Characterization of Postprandial Effects on CSF Metabolomics: A Pilot Study with Parallel Comparison to Plasma
Abstract
:1. Introduction
2. Results
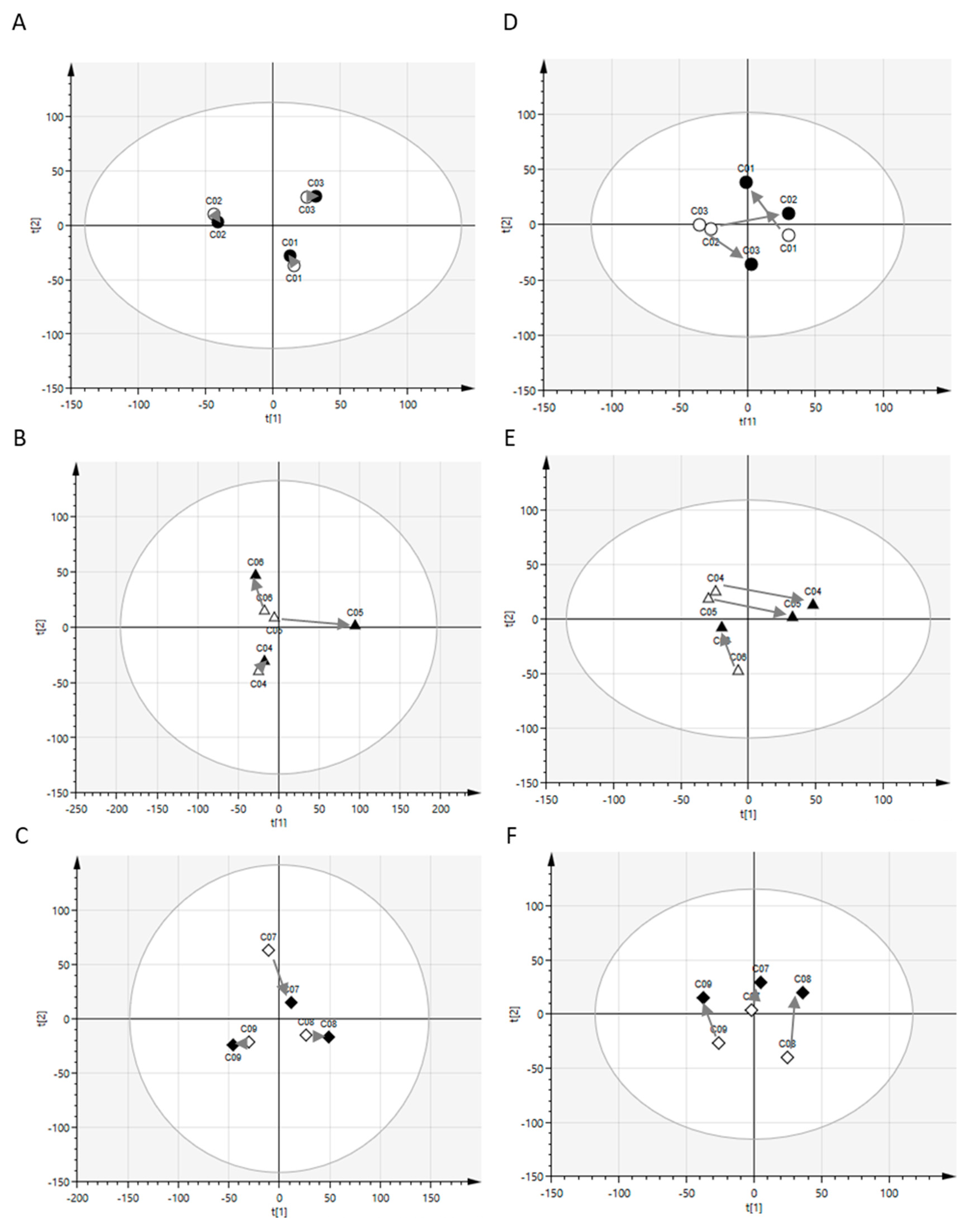
2.1. Postprandial Effects on Global Profiles of CSF Metabolites
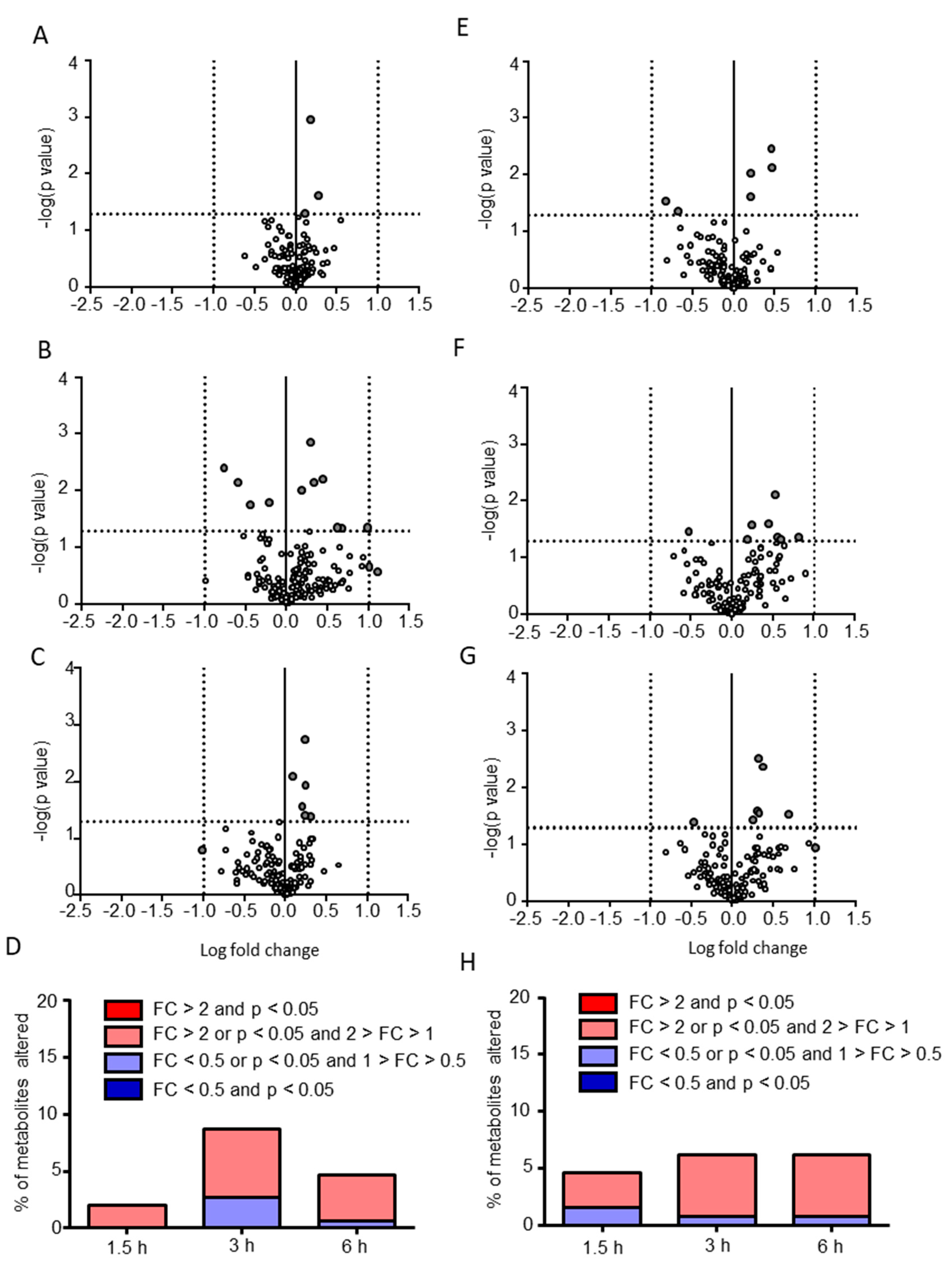
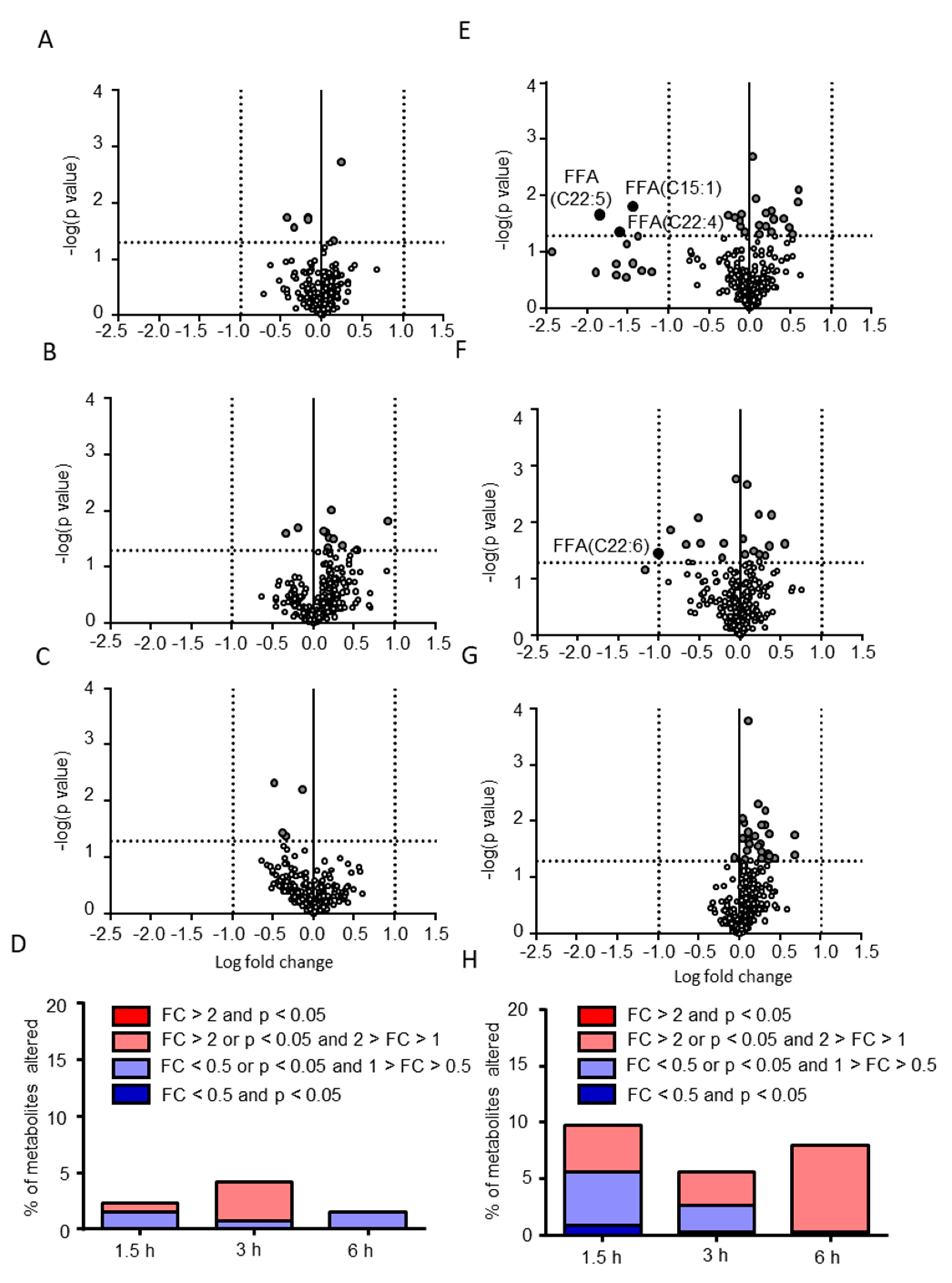
2.2. Postprandial Effects on Individual CSF Metabolites
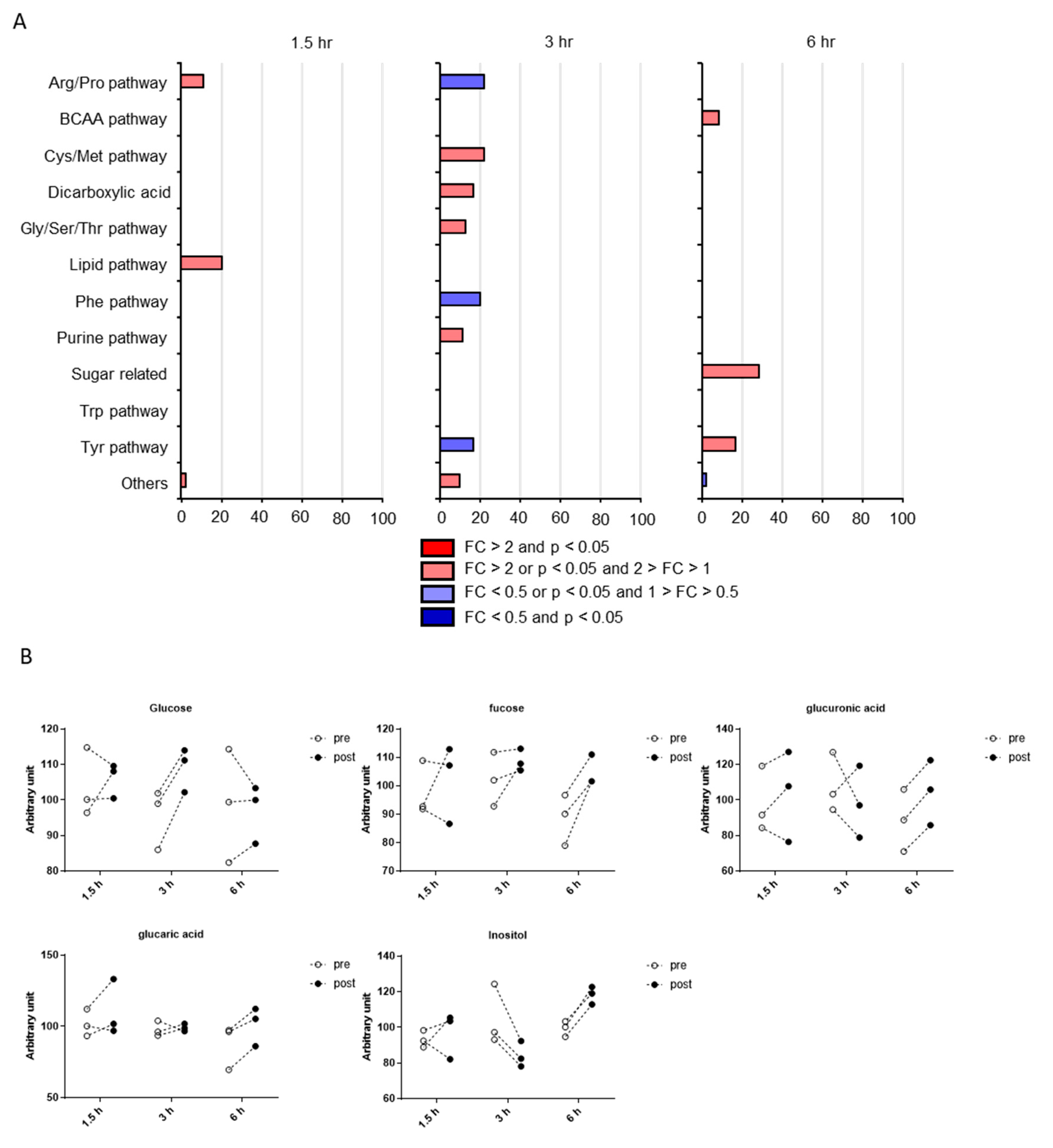
2.3. Classification of Putative CSF Metabolites Altered by Postprandial Effects
3. Discussion
4. Materials and Methods
4.1. Subjects and Sample Collection
4.2. Metabolomic Analysis
4.3. Data Cleaning and Statistical Analyses
4.4. Principal Component Analysis (PCA)
4.5. Pathway Occupancy Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Watkins, S.M.; German, J.B. Metabolomics and biochemical profiling in drug discovery and development. Curr. Opin. Mol. Ther. 2002, 4, 224–228. [Google Scholar] [PubMed]
- Hollywood, K.; Brison, D.R.; Goodacre, R. Metabolomics: Current technologies and future trends. Proteomics 2006, 6, 4716–4723. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Current progress in computational metabolomics. Brief. Bioinform. 2007, 8, 279–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabatine, M.S.; Liu, E.; Morrow, D.A.; Heller, E.; McCarroll, R.; Wiegand, R.; Berriz, G.F.; Roth, F.P.; Gerszten, R.E. Metabolomic identification of novel biomarkers of myocardial ischemia. Circulation 2005, 112, 3868–3875. [Google Scholar] [CrossRef] [Green Version]
- Xue, R.; Lin, Z.; Deng, C.; Dong, L.; Liu, T.; Wang, J.; Shen, X. A serum metabolomic investigation on hepatocellular carcinoma patients by chemical derivatization followed by gas chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 2008, 22, 3061–3068. [Google Scholar] [CrossRef]
- He, Y.; Yu, Z.; Giegling, I.; Xie, L.; Hartmann, A.M.; Prehn, C.; Adamski, J.; Kahn, R.; Li, Y.; Illig, T.; et al. Schizophrenia shows a unique metabolomics signature in plasma. Transl. Psychiatry 2012, 2, e149. [Google Scholar] [CrossRef] [Green Version]
- Saito, K.; Maekawa, K.; Pappan, K.L.; Urata, M.; Ishikawa, M.; Kumagai, Y.; Saito, Y. Differences in metabolite profiles between blood matrices, ages, and sexes among Caucasian individuals and their interindividual variations. Metabolomics 2014, 10, 402–413. [Google Scholar] [CrossRef]
- Saito, K.; Maekawa, K.; Kinchen, J.M.; Tanaka, R.; Kumagai, Y.; Saito, Y. Gender- and age-associated differences in serum metabolite profiles among Japanese populations. Biol. Pharm. Bull. 2016, 39, 1179–1186. [Google Scholar] [CrossRef] [Green Version]
- Mittelstrass, K.; Ried, J.S.; Yu, Z.; Krumsiek, J.; Gieger, C.; Prehn, C.; Roemisch-Margl, W.; Polonikov, A.; Peters, A.; Theis, F.J.; et al. Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS Genet. 2011, 7, e1002215. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, M.; Maekawa, K.; Saito, K.; Senoo, Y.; Urata, M.; Murayama, M.; Tajima, Y.; Kumagai, Y.; Saito, Y. Plasma and serum lipidomics of healthy white adults shows characteristic profiles by subjects’ gender and age. PLoS ONE 2014, 9, e91806. [Google Scholar] [CrossRef] [Green Version]
- Takehana, S.; Yoshida, H.; Ozawa, S.; Yamazaki, J.; Shimbo, K.; Nakayama, A.; Mizukoshi, T.; Miyano, H. The effects of pre-analysis sample handling on human plasma amino acid concentrations. Clin. Chim. Acta 2016, 455, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Ueno, S.; Nakayama, A.; Nitta, S.I.; Arai, K.; Hasunuma, T.; Saito, Y. Overall similarities and a possible factor affecting plasma metabolome profiles between venous and capillary blood samples from 20 healthy human males. J. Pharm. Sci. 2019, 108, 3737–3744. [Google Scholar] [CrossRef] [PubMed]
- Pellis, L.; van Erk, M.J.; van Ommen, B.; Bakker, G.C.; Hendriks, H.F.; Cnubben, N.H.; Kleemann, R.; van Someren, E.P.; Bobeldijk, I.; Rubingh, C.M.; et al. Plasma metabolomics and proteomics profiling after a postprandial challenge reveal subtle diet effects on human metabolic status. Metabolomics 2012, 8, 347–359. [Google Scholar] [CrossRef] [Green Version]
