HPLC-HRMS Global Metabolomics Approach for the Diagnosis of “Olive Quick Decline Syndrome” Markers in Olive Trees Leaves
Abstract
:1. Introduction
2. Results
3. Discussion
3.1. HPLC-HRMS Analytical Method Development
3.2. Sample Two-Dimensional Clustering
3.3. Features Annotation
3.4. Feature Abundance Class-Related Variability
4. Materials and Methods
4.1. Chemicals and Materials
4.2. Sampling Protocol of Plant Samples
4.3. Quality Controls Setting
4.4. Sample Harvesting and Preparation
4.5. Extraction Protocol of Plant Samples and QCs Sample Generation
4.6. HPLC-ESI-LTQ Orbitrap Parameters
4.7. Data Processing and Statistical Analysis
- 1.
- Kruskall–Wallis test was applied to identify the features showing a significant difference between HP and OP samples (p < 0.01 after Bonferroni correction).
- 2.
- Significant features were then ranked on the bases of their median intensity in the two sample classes.
- 3.
- The potential list of infection biomarkers was selected:
- a-
- By considering the features present in the top quartile of the ranked list for OP and in the lower quartile of the HP list.
- b-
- By considering the features present in the top quartile of the ranked list for HP and in the lower quartile of the OP list.
4.8. Metabolites Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- D’Onghia, A.M.; Digiaro, M.; Brunel, S.; Valentini, F. Xylella Fastidiosa & the Olive Quick Decline Syndrome (OQDS) A Serious Worldwide Challenge for the Safeguard of Olive Trees; CIHEAM: Valenzano, Italia, 2017. [Google Scholar]
- Martelli, G.P.; Boscia, D.; Porcelli, F.; Saponari, M. The olive quick decline syndrome in south-east Italy: A threatening phytosanitary emergency. Eur. J. Plant Pathol. 2016, 144, 235–243. [Google Scholar] [CrossRef]
- Nigro, F.; Boscia, D.; Antelmi, I.; Ippolito, A. Fungal species associated with a severe decline of olive in southern Italy. J. Plant Pathol. 2013, 95, 668. [Google Scholar] [CrossRef]
- Saponari, M.; Loconsole, G.; Cornara, D.; Yokomi, R.K.; De Stradis, A.; Boscia, D.; Bosco, D.; Martelli, G.P.; Krugner, R.; Porcelli, F. Infectivity and Transmission of Xylella fastidiosa by Philaenus spumarius (Hemiptera: Aphrophoridae) in Apulia, Italy. J. Econ. Entomol. 2014, 107, 1316–1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cariddi, C.; Saponari, M.; Boscia, D.; De Stradis, A.; Loconsole, G.; Nigro, F.; Porcelli, F.; Potere, O.; Martelli, G.P. Isolation of Xylella fastidiosa strain infecting olive and oleander in Apulia, Italy. J. Plant Pathol. 2014, 96, 425–429. [Google Scholar] [CrossRef]
- Colnaghi Simionato, A.V.; da Silva, D.S.; Lambais, M.R.; Carrilho, E. Characterization of a putativeXylella fastidiosa diffusible signal factor by HRGC-EI-MS. J. Mass Spectrom. 2007, 42, 1375–1381. [Google Scholar] [CrossRef] [PubMed]
- Marques, L.L.R.; Ceri, H.; Manