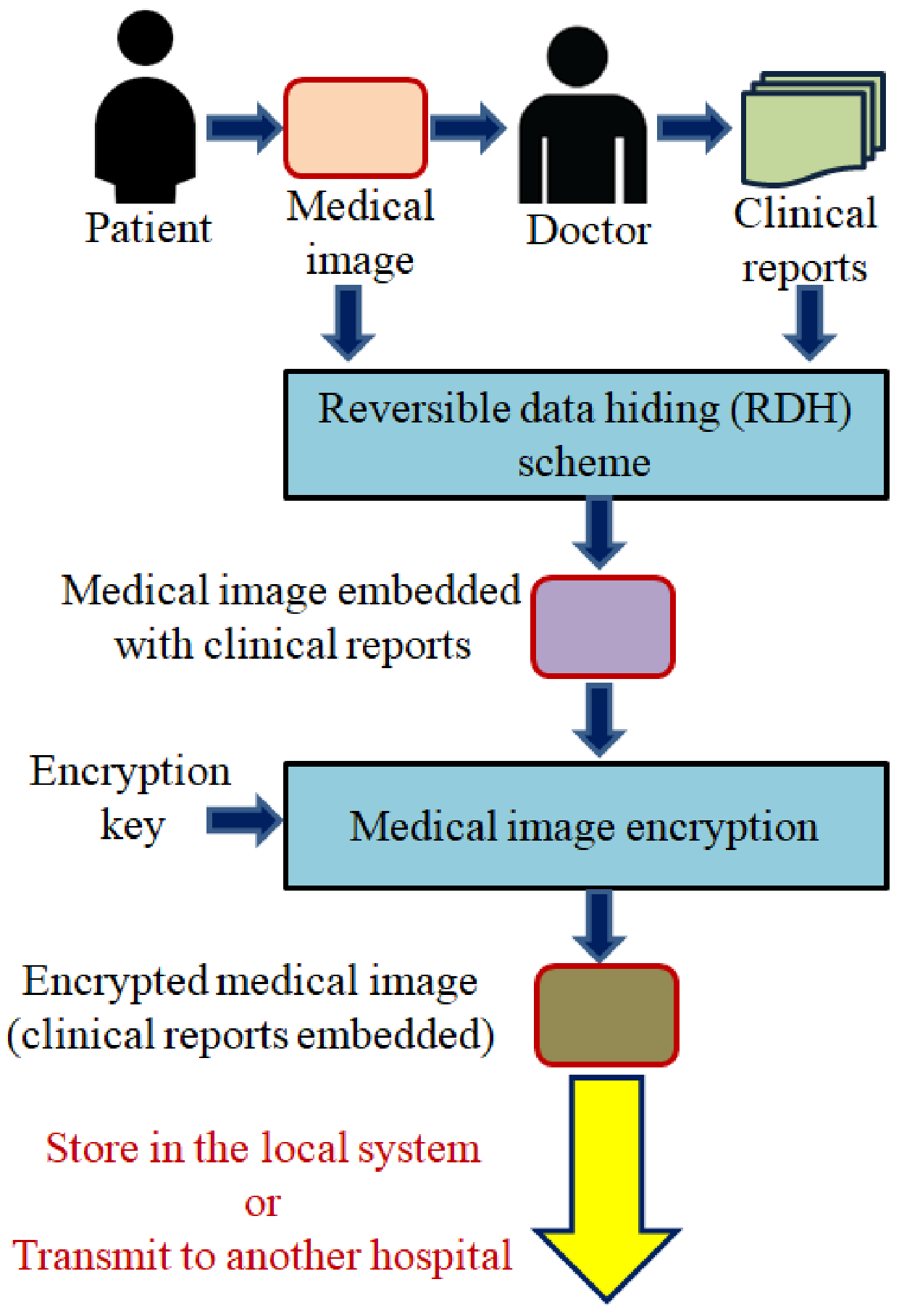
This research focuses on the design and implementation of a new RDH scheme suited for the healthcare domain, especially for the transmission of clinical reports and medical images. The overview of a RDH scheme in a medical image transmission is shown in Figure 2. On the receiver side, firstly, the received medical image will be decrypted. Further, the hidden clinical reports will be extracted while recovering the original medical image. The extracted clinical report and the restored medical image will be used for further diagnosis or treatment.
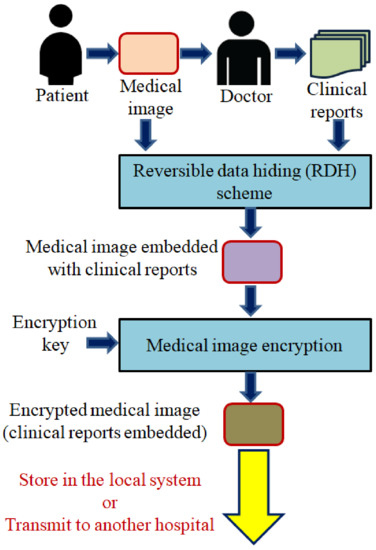
Figure 2.
Overview of RDH scheme to embed clinical reports in medical images.
An RDH method to use in this domain should have the following properties:
- High embedding capacity to ensure that lengthy clinical reports can be embedded in the medical image.
- The RDH scheme in this domain must ensure that the original image will be recovered without any loss of information.
- The scheme should allow the error-free extraction of the clinical reports from the medical image.
- The RDH scheme should provide real-time performance to ensure the availability of medical images and reports in time during diagnosis.
We propose a new RDH scheme in this manuscript by considering all the key objectives of RDH schemes in medical image transmission. The major contribution of the manuscripts are listed below:
- We propose a novel bit plane compression-based RDH scheme.
- The proposed scheme provides high embedding capacity without compromising image recovery and message extraction.
- Elias gamma is a well-known variable-length encoding scheme in practice, and it is suited for encoding the run-length sequences during compression. This manuscript introduces a modified Elias gamma scheme, which helps us to embed additional bits during the encoding process.
- An experimental study of the proposed scheme is carried out on natural images and medical images. The results show that the proposed system performs better than the well-known existing schemes.
Novelty of the Proposed Approach
The novelty of the proposed scheme is listed below:
- We propose a new RDH scheme suitable for the transmission of electronic patient reports along with medical images.
- The final image in the proposed process is in encrypted form, which ensures the security of the medical image and the patient reports.
- We introduced a bit plane compression-based RDH scheme, which can provide a high embedding rate with a 0 bit error rate during image recovery and data extraction.
- The well-known Elias gamma encoding process is modified to embed additional bits during the encoding process.
The remaining sections of this manuscript discuss related works, the proposed algorithms, the illustrative example, the experimental setup, and the results.
2. Related Work
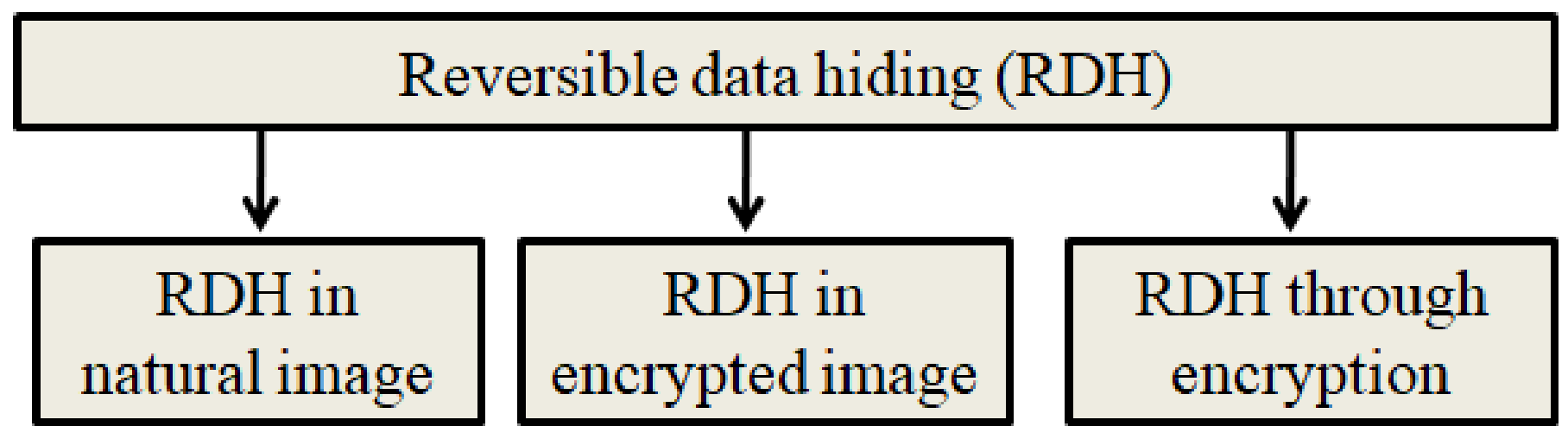
The existing RDH schemes can be broadly classified into three categories, and they are graphically represented in Figure 3.
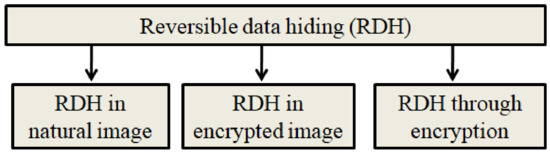
Figure 3.
Types of RDH schemes.
- RDH in original image: The original image will be considered during the RDH process. The quality of the stego image (image obtained after data hiding) is a major concern here.
- RDH in the encrypted image: In this, the RDH algorithm will consider the encrypted image. The embedding rate and the security are the two concerns when designing a new RDH scheme in encrypted images.
- RDH through encryption: This scheme combines image encryption and data hiding.
This section reviews a few well-known RDH schemes introduced for clinical data transmission during medical image transmission.
The existing RDH schemes mainly explored key concepts such as histogram shifting, difference expansion, lossless compression, and image scaling. Of course, many other approaches in the literature utilize entirely different ideas for RDH.
The basic version of the histogram-shifting approach is introduced in [25]. The scheme followed a RDH process in a natural image. The histogram of the cover image is computed first, and the grayscale value corresponding to the peak in the histogram is taken as the reference pixel value. Let us assume the peak intensity value is , and the approach discussed in [25] increments all the pixel values by one if the grayscale value in the cover image is greater than . This process will create a gap in the histogram just after the grayscale value . The maximum embedding capacity possible from this approach will be , where is the number of pixels with grayscale value . As per this scheme, the secret message bits can be embedded in the pixels with grayscale value . To embed an extra bit B in a cover image pixel with grayscale value , we need to add the B value with . The embedding capacity of the scheme is purely dependent on the peak in the histogram of the cover image. One of the major concerns of the histogram-shifting RDH approach is overflow or underflow during the histogram shifting.
In 2020, an efficient method to handle overflow or underflow in the histogram-shifting RDH scheme was discussed [26]. The system ensures an efficient representation of the location map to distinguish overflow/underflow pixels from original pixels. The method claims that the total length of the location map will be the number of 254 s and 255 s in the cover image.
Later, in 2021, a block-wise histogram shifting RDH with overflow handling was discussed in [27]. In this scheme, the histogram is computed from the non-overlapping image blocks of size pixels. The block-wise histogram RDH scheme helps to improve the embedding rate. Many other things need to be handled in the block-wise histogram RDH, such as the peak intensity from each block, the blocks used for RDH purposes, etc. The marker information (which is used to distinguish the blocks that are used for RDH from other blocks) is compressed using run-length encoding. Overall results show an excellent, effective embedding rate as compared to the existing approaches, even after embedding the additional overhead details in the image itself.
A number of approaches based on prediction error histogram processing were also reported in the literature [28,29]. In these schemes, the histogram is generated from the prediction error instead of grayscale pixel values from the image. In a prediction error histogram-based RDH, a predefined prediction approach will be used to predict each of the pixels in the original image. Then, the prediction error will be computed to compute the prediction error histogram. The prediction error computation is defined in Equation (1).
where is the prediction error corresponding to the original pixel when is the predicted pixel value. Later, pair-wise prediction error and machine learning-based prediction approaches were widely studied to improve the embedding rate [30,31,32].
Difference expansion is another major approach exploited for RDH in natural images and medical images [33]. This scheme is also concerned with overflow or underflow issues. An additional location map is required to handle overflow and underflow to ensure the lossless recovery of the original image. The well-known difference expansion RDH schemes are available in [34,35,36].
A RDH scheme capable of providing high embedding capacity through an image scaling approach is discussed in [37]. In this approach, the original image of size pixels is scaled up to a size of . The new pixels are introduced in the scaled-up image by replicating the pixels. The newly introduced pixel through the pixel-to-block conversion process is used to embed the clinical data and the additional authentication details for tamper detection. In every block of size pixels, every pixel except one pixel (pixel at the location (0, 0)) is used for embedding. The system claimed a high embedding rate, but the primary concern is that the bandwidth requirement for transmission is four times greater than the bandwidth requirement for transmitting the original image. However, the scheme allows a concealed transmission of clinical data and the medical image.
The scheme discussed in [37] was considered for further study by researchers in 2021, and an extended version was proposed in [38]. This paper discussed a scheme that can facilitate patient report transmission along with tamper detection capability. An authentication code is generated from the medical image, and the same is embedded in the image for tamper detection. Similar to the scheme discussed in [37], the original image of size pixels is scaled up to a size of . The new pixels in the scaled-up image are estimated based on a set of new predefined rules, which helps obtain scaled-up (watermarked) images with better visual quality. The readers may note that the key concern of the scheme [37] remains in the scheme reported in [38].
A Reed–Solomon code-based RDH scheme was introduced in [39]. The key benefit of the scheme discussed here is the resistance of the data-hiding scheme against salt and pepper noise. This scheme also ensures a high-quality stego image after performing data hiding.
An improved chaos encryption (ICE)-based approach was introduced in [40] to improve the security and the quality of the stego image. An enhanced modified signed digit (EMSD) and least significant substitution-based algorithm were discussed in [41]. This scheme ensures a high embedding rate, low computational complexity and high visual quality. Since this is a hybrid approach, steganalysis and decoding of the secret message is quite difficult in this approach.
The literature review in this domain led us to the following conclusion:
- The embedding capacity is a major concern in the RDH domain, and all researchers are working towards this. Note that lossless recovery of the original image and the message extraction should not be compromised while achieving a better embedding rate.
- An image scaling-based approach is not a good solution for the RDH since it leads to high bandwidth requirements and storage space that we do not want to entertain.
- The histogram-based RDH schemes and difference expansion-based RDH schemes need to consider the location maps to ensure lossless recovery due to overflow and underflow.
- The RDH schemes should be lightweight enough to provide real-time performance during data hiding and image recovery.
All the points mentioned above were kept in mind while proposing a new bit plane compression-based RDH scheme with the modified Elias gamma encoding approach.
3. Preliminaries
The proposed scheme uses mainly two concepts: run-length encoding and Elias gamma encoding. These two are detailed in this section to understand the proposed method in a better way.
3.1. Run-Length Encoding
Run-length encoding (RLE) is a lossless data compression method that analyzes a sequence with the same value occurring many consecutive times. It helps to compress the data by encoding the sequence by storing a single data value and its count.
Two examples are given below to understand the RLE process. The first example considers a sequence of ASCII characters, and the second example assumes a bit sequence.
Let us assume that the given character sequence C is as follows:
The RLE sequence that corresponds to C is given below:
Similarly, let us assume that we have a binary sequence B, as given below:
The RLE sequence that corresponds to B is given below:
The RLE for the bit sequence is slightly different when we compare it with the RLE of ASCII characters. While applying RLE for ASCII characters, we need to keep each character in the run and its count. But while performing RLE in a binary sequence, we need to keep the first bit of the original arrangement; then, we need to keep counts. Let us denote the where indicates the starting bit in the original series (0 or 1); then, indicates how many times consecutively appears in the original series, and indicates how many times occurs in consecutive positions and so on.
3.2. Elais Gamma Encoding
The run-length sequence that we obtain through the RLE process cannot be stored or transmitted since we need to convert the same to a sequence of bits. We can use a fixed-length encoding approach or a variable-length approach to transform the RLE sequence into a series of bits. The number of bits used for representing each of the sequences in RLE will be determined by the number of bits required to represent the maximum value in the sequence. For example, if Q is the maximum run-length size, then number of bits should be used to store each value in the RLE sequence. The fixed-length encoding is not preferred since we may be wasting several bits while encoding small integers. This motivated us to use some variable-length encoding approaches in the RLE encoding process. The Elias gamma encoding process (EGE) is one well-known variable-length encoding approach.
Let us assume that we have an integer value G, the corresponding binary value is , where . In this case, if we use Elias gamma encoding, we will embed number of 0s as the prefix of to obtain the corresponding Elias gamma-encoded sequence.
Consider the RLE sequence given in Equation (5); the binary values correspondiong to 6 is 110, 5 is 101 and 10 is 1010. Therefore, the Elias gamma encoded sequence will be as follows:
The green-colored bit sequence indicates the Elias gamma code corresponding to the count of 1 s, and the red-colored bit sequence shows the Elias gamma code corresponding to the count of 0 s.
The readers may note that we have modified the existing Elias gamma encoding process in this work so that we shall embed a sequence of bits in the Elias gamma-encoded sequence itself.
4. Proposed Scheme
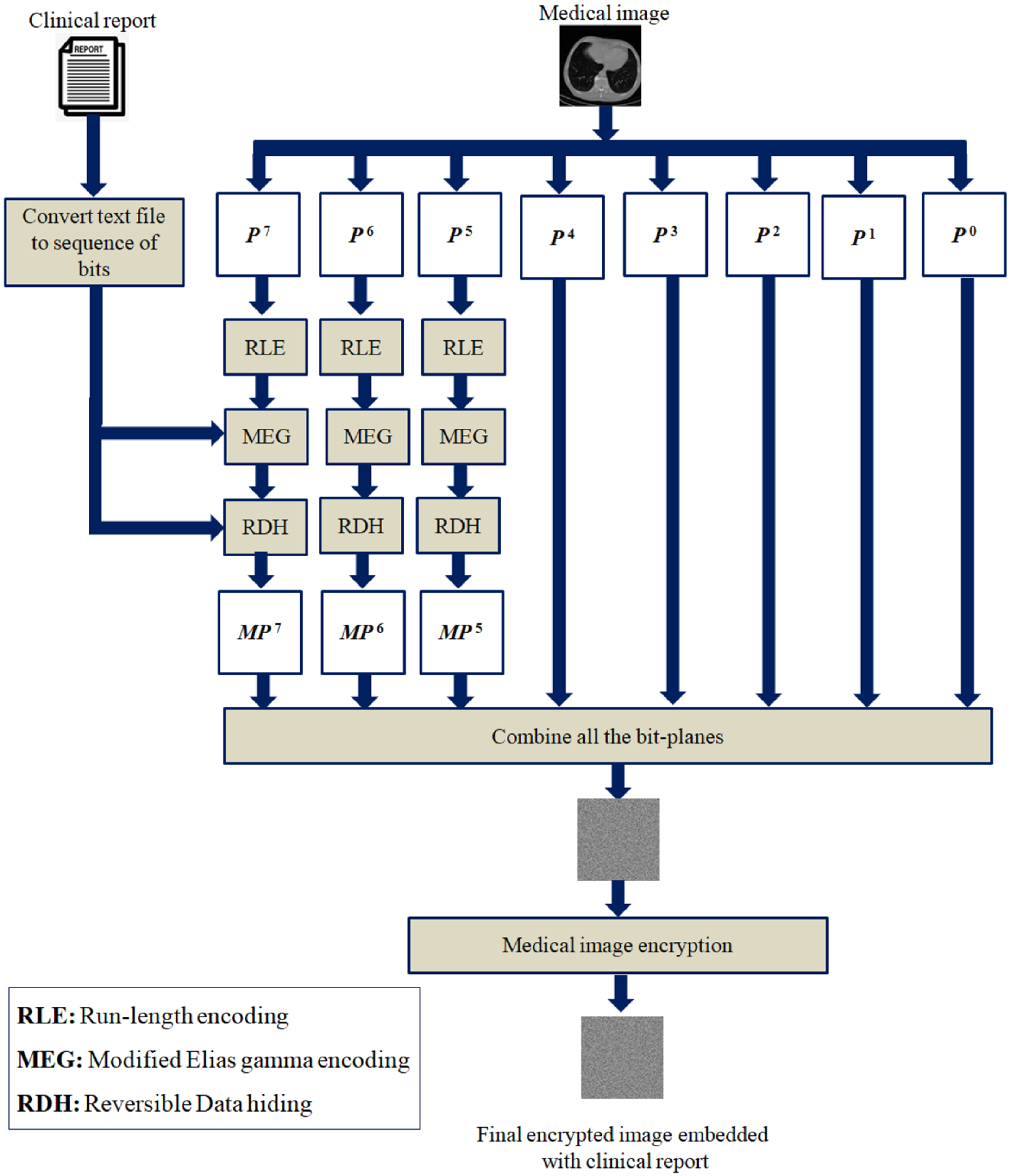
The proposed scheme is detailed in this section. The proposed reversible data hiding algorithm for embedding clinical data in the medical image is given in Algorithm 1. Algorithm 2 shows the newly modified Elias gamma encoding procedure to embed additional message bits during the encoding process. The key steps required to extract the clinical report and restore the original medical image are given in Algorithm 3. Figure 4 depicts the proposed scheme.
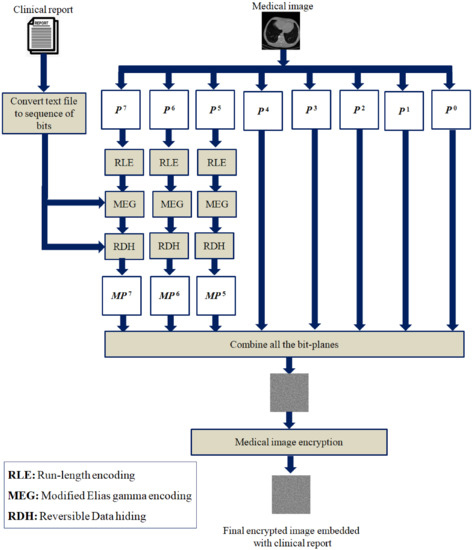
Figure 4.
Illustrative diagram of the proposed scheme.
| Algorithm 1: Proposed bit plane compression-based RDH scheme using modified Elias gamma encoding |
 |
| Algorithm 2: Modified Elias gamma approach for variable length encoding. |
 |
| Algorithm 3: Clinical report extraction and image recovery from the proposed scheme. |
 |
Illustrative Example
For a better understanding of the proposed scheme, an illustrative example is discussed in this section. As per the proposed RDH scheme, a medical image I of size and a clinical report of T are considered as the input. The medical images may use 8-bit, 12-bit, or 16-bit grayscale representation for capturing more gray levels, which may help the doctors to make better diagnoses. Our experimental study considered 8-bit medical images, and we assume that the clinical report is a text file. The clinical report content should be converted to the corresponding bit representation to perform RDH.
In the second step, the given medical image I is divided into 8 different bit planes. If we analyze the bit planes, it can be observed that the bits in the most significant bit planes will be highly correlated. Since the few most significant planes are highly correlated, those planes can be compressed with a high compression ratio. We have considered the Q number of most significant bit planes for compression using RLE. To encode RLE, we have introduced a modified version of the Elias gamma encoding technique with an RDH capability. RDH we will achieve in two ways: the RDH during the modified Elias gamma encoding procedure and the other through the embedding of data in the space created through lossless compression.
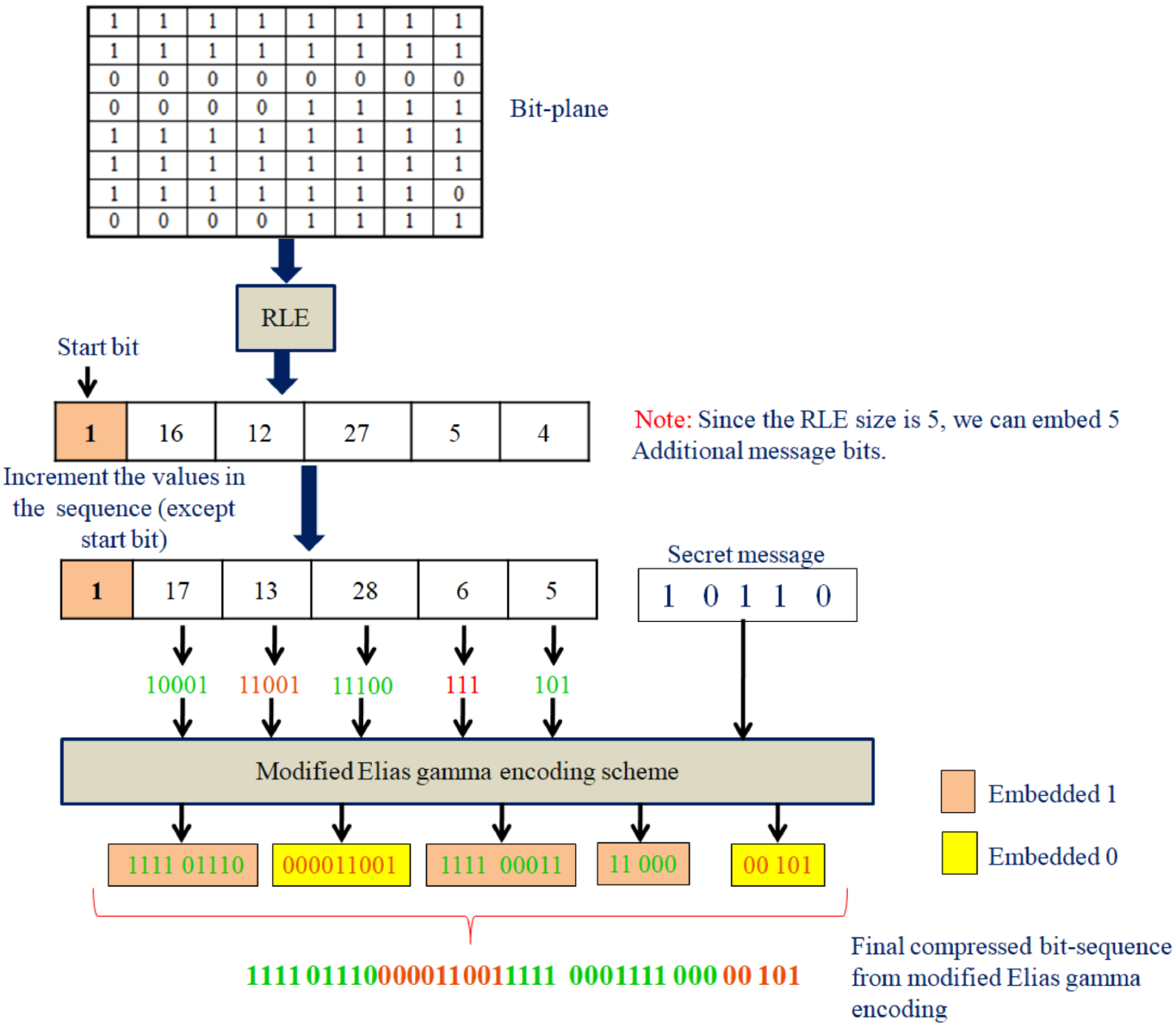
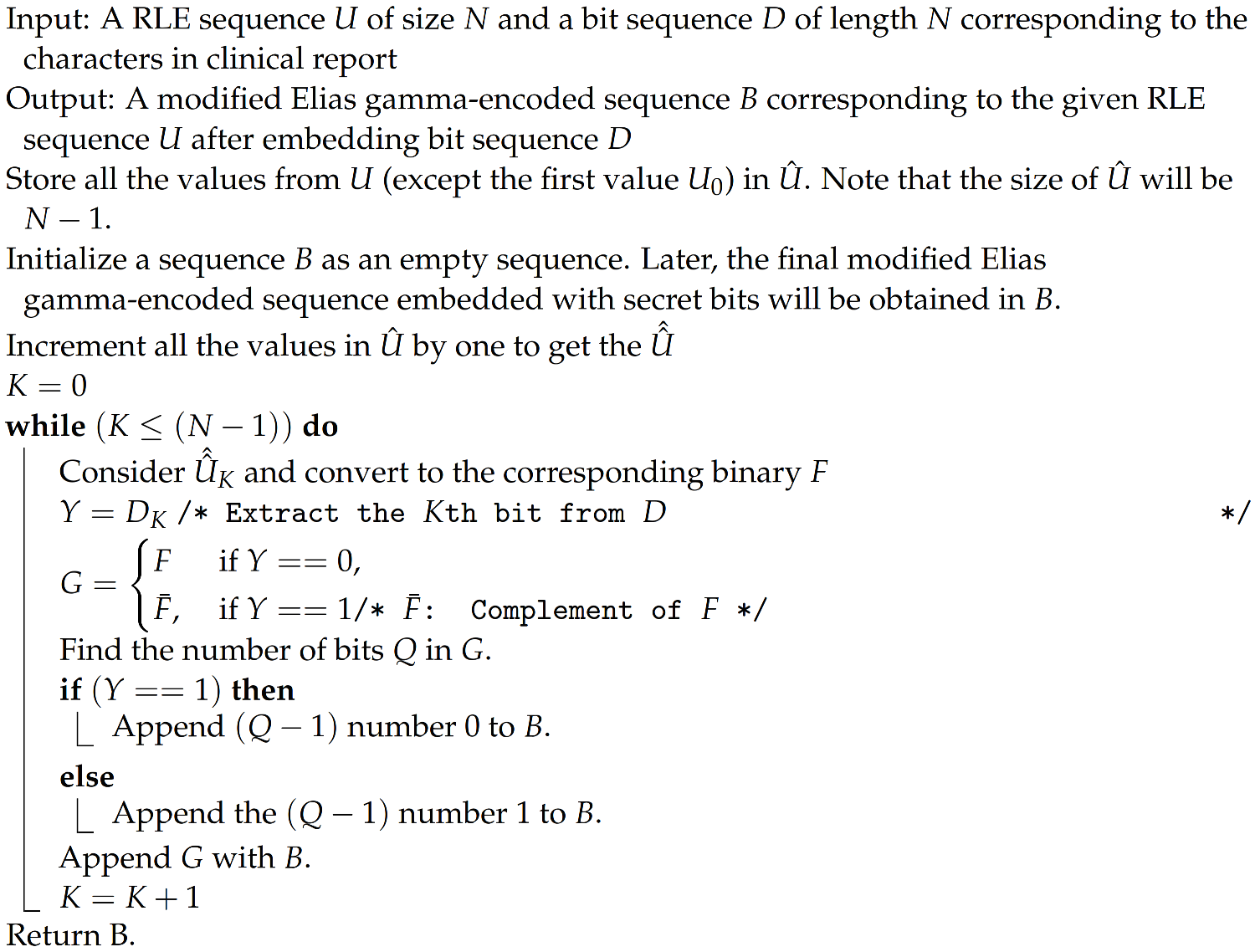
An illustrative example to demonstrate the modified Elias gamma encoding approach is given in Figure 5.
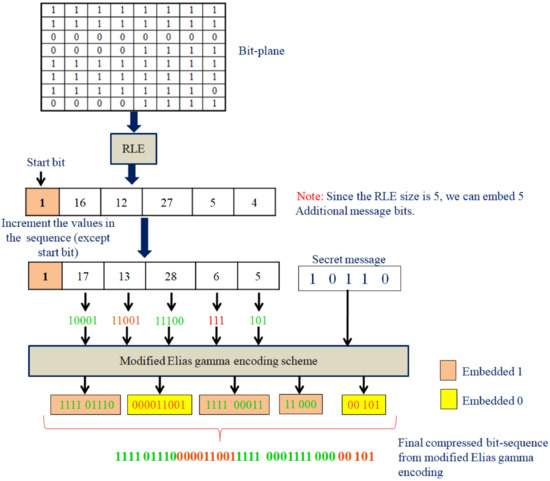
Figure 5.
An illustrative example for modified Elias gamma encoding.
Figure 5 shows one bit plane for demonstration purposes. Firstly, the RLE sequence is identified from the bit plane. Then, we used the modified Elias gamma encoding method to convert the run-length sequence to a sequence of bits. For this, the RLE sequence that we computed was incremented by one. Note that this increment process may lead to an increase in the number of bits in the corresponding binary representation if the integer value is , where J is a positive integer. Let us denote the RLE sequence obtained after incrementing by one with (except the first value, which is the starting bit).
Each of the run-length values will be considered from , and the number of bits that we can embed through the modified Elias gamma encoding process will be dependent on the number of integers in the run-length sequence. In the illustrative example, there are five run-length sequences, so we shall embed 5 bits during the modified Elias gamma encoding process. Let us assume the sequence of bits that we need to embed is .
Now, let us consider the first sequence in the run-length sequence, which is 16. After incrementing 16 by 1, we will get 17. The binary value corresponding to 17 is 10001. Let us assume that we need to embed the first bit from the secret message 1 in the run-length value 17. Since we need to embed a bit value, find the bit inverse of 10001, which is 01110. Now, adding 1111 as the prefix of 01110 will result in 111101110. If we need to embed a bit value 0, in that case, we will consider the binary value as it is, and we need to add number of 0s as a prefix, where Q is the number of bits in the binary representation of the selected run-length count; note that this is one part of RDH in the proposed scheme. In addition to this, we will make use of the space created through the lossless compression to store the secret message bits.
In the illustrative example, we shall see that the bit plane size is pixels, which requires 64 bits for its actual representation. Now we shall see that the final bit sequence obtained from the last stage of modified Elias gamma encoding consists of 37. Note that we already embedded a 5-bit secret message during the modified Elias gamma process. If we analyze the vacant space created through compression, it will be . The final bit plane of 64 bits will be generated by concatenating 27 bits from the clinical report (secret message) with the bit sequence obtained from the modified Elias gamma encoding procedure. This will be converted to a 2D array of size and considered the modified bit plane.
This process will be continued for a few numbers of the most significant bit planes. Note that the RLE compression can create some vacant spaces only when there is a high correlation between the adjacent bits. If we see the bit planes in the least significant positions, they will be highly random, and we cannot achieve compression over there with RLE. Therefore, those planes should remain the same in the final stego image.
Once we perform compression and data hiding on a few of the most significant selected bit planes, then we will combine all those planes along with the unmodified least significant planes to obtain the final image . Note that will not have any natural image characteristics since we completely modified the most significant planes. The image we will encrypt using a secure image encryption method using the encryption key K before it is transmitted through the network.
On the receiver side, the receiver will decrypt the image using the decryption key K. Further, the decoding process will be conducted on the bit planes. Note that the receiver should know the number of bit planes used for the RDH process. The modified Elias gamma decoding and run-length decoding will be applied on all the most significant bit planes that are used for RDH. During the decoding process, we will extract a few secret message bits. The decoding process will be stopped once we get the number of bits which will be bits of the original bit plane.
5. Experimental Study and Result Analysis
The proposed RDH scheme, data extraction, and image restoration algorithms are implemented using Matlab 2020. We have considered natural images from the USC-SIPI image dataset [42] and medical images from [43].
This section mainly discusses the experimental results that we observed during the study. The following efficiency parameters are considered during the analysis:
- Embedding rate;
- Security analysis;
- Analysis of image recovery;
- Time complexity analysis.
5.1. Embedding Rate
The embedding rate is one of the key parameters that we need to consider when analyzing an RDH scheme. In a medical image transmission context, this defines the number of bits (from the clinical report) that we can embed in the medical image so that we shall restore the original image during the extraction of hidden bits.
Mathematically, the embedding rate is defined as
where is the embedding rate, is the number of bits embedded in the medical image, and is the total number of pixels in the medical image.
The embedding rate from the proposed RDH scheme relies on the bit plane compression that we can achieve from the bit planes.
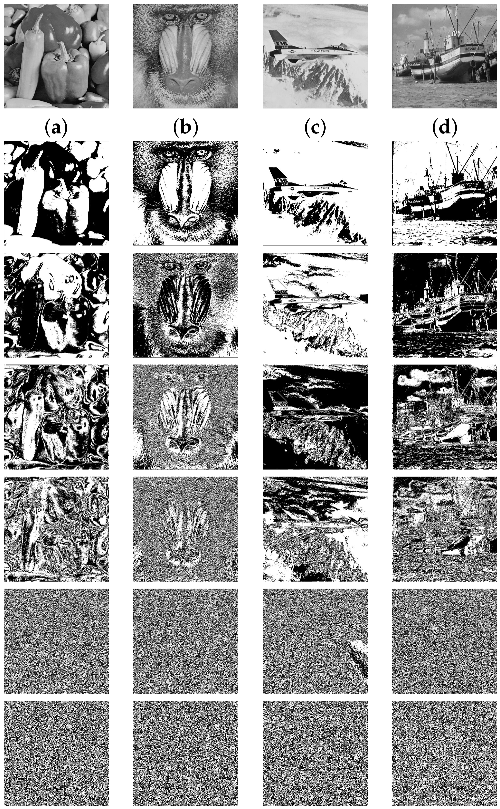
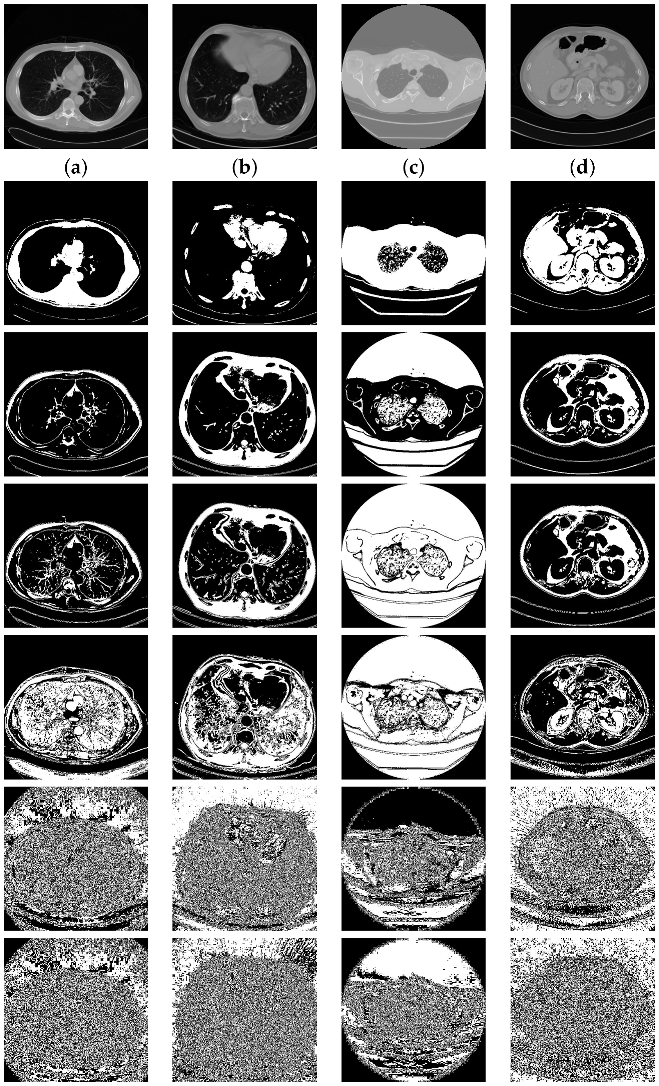
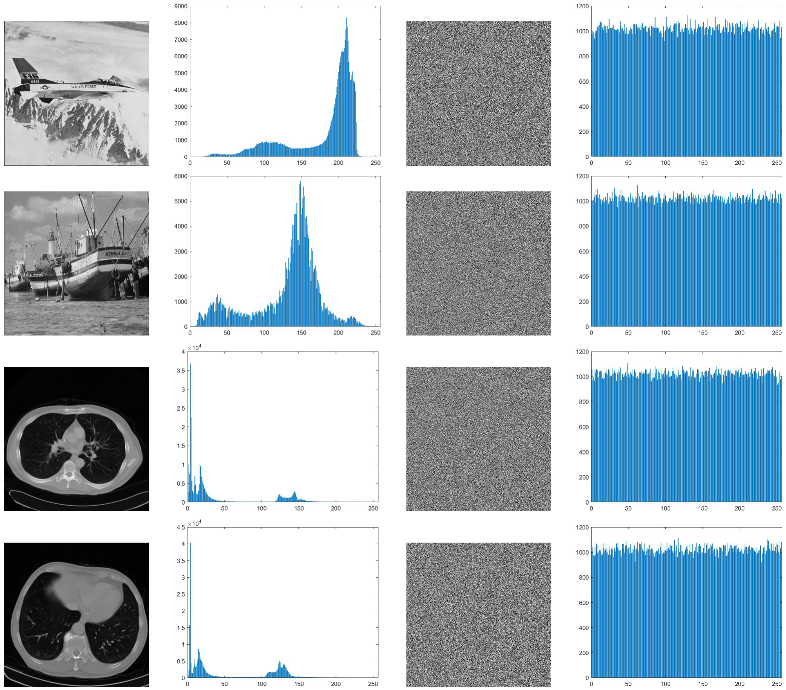
The compression ratio obtained from each of the bit planes of the eight images (four natural images and four medical images) is given in Table 1. Table 1 gives an overall idea regarding the compression ratio and its relationship with the bit plane. The RLE and modified Elias gamma encoding cannot always ensure the compression of the data. The compression method will provide compression only when lengthy runs of bits are in the bit plane. The medical images are generally very smooth with smooth textures. Hence, we shall see that several most significant bit planes give high compression (compression ratio greater than 1). In Table 1, the cells are marked with two different colors (green/red). The green color indicates that the corresponding bit planes are compressed with a compression ratio greater than 1. This indicates that we shall obtain some free space to embed additional data. The cells colored with red indicate that those planes are compressed with a compression ratio of more than 1 (the size of the compressed data is greater than the size of the actual data). Such planes we cannot use for the data-hiding process. Figure 6 depicts bit planes extracted from the natural images, and Figure 7 depicts bit planes extracted from the medical images.