- Karimpour, M.; Surowiec, I.; Wu, J.; Gouveia-Figueira, S.; Pinto, R.; Trygg, J.; Zivkovic, A.M.; Nording, M.L. Postprandial metabolomics: A pilot mass spectrometry and NMR study of the human plasma metabolome in response to a challenge meal. Anal. Chim. Acta 2016, 908, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Moriya, T.; Satomi, Y.; Kobayashi, H. Metabolomics of postprandial plasma alterations: A comprehensive Japanese study. J. Biochem. 2018, 163, 113–121. [Google Scholar] [CrossRef]
- Mouton-Barbosa, E.; Roux-Dalvai, F.; Bouyssié, D.; Berger, F.; Schmidt, E.; Righetti, P.G.; Guerrier, L.; Boschetti, E.; Burlet-Schiltz, O.; Monsarrat, B.; et al. In-depth exploration of cerebrospinal fluid by combining peptide ligand library treatment and label-free protein quantification. Mol. Cell. Proteom. 2010, 9, 1006–1021. [Google Scholar] [CrossRef] [Green Version]
- Hühmer, A.F.; Biringer, R.G.; Amato, H.; Fonteh, A.N.; Harrington, M.G. Protein analysis in human cerebrospinal fluid: Physiological aspects, current progress and future challenges. Dis. Markers 2006, 22, 3–26. [Google Scholar] [CrossRef]
- Wallin, A.K.; Blennow, K.; Zetterberg, H.; Londos, E.; Minthon, L.; Hansson, O. CSF biomarkers predict a more malignant outcome in Alzheimer disease. Neurology 2010, 74, 1531–1537. [Google Scholar] [CrossRef]
- Wuolikainen, A.; Andersen, P.M.; Moritz, T.; Marklund, S.L.; Antti, H. ALS patients with mutations in the SOD1 gene have an unique metabolomic profile in the cerebrospinal fluid compared with ALS patients without mutations. Mol. Genet. Metab. 2012, 105, 472–478. [Google Scholar] [CrossRef]
- Blasco, H.; Corcia, P.; Pradat, P.F.; Bocca, C.; Gordon, P.H.; Veyrat-Durebex, C.; Mavel, S.; Nadal-Desbarats, L.; Moreau, C.; Devos, D.; et al. Metabolomics in cerebrospinal fluid of patients with amyotrophic lateral sclerosis: An untargeted approach via high-resolution mass spectrometry. J. Proteome Res. 2013, 12, 3746–3754. [Google Scholar] [CrossRef]
- Trushina, E.; Dutta, T.; Persson, X.M.; Mielke, M.M.; Petersen, R.C. Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimer’s disease using metabolomics. PLoS ONE 2013, 20, e63644. [Google Scholar]
- Willkommen, D.; Lucio, M.; Moritz, F.; Forcisi, S.; Kanawati, B.; Smirnov, K.S.; Schroeter, M.; Sigaroudi, A.; Schmitt-Kopplin, P.; Michalke, B. Metabolomic investigations in cerebrospinal fluid of Parkinson’s disease. PLoS ONE 2018, 13, e0208752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rustan, A.C.; Hustvedt, B.E.; Drevon, C.A. Postprandial decrease in plasma unesterified fatty acids during n-3 fatty acid feeding is not caused by accumulation of fatty acids in adipose tissue. Biochim. Biophys. Acta 1998, 1390, 245–257. [Google Scholar] [CrossRef]
- Podlecka-Piętowska, A.; Kacka, A.; Zakrzewska-Pniewska, B.; Nojszewska, M.; Zieminska, E.; Chalimoniuk, M.; Toczylowska, B. Altered cerebrospinal fluid concentrations of hydrophobic and hydrophilic compounds in early stages of multiple sclerosis-metabolic profile analyses. J. Mol. Neurosci. 2019, 69, 94–105. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.X.; Chen, K.; Huang, F.Q.; Alolga, R.N.; Ma, J.; Wu, Z.X.; Fan, Y.; Ma, G.; Guan, M. Cerebrospinal fluid-based metabolomics to characterize different types of brain tumors. J. Neurol. 2019. [Google Scholar] [CrossRef]
- Bergau, N.; Maul, S.; Rujescu, D.; Simm, A.; Navarrete Santos, A. Reduction of glycolysis intermediate concentrations in the cerebrospinal fluid of Alzheimer’s disease patients. Front. Neurosci. 2019, 13, 871. [Google Scholar] [CrossRef]
- Geracioti, T.D.; Loosen, P.T.; Ebert, M.H.; Schmidt, D.; Ekhator, N.N. Fasting and postprandial cerebrospinal fluid glucose concentrations in healthy women and in an obese binge eater. Int. J. Eat. Disord. 1995, 18, 365–369. [Google Scholar] [CrossRef]