Table 1.
Compression ratio obtained from sample images.

Figure 6.
Bit planes extracted from the natural images (Row 1: original images. (a) Peppers, (b) baboon, (c) airplane, (d) boat. Row 2: plane 7; row 3: plane 6; row 4: plane 5; row 5: plane 4; row 6: plane 1; row 7: plane 0).
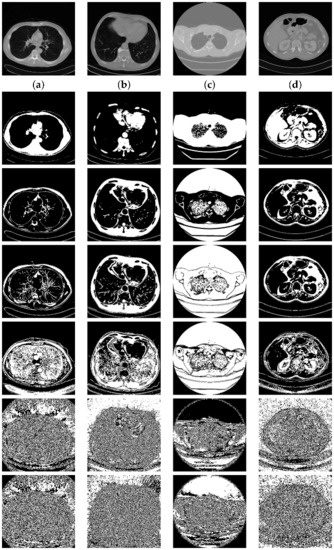
Figure 7.
Bit planes extracted from the medical images (Row 1: original images; (a) medical image-1; (b) medical image-2; (c) medical image-3; (d) medical image-4. Row 2: plane 7; row 3: plane 6; row 4: plane 5; row 5: plane 4; row 6: plane 1; Row 7: Plane 0).
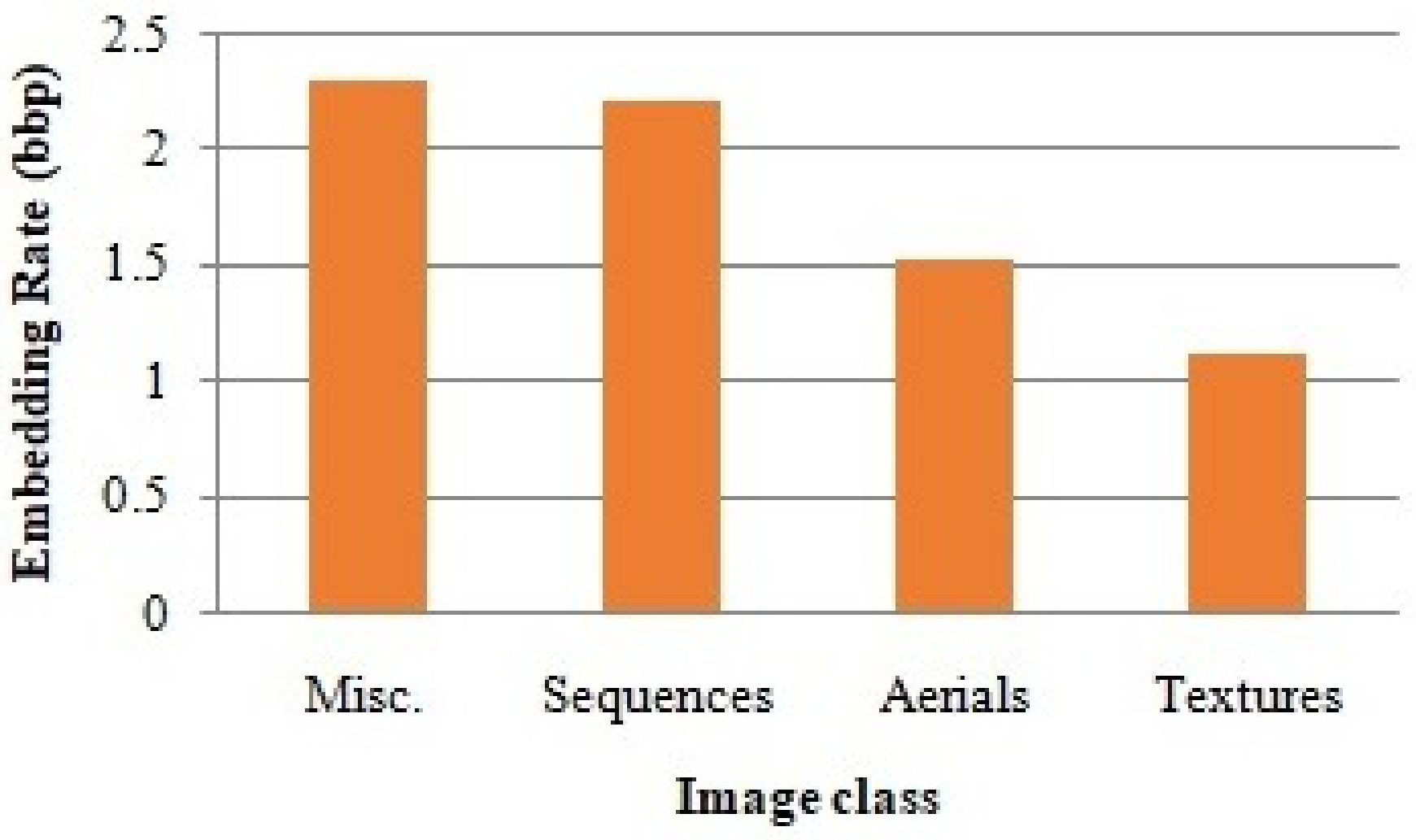
The average embedding rate obtained from the four natural image classes is shown in Figure 8. Note that the four natural image classes have their own features. The average embedding rate from and are high compared to the two other classes since the images in the first two classes are so smooth, which will lead to high compression.
Figure 8.
Embedding rate obtained from natural image classes.
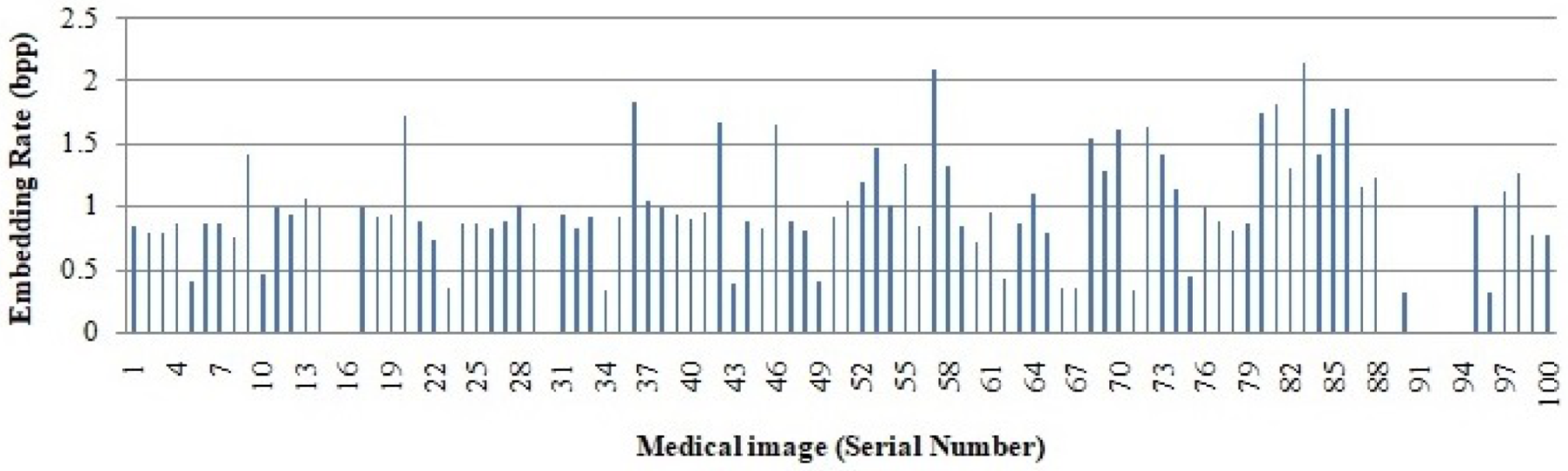
The embedding rate obtained from all 100 medical images is graphically represented in Figure 9. Since the medical images in the dataset have entirely different characteristics, the embedding rate varies. The results show that from most of the images, we obtain an embedding rate of more than 1.
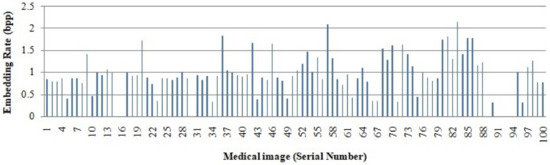
Figure 9.
Embedding rate of 100 medical images.
The proposed approach is also compared with the conventional RLE with the Elias gamma scheme to demonstrate the improvement of the proposed modified Elias gamma encoding approach. In Table 2, the RLE + EG indicates the use of conventional run-length encoding with the Elias gamma encoding approach. Similarly, RLE + MEG indicates the proposed method based on run-length encoding with a newly modified Elias gamma approach.

Table 2.
Comparison of embedding rate (in bpp) with the RLE + EG approach.
5.2. Analysis of Embedding Capacity through Modified Elias Gamma Encoding Process
Let us assume that we have a run-length encoding sequence obtained as follows:
where B indicates the starting bit in the plane, and indicates the run-length counts in which . Let us denote the number of bits required to represent by using .
Note that during the modified Elias gamma encoding process, the run-length sequence values will be incremented by one. Let us denote the run-length sequence obtained after incrementing the original sequence by .
Let us denote the number of bits required to represent by using .
While incrementing the actual run-length sequence by one, there are two possibilities:
- Case 1 (best case):In this case, we will obtain the maximum embedding capacity through the modified Elias gamma encoding process. Note that in this case, we shall embed N additional bits through the encoding process without any change in the length of the binary sequence obtained after encoding. This is the best possible embedding capacity that we can additionally achieve through a modified Elias gamma encoding scheme. Note that in addition to this, we will use the empty space created through compression also to keep additional messages.
- Case 2 (worst case):In this case, every run-length sequence needs one extra bit for representation. Therefore, the overall overhead will become during the modified Elias gamma encoding process. However, we will have an opportunity to embed one-bit additional data on all the N sequences. Hence, the extra overhead will become N bits.However, note that the probability of happening this situation is very low. This situation will happen only when all the run-length sequences are from , where K can be any positive integer.If we consider an image of , there will be 262,144 pixels. The probability that one from the set (where 262,144) is . This shows that the probability to obtain the value, which will lead to the worst-case embedding capacity, is very close to 0.
5.3. Analysis of Image Recovery
The lossless restoration of the original image is the critical concern of any RDH scheme. The recovery of the original image can be analyzed using reference image quality measures such as peak-to-signal noise ratio (PSNR) [44] and the structural similarity index (SSIM) [44]. The PSNR between the original image I and the restored image G of size can be defined as follows:
where E is the mean square error between two images. If both the original image I and the restored image G are the same, then E will be 0. Otherwise, it will be a positive value. To find the PSNR value P between two images, we have to use E in the following way:
The P value will be ∞ if the original image I and the restored image G are the same. Note that if both the images are the same, the E value will be 0, which will lead to .
Similarly, SSIM is another measure that we can use to analyze the quality degradation between the original image I and the restored image G [44].
To ensure the image recoverability of the proposed scheme, we have analyzed the PSNR and SSIM. The PSNR and SSIM measure observed during the recovery analysis of the proposed scheme is given in Table 3.

Table 3.
PSNR and SSIM observed during image recovery.
The results shown in Table 3 demonstrate that the proposed scheme losslessly restores the image without any recovery error.
5.4. Security Analysis
Note that the final image we obtain after performing RDH through the proposed scheme will be encrypted using a secure image encryption scheme. The RDH scheme does not directly provide any data security for the image or hidden clinical report. However, the use of secure image encryption will protect the confidentiality of the medical image and confidential information from attackers.
During the simulation of the proposed scheme, we used the AES image encryption technique, which is very popular for the secure transmission of data in all domains. Entropy and histogram analysis are the two efficiency parameters helpful in analyzing the randomness of the final encrypted image.
Entropy is a measure that analyzes the data to identify randomness [45]. Entropy from an 8-bit grayscale image can be computed in the following way:
where E indicates the entropy measure of the image given image, and X is the different grayscale values that can be present in the given image. In an 8-bit grayscale image, X will be from the set {0, 1,…,255}. indicates the probability of the grayscale value X in the given image.
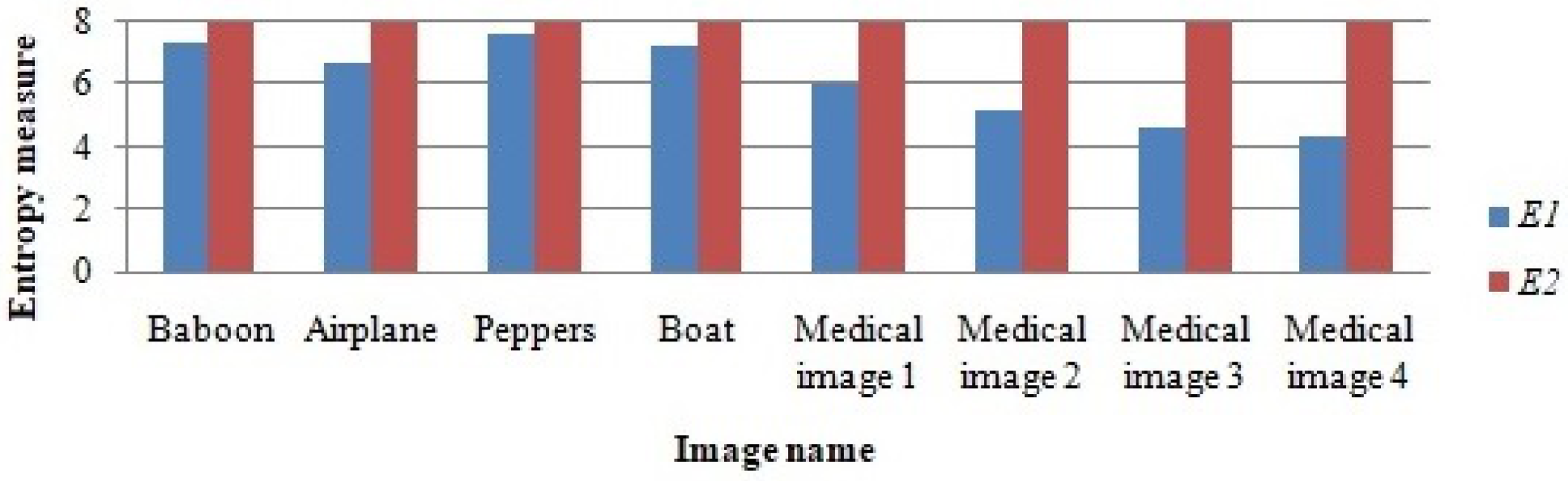
The entropy observed from the selected images before RDH (), the encryption process, and the entropy observed in the encrypted image () is demonstrated in the bar graph given in Figure 10.
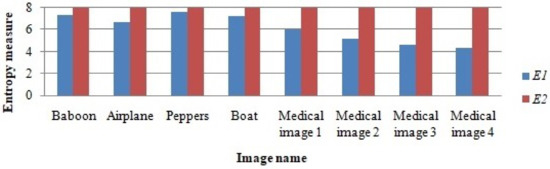
Figure 10.
Entropy measure observed from original image () and from the final encrypted image with hidden data ().
From Figure 10, it can be seen that the data hiding process does not affect the entropy from the encrypted image. The final entropy measure () is very close to 8, and this indicates that the pixels in the final encrypted image are uniformly distributed, which led to the entropy measure of 8.
In addition to this, we also analyzed the histogram of the original image and the final encrypted image to show that the proposed RDH scheme does not affect the encryption efficiency. The histograms obtained from the original images and the encrypted images are given in Figure 11. From the histogram analysis given in Figure 11, it can be seen that the histogram obtained from the final encrypted images looks flat since the pixel values are uniformly distributed. The RDH process does not affect encryption efficiency.
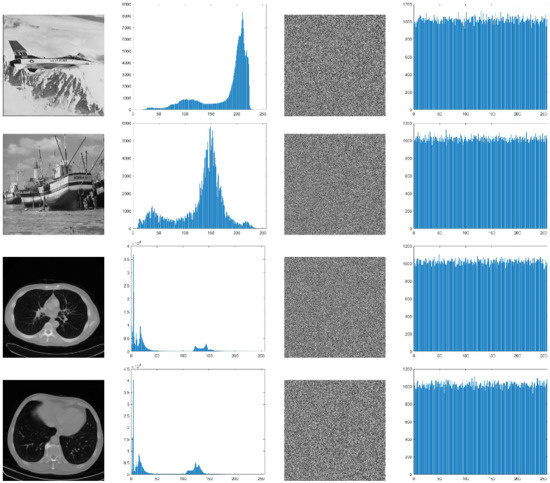
Figure 11.
Histogram analysis of the final encrypted images.
5.5. Time Complexity Analysis
We have conducted a detailed time complexity analysis of the proposed RDH algorithm. The critical activity during the RDH process is the bit plane compression, for which we need to access all the bits from all K numbers of the most significant bit planes. Let us assume that the original image size is ; in that case, we need to access K times the bit planes bits. Thus, the overall operations will be . Since K is a constant value from , the final time complexity of the proposed RDH process will be .