- Kawai, K.; Murayama, Y.; Okuda, Y.; Yamashita, K. Postprandial glucose, insulin and glucagon responses to meals with different nutrient compositions in non-insulin-dependent diabetes mellitus. Endocrinol. Jpn. 1987, 34, 745–753. [Google Scholar] [CrossRef] [Green Version]
- Robins, D.C. Phosphatidylethanolamine and lysophosphatidylethanolamine. J. Pharm. Pharmacol. 1963, 15, 701–722. [Google Scholar] [CrossRef]
- Patel, D.; Witt, S.N. Ethanolamine and phosphatidylethanolamine: Partners in health and disease. Oxid. Med. Cell. Longev. 2017, 2017, 4829180. [Google Scholar] [CrossRef] [Green Version]
- Meikle, P.J.; Barlow, C.K.; Mellett, N.A.; Mundra, P.A.; Bonham, M.P.; Larsen, A.; Cameron-Smith, D.; Sinclair, A.; Nestel, P.J.; Wong, G. Postprandial plasma phospholipids in men are influenced by the source of dietary fat. J. Nutr. 2015, 145, 2012–2018. [Google Scholar] [CrossRef] [Green Version]
- Averill, M.; Rubinow, K.B.; Cain, K.; Wimberger, J.; Babenko, I.; Becker, J.O.; Foster-Schubert, K.E.; Cummings, D.E.; Hoofnagle, A.N.; Vaisar, T. Postprandial remodeling of high-density lipoprotein following high saturated fat and high carbohydrate meals. J. Clin. Lipidol. 2019, 14, 66–76.e11. [Google Scholar] [CrossRef]
- Klein, J.D.; Sherrill, J.B.; Morello, G.M.; San Miguel, P.J.; Ding, Z.; Liangpunsakul, S.; Liang, T.; Muir, W.M.; Lumeng, L.; Lossie, A.C. A snapshot of the hepatic transcriptome: Ad libitum alcohol intake suppresses expression of cholesterol synthesis genes in alcohol-preferring (P) rats. PLoS ONE 2014, 9, e110501. [Google Scholar] [CrossRef] [Green Version]
- De Wit, N.J.; Afman, L.A.; Mensink, M.; Müller, M. Phenotyping the effect of diet on non-alcoholic fatty liver disease. J. Hepatol. 2012, 57, 1370–1373. [Google Scholar] [CrossRef]
- Zarrinpar, A.; Chaix, A.; Panda, S. Daily Eating Patterns and Their Impact on Health and Disease. Trends Endocrinol. Metab. 2016, 27, 69–83. [Google Scholar] [CrossRef] [Green Version]