Similarly, during the data extraction and image restoration phase, we need to access the bits from the bit planes and want to do the run-length decoding procedure. Again, in this phase, we need to access a constant number of bit planes for image restoration and data extraction and note that each plane has a size of bits. Hence, the time complexity of the data extraction and image recovery process is .
Note that the time complexity of the proposed method is linear to the input size (total number of pixels in the given image).
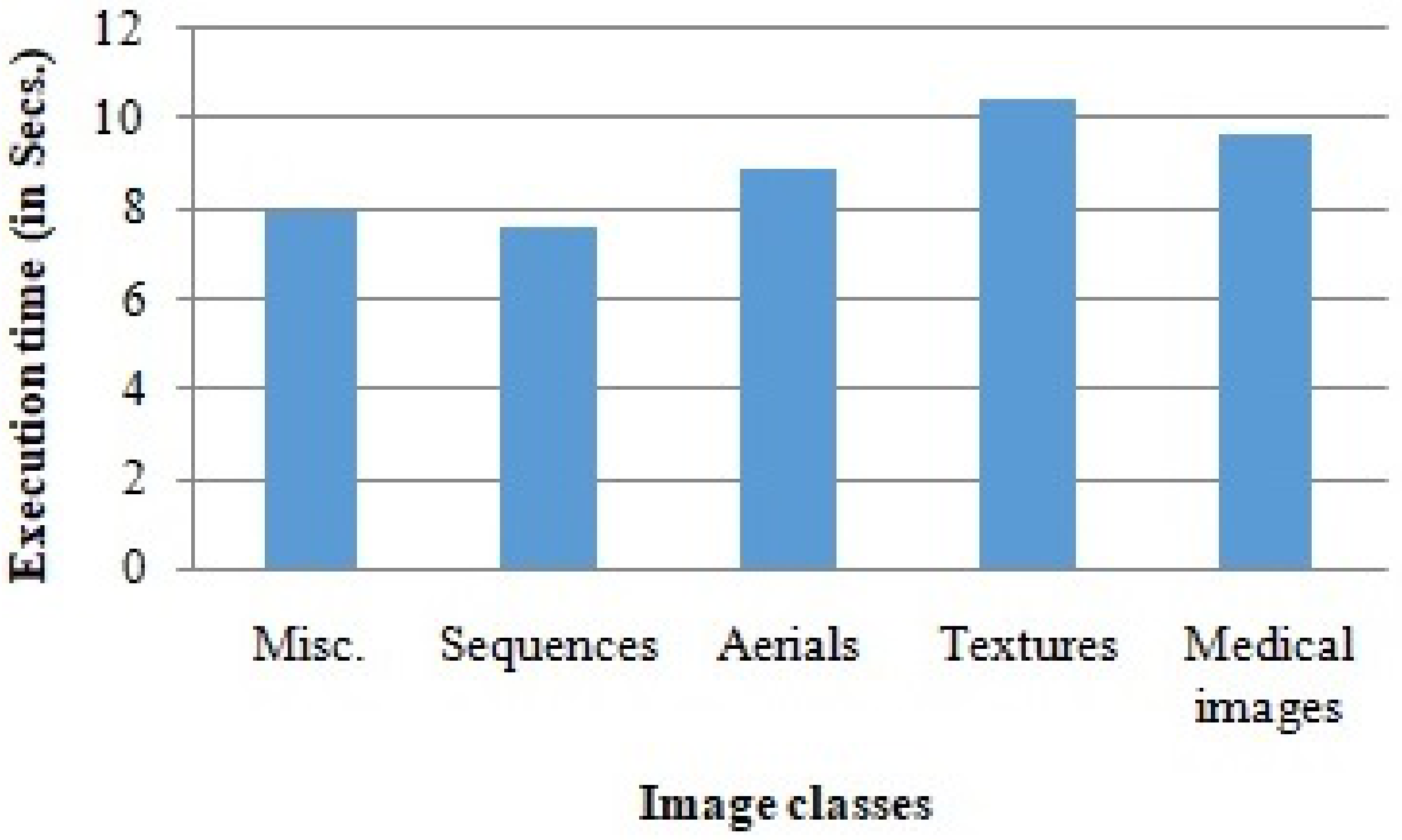
We used Matlab 2020 to implement the proposed algorithms, and the simulations were carried out on a machine with an Intel I5 8th generation processor. The average execution time observed during the experimental study of the proposed algorithms is demonstrated in Figure 12. The results show that the proposed scheme can be used in real-time applications.
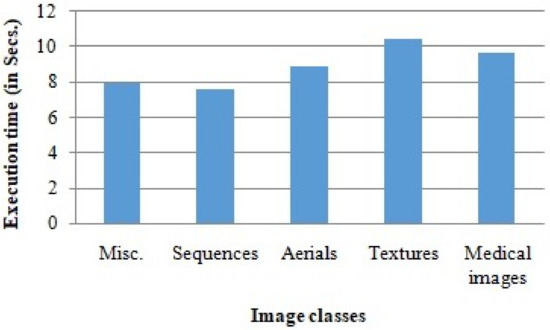
Figure 12.
Average execution time of the proposed scheme in various image classes.
6. Comparative Study
The embedding rate from the proposed scheme for a few well-known images is compared with the embedding rate from a few well-known existing methods in Table 4. It can be seen that the embedding rate from the proposed scheme is far better than the embedding rate from the existing methods.

Table 4.
Comparison of embedding rate for the well-known images.
7. Conclusions
A high-capacity reversible data hiding scheme based on bit plane compression using run-length encoding and a modified Elias gamma encoding process is introduced in this manuscript. The well-known variable-length encoding scheme, the Elias gamma encoding method, is modified and presented as a new approach capable of embedding additional secret message bits during the encoding process. An experimental study is carried out on standard natural images and medical images to analyze the efficiency of the proposed scheme. We have observed an excellent embedding rate for the medical images and a sufficiently good embedding rate for other natural images. The average embedding rate from the proposed scheme for the medical images is 0.75 bits per pixel. The results show that the proposed scheme can be used in the healthcare domain to transmit clinical reports by embedding the same in the respective medical images. We assumed that the final image would be transmitted to the receiver without any error in the proposed scheme. Future works can be focused on designing and implementing a robust RDH scheme capable of resisting attacks.
Author Contributions
Conceptualization, B.S. and S.H.; Data curation, V.M.M.; Formal analysis, K.S.R.M., B.S. and S.H.; Investigation, K.S.R.M.; Methodology, N.V., P.K.R.M. and S.H.; Resources, N.V.; Software, V.M.M.; Supervision, V.M.M.; Visualization, N.V.; Writing—original draft, V.M.M., B.S. and P.K.R.M.; Writing—review & editing, V.M.M., P.K.R.M. and S.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chengoden, R.; Victor, N.; Huynh-The, T.; Yenduri, G.; Jhaveri, R.H.; Alazab, M.; Bhattacharya, S.; Hegde, P.; Maddikunta, P.K.R.; Gadekallu, T.R. Metaverse for Healthcare: A Survey on Potential Applications, Challenges and Future Directions. arXiv 2022, arXiv:2209.04160. [Google Scholar]
- Bhattacharya, S.; Maddikunta, P.K.R.; Pham, Q.V.; Gadekallu, T.R.; Chowdhary, C.L.; Alazab, M.; Piran, M.J. Deep learning and medical image processing for coronavirus (COVID-19) pandemic: A survey. Sustain. Cities Soc. 2021, 65, 102589. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.N.; Singh, A.K. Towards Integrating Image Encryption with Compression: A Survey. ACM Trans. Multimed. Comput. Commun. Appl. (TOMM) 2022, 18, 89. [Google Scholar] [CrossRef]
- Rupa, C.; Harshita, M.; Srivastava, G.; Gadekallu, T.R.; Maddikunta, P.K.R. Securing Multimedia using a Deep Learning based Chaotic Logistic Map. IEEE J. Biomed. Health Inform. 2022. [Google Scholar] [CrossRef]
- Abd el Samie, F.E.; Ahmed, H.E.H.; Elashry, I.F.; Shahieen, M.H.; Faragallah, O.S.; El-Rabaie, E.S.M.; Alshebeili, S.A. Image Encryption: A Communication Perspective; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Srivastava, G.; Jhaveri, R.H.; Bhattacharya, S.; Pandya, S.; Maddikunta, P.K.R.; Yenduri, G.; Hall, J.G.; Alazab, M.; Gadekallu, T.R. XAI for Cybersecurity: State of the Art, Challenges, Open Issues and Future Directions. arXiv 2022, arXiv:2206.03585. [Google Scholar]
- Manikandan, V.; Masilamani, V. An efficient visually meaningful image encryption using Arnold transform. In Proceedings of the IEEE Students’ Technology Symposium (TechSym), Kharagpur, India, 30 September–2 October 2016; pp. 266–271. [Google Scholar]
- Dolendro Singh, L.; Manglem Singh, K. Visually meaningful multi-image encryption scheme. Arab. J. Sci. Eng. 2018, 43, 7397–7407. [Google Scholar] [CrossRef]
- Bender, W.; Gruhl, D.; Morimoto, N.; Lu, A. Techniques for data hiding. IBM Syst. J. 1996, 35, 313–336. [Google Scholar] [CrossRef]
- Alazab, M.; RM, S.P.; Parimala, M.; Maddikunta, P.K.R.; Gadekallu, T.R.; Pham, Q.V. Federated Learning for Cybersecurity: Concepts, Challenges, and Future Directions. IEEE Trans. Ind. Inform. 2021, 18, 3501–3509. [Google Scholar] [CrossRef]
- Taheri, R.; Shojafar, M.; Alazab, M.; Tafazolli, R. FED-IIoT: A robust federated malware detection architecture in industrial IoT. IEEE Trans. Ind. Inform. 2020, 17, 8442–8452. [Google Scholar] [CrossRef]
- Singh, A.K.; Lv, Z.; Lu, H.; Chang, X. Guest editorial: Recent trends in multimedia data-hiding: A reliable mean for secure communications. J. Ambient Intell. Humaniz. Comput. 2020, 11, 1795–1797. [Google Scholar] [CrossRef]
- Liu, L.; Meng, L.; Zheng, W.; Peng, Y.; Wang, X. A Larger Capacity Data Hiding Scheme Based on DNN. Wirel. Commun. Mob. Comput. 2022, 2022, 5425674. [Google Scholar] [CrossRef]
- Mohanarathinam, A.; Kamalraj, S.; Prasanna Venkatesan, G.; Ravi, R.V.; Manikandababu, C. Digital watermarking techniques for image security: A review. J. Ambient Intell. Humaniz. Comput. 2020, 11, 3221–3229. [Google Scholar] [CrossRef]
- Alazab, M.; Tang, M. Deep Learning Applications for Cyber Security; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Jose, A.; Subramaniam, K. Comparative analysis of reversible data hiding schemes. IET Image Process. 2020, 14, 2064–2073. [Google Scholar] [CrossRef]
- Kumar, S.; Gupta, A.; Walia, G.S. Reversible data hiding: A contemporary survey of state-of-the-art, opportunities and challenges. Appl. Intell. 2021, 52, 7373–7406. [Google Scholar] [CrossRef]
- Singh, P.; Raman, B. Reversible data hiding based on Shamir’s secret sharing for color images over cloud. Inf. Sci. 2018, 422, 77–97. [Google Scholar] [CrossRef]
- Senthilnathan, T.; Prabu, P.; Sivakumar, R.; Sakthivel, S. An enhancing reversible data hiding for secured data using shuffle block key encryption and histogram bit shifting in cloud environment. Clust. Comput. 2019, 22, 12839–12847. [Google Scholar] [CrossRef]
- Liu, X.; Lou, J.; Fang, H.; Chen, Y.; Ouyang, P.; Wang, Y.; Zou, B.; Wang, L. A novel robust reversible watermarking scheme for protecting authenticity and integrity of medical images. IEEE Access 2019, 7, 76580–76598. [Google Scholar] [CrossRef]
- Zhong, X.; Shih, F.Y. A high-capacity reversible watermarking scheme based on shape decomposition for medical images. Int. J. Pattern Recognit. Artif. Intell. 2019, 33, 1950001. [Google Scholar] [CrossRef]
- Panchikkil, S.; Manikandan, V.; Zhang, Y.D. A pseudo-random pixel mapping with weighted mesh graph approach for reversible data hiding in encrypted image. Multimed. Tools Appl. 2022, 81, 16279–16307. [Google Scholar] [CrossRef]
- Kaw, J.A.; Loan, N.A.; Parah, S.A.; Muhammad, K.; Sheikh, J.A.; Bhat, G.M. A reversible and secure patient information hiding system for IoT driven e-health. Int. J. Inf. Manag. 2019, 45, 262–275. [Google Scholar] [CrossRef]
- Mansour, R.F.; Parah, S.A. Reversible Data Hiding for Electronic Patient Information Security for Telemedicine Applications. Arab. J. Sci. Eng. 2021, 46, 9129–9144. [Google Scholar] [CrossRef]
- Ni, Z.; Shi, Y.Q.; Ansari, N.; Su, W. Reversible data hiding. IEEE Trans. Circuits Syst. Video Technol. 2006, 16, 354–362. [Google Scholar]
- Manikandan, V.; Renjith, P. An efficient overflow handling technique for histogram shifting based reversible data hiding. In Proceedings of the 2020 International Conference on Innovative Trends in Information Technology (ICITIIT), Kottayam, India, 13–14 February 2020; pp. 1–6. [Google Scholar]
- Kandala Sree Rama Murthy, V.M.M. Reversible Data Hiding using Block-wise Histogram Shifting and Run-length Encoding. Int. J. Adv. Comput. Sci. Appl. 2021, 12, 74–80. [Google Scholar]
- Li, X.; Yang, B.; Zeng, T. Efficient reversible watermarking based on adaptive prediction-error expansion and pixel selection. IEEE Trans. Image Process. 2011, 20, 3524–3533. [Google Scholar]
- Fu, D.S.; Jing, Z.J.; Zhao, S.G.; Fan, J. Reversible data hiding based on prediction-error histogram shifting and EMD mechanism. AEU-Int. J. Electron. Commun. 2014, 68, 933–943. [Google Scholar] [CrossRef]
- Hou, J.; Ou, B.; Tian, H.; Qin, Z. Reversible data hiding based on multiple histograms modification and deep neural networks. Signal Process. Image Commun. 2021, 92, 116118. [Google Scholar] [CrossRef]
- Wang, J.; Mao, N.; Chen, X.; Ni, J.; Wang, C.; Shi, Y. Multiple histograms based reversible data hiding by using FCM clustering. Signal Process. 2019, 159, 193–203. [Google Scholar] [CrossRef]
- He, W.; Xiong, G.; Wang, Y. Reversible Data Hiding Based on Multiple Pairwise PEE and Two-Layer Embedding. Secur. Commun. Netw. 2022, 2022, 2051058. [Google Scholar] [CrossRef]
- Chang, C.C.; Lu, T.C. A difference expansion oriented data hiding scheme for restoring the original host images. J. Syst. Softw. 2006, 79, 1754–1766. [Google Scholar] [CrossRef]
- Natarajan, V. Hybrid local prediction error-based difference expansion reversible watermarking for medical images. Comput. Electr. Eng. 2016, 53, 333–345. [Google Scholar]
- Lakshmanan, S.; Rani, M. Reversible data hiding in medical images using edge detection and difference expansion technique. J. Comput. Theor. Nanosci. 2018, 15, 2400–2404. [Google Scholar] [CrossRef]
- Manikandan, V.M.; Prasad, N.L.; Vedhanayagam, M. Half difference expansion based reversible data hiding scheme for medical image forensics. Curr. Med. Imaging 2020, 16, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Parah, S.A.; Ahad, F.; Sheikh, J.A.; Bhat, G.M. Hiding clinical information in medical images: A new high capacity and reversible data hiding technique. J. Biomed. Inform. 2017, 66, 214–230. [Google Scholar] [CrossRef] [PubMed]
- Malayil, M.V.; Vedhanayagam, M. A novel image scaling based reversible watermarking scheme for secure medical image transmission. ISA Trans. 2021, 108, 269–281. [Google Scholar] [CrossRef]
- Konyar, M.Z.; Öztürk, S. Reed Solomon coding-based medical image data hiding method against salt and pepper noise. Symmetry 2020, 12, 899. [Google Scholar] [CrossRef]
- Rashmi, P.; Supriya, M.; Hua, Q. Enhanced Lorenz-Chaotic Encryption Method for Partial Medical Image Encryption and Data Hiding in Big Data Healthcare. Secur. Commun. Netw. 2022, 2022, 9363377. [Google Scholar] [CrossRef]
- Solak, S. High embedding capacity data hiding technique based on EMSD and LSB substitution algorithms. IEEE Access 2020, 8, 166513–166524. [Google Scholar] [CrossRef]
- USC. Image Database. Available online: http://sipi.usc.edu/database/ (accessed on 1 May 2020).
- CT Medical Images. Available online: https://www.kaggle.com/datasets/kmader/siim-medical-images (accessed on 31 August 2022).
- Setiadi, D.R.I.M. PSNR vs. SSIM: Imperceptibility quality assessment for image steganography. Multimed. Tools Appl. 2021, 80, 8423–8444. [Google Scholar] [CrossRef]
- Panchikkil, S.; Manikandan, V.; Zhang, Y.D. An efficient spatial transformation-based entropy retained reversible data hiding scheme in encrypted images. Optik 2022, 261, 169211. [Google Scholar] [CrossRef]
- Zhang, X. Reversible data hiding in encrypted image. IEEE Signal Process. Lett. 2011, 18, 255–258. [Google Scholar] [CrossRef]
- Hong, W.; Chen, T.S.; Wu, H.Y. An improved reversible data hiding in encrypted images using side match. IEEE Signal Process. Lett. 2012, 19, 199–202. [Google Scholar] [CrossRef]
- Zhang, X. Separable reversible data hiding in encrypted image. IEEE Trans. Inf. Forensics Secur. 2011, 7, 826–832. [Google Scholar] [CrossRef]
- Chen, Y.C.; Shiu, C.W.; Horng, G. Encrypted signal-based reversible data hiding with public key cryptosystem. J. Vis. Commun. Image Represent. 2014, 25, 1164–1170. [Google Scholar] [CrossRef]
- Shiu, C.W.; Chen, Y.C.; Hong, W. Encrypted image-based reversible data hiding with public key cryptography from difference expansion. Signal Process. Image Commun. 2015, 39, 226–233. [Google Scholar] [CrossRef]
- Zhang, X.; Long, J.; Wang, Z.; Cheng, H. Lossless and reversible data hiding in encrypted images with public-key cryptography. IEEE Trans. Circuits Syst. Video Technol. 2015, 26, 1622–1631. [Google Scholar] [CrossRef]
- Li, M.; Li, Y. Histogram shifting in encrypted images with public key cryptosystem for reversible data hiding. Signal Process. 2017, 130, 190–196. [Google Scholar] [CrossRef]
- Agrawal, S.; Kumar, M. Mean value based reversible data hiding in encrypted images. Opt.-Int. J. Light Electron Opt. 2017, 130, 922–934. [Google Scholar] [CrossRef]
- Xiong, L.; Xu, Z.; Shi, Y.Q. An integer wavelet transform based scheme for reversible data hiding in encrypted images. Multidimens. Syst. Signal Process. 2018, 29, 1191–1202. [Google Scholar] [CrossRef]
- Tang, Z.; Xu, S.; Ye, D.; Wang, J.; Zhang, X.; Yu, C. Real-time reversible data hiding with shifting block histogram of pixel differences in encrypted image. J. Real-Time Image Process. 2019, 16, 709–724. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).