- Breier, A.; Wolkowitz, O.M.; Doran, A.R.; Bellar, S.; Pickar, D. Neurobiological effects of lumbar puncture stress in psychiatric patients and healthy volunteers. Psychiatry Res. 1988, 25, 187–194. [Google Scholar] [CrossRef]
- Hattori, K.; Ota, M.; Sasayama, D.; Yoshida, S.; Matsumura, R.; Miyakawa, T.; Yokota, Y.; Yamaguchi, S.; Noda, T.; Teraishi, T.; et al. Increased cerebrospinal fluid fibrinogen in major depressive disorder. Sci. Rep. 2015, 5, 11412. [Google Scholar] [CrossRef]
- Yuan, M.; Breitkopf, S.B.; Yang, X.; Asara, J.M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012, 7, 872–881. [Google Scholar] [CrossRef] [Green Version]
- Satomi, Y.; Hirayama, M.; Kobayashi, H. One-step lipid extraction for plasma lipidomics analysis by liquid chromatography mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017, 1063, 93–100. [Google Scholar] [CrossRef]


| Group | Name | p-Value | Fold Change | ||||
|---|---|---|---|---|---|---|---|
| 1.5 h | 3 h | 6 h | 1.5 h | 3 h | 6 h | ||
| Amino sugar | N-Acetylneuraminic acid | 0.53 | <0.01 | 0.48 | 0.9 | 1.36 | 1.18 |
| Arg/Pro pathway | 5-Aminovaleric acid | 0.02 | 0.14 | 0.64 | 1.22 | 1.71 | 0.83 |
| Arg/Pro pathway | Arginine | 0.97 | <0.01 | 0.75 | 1.01 | 0.59 | 1.08 |
| Arg/Pro pathway | Creatine | 0.66 | 0.02 | 0.95 | 1.04 | 0.73 | 1.01 |
| BCAA pathway | 3-Hydroxy-2-methylbutanoic acid | 0.61 | 0.07 | 0.04 | 1.1 | 0.84 | 1.24 |
| C5-Branched dibasic acid pathway | Citramalic acid | 0.28 | 0.4 | 0.16 | 0.65 | 1.39 | 0.49 |
| Cholesterol pathway | Mevalonic acid | 0.93 | <0.01 | 0.69 | 0.99 | 1.27 | 0.94 |
| Cys/Met pathway | Cystine | 0.54 | 0.22 | 0.61 | 1.05 | 2.01 | 1.08 |
| Cys/Met pathway | N-Formylmethionine | 0.72 | <0.01 | 0.98 | 1.06 | 1.23 | 1 |
| Dicarboxylic acid | 2-Hydroxyglutaric acid | 0.14 | 0.05 | 0.38 | 1.1 | 1.54 | 0.7 |
| Gly/Ser/Thr pathway | Dimethylglycine | 0.47 | 0.27 | 0.75 | 1.03 | 2.16 | 0.92 |
| Glycolysis | Glucose | 0.69 | <0.01 | 0.76 | 1.02 | 1.14 | 0.98 |
| Lipid pathway | Acetoacetic acid | 0.05 | 0.57 | 0.72 | 1.09 | 1.21 | 1.02 |
| N-acetyl-amino acid | N-Acetyltyrosine ethyl ester | <0.01 | 0.77 | 0.26 | 1.14 | 0.91 | 0.77 |
| Nicotinate and nicotinamide pathway | Nicotinamide | 0.22 | 0.05 | 0.55 | 1.29 | 1.99 | 0.8 |
| Phe pathway | 4-Hydroxybenzoic acid | 0.53 | <0.01 | 0.76 | 0.84 | 0.67 | 1.07 |
| Purine pathway | Phosphoric acid | 0.94 | 0.05 | 0.13 | 0.98 | 1.6 | 0.82 |
| Sugar related | Fucose | 0.63 | 0.26 | 0.04 | 1.05 | 1.06 | 1.18 |
| Sugar related | Glucaric acid | 0.34 | 0.8 | 0.03 | 1.09 | 1.01 | 1.15 |
| Sugar related | Glucuronic acid | 0.53 | 0.54 | <0.01 | 1.05 | 0.91 | 1.18 |
| Sugar related | Inositol | 0.68 | 0.07 | 0.01 | 1.04 | 0.8 | 1.19 |
| Tyr pathway | p-Hydroxyphenylpyruvic acid | 0.3 | 0.02 | 0.6 | 0.87 | 0.87 | 1.14 |
| Tyr pathway | Tyramine | 0.87 | 0.34 | <0.01 | 0.99 | 1.06 | 1.07 |
| Group | Name | p-Value | Fold Change | ||||
|---|---|---|---|---|---|---|---|
| 1.5 h | 3 h | 6 h | 1.5 h | 3 h | 6 h | ||
| Ceramide | Cer(18:0) | 0.13 | 0.02 | 0.66 | 1.09 | 1.11 | 1.01 |
| Cholesteryl esters(acyl) | CE(a-22:6) | 0.97 | 0.02 | 0.27 | 1.01 | 1.09 | 0.80 |
| Free fatty acid | FFA(C14:0) | 0.02 | 0.56 | 0.75 | 0.89 | 1.14 | 1.05 |
| Free fatty acid | FFA(C14:1) | 0.88 | 0.82 | <0.01 | 1.06 | 1.03 | 0.72 |
| Free fatty acid | FFA(C18:1) | 0.13 | 0.05 | 0.41 | 0.65 | 1.45 | 1.20 |
| Free fatty acid | FFA(C20:0) | 0.03 | 0.12 | 0.43 | 0.79 | 1.48 | 1.29 |
| Free fatty acid | FFA(C22:0) | 0.02 | 0.11 | 0.59 | 0.75 | 1.23 | 1.25 |
| Lysophosphatidylethanolamine(acyl) | LPE(a-18:0) | 0.13 | 0.02 | 0.85 | 1.33 | 1.89 | 1.05 |
| Lysophosphatidylethanolamine(acyl) | LPE(a-18:1) | 0.25 | 0.03 | 0.46 | 1.26 | 0.79 | 0.97 |
| Phosphatidylcholine(acyl) | PC(aa-40:5) | 0.91 | 0.03 | 0.84 | 1.00 | 1.19 | 0.98 |
| Phosphatidylcholine(acyl) | PC(aa-42:7) | 0.27 | 0.05 | 0.25 | 1.03 | 1.13 | 1.08 |
| Phosphatidylcholine(alk) | PC(ae-38:5) | 0.66 | <0.01 | 0.47 | 1.01 | 1.16 | 0.90 |
| Phosphatidylcholine(alk) | PC(ae-38:6) | 0.42 | 0.03 | 0.58 | 0.98 | 1.14 | 0.94 |
| Phosphatidylethanolamine(acyl) | PE(aa-34:1) | 0.02 | 0.50 | 0.54 | 0.89 | 1.08 | 0.91 |
| Phosphatidylethanolamine(acyl) | PE(aa-38:4) | 0.05 | 0.08 | 0.49 | 1.11 | 1.35 | 1.09 |
| Phosphatidylethanolamine(alk) | PE(ae-34:2) | 0.39 | 0.33 | <0.01 | 1.06 | 1.15 | 0.91 |
| Phosphatidylethanolamine(alk) | PE(ae-36:4) | 0.63 | 0.43 | 0.04 | 1.05 | 1.11 | 0.77 |
| Phosphatidylethanolamine(alk) | PE(ae-36:5) | 0.97 | 0.04 | 0.09 | 1.00 | 1.28 | 0.85 |
| Sphingomyelin | SM(23:2) | <0.01 | 0.86 | 0.34 | 1.18 | 0.93 | 0.76 |
| Sulfatide-Hex Ganglioside | Sulfatide(Hex/16:0) | 0.56 | 0.73 | 0.04 | 0.94 | 0.95 | 0.79 |
| Sulfatide-Hex Ganglioside | Sulfatide(Hex/20:0) | 0.85 | 0.02 | 0.20 | 0.99 | 0.88 | 0.84 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saito, K.; Hattori, K.; Andou, T.; Satomi, Y.; Gotou, M.; Kobayashi, H.; Hidese, S.; Kunugi, H. Characterization of Postprandial Effects on CSF Metabolomics: A Pilot Study with Parallel Comparison to Plasma. Metabolites 2020, 10, 185. https://doi.org/10.3390/metabo10050185
Saito K, Hattori K, Andou T, Satomi Y, Gotou M, Kobayashi H, Hidese S, Kunugi H. Characterization of Postprandial Effects on CSF Metabolomics: A Pilot Study with Parallel Comparison to Plasma. Metabolites. 2020; 10(5):185. https://doi.org/10.3390/metabo10050185
Chicago/Turabian StyleSaito, Kosuke, Kotaro Hattori, Tomohiro Andou, Yoshinori Satomi, Masamitsu Gotou, Hiroyuki Kobayashi, Shinsuke Hidese, and Hiroshi Kunugi. 2020. "Characterization of Postprandial Effects on CSF Metabolomics: A Pilot Study with Parallel Comparison to Plasma" Metabolites 10, no. 5: 185. https://doi.org/10.3390/metabo10050185
APA StyleSaito, K., Hattori, K., Andou, T., Satomi, Y., Gotou, M., Kobayashi, H., Hidese, S., & Kunugi, H. (2020). Characterization of Postprandial Effects on CSF Metabolomics: A Pilot Study with Parallel Comparison to Plasma. Metabolites, 10(5), 185. https://doi.org/10.3390/metabo10050185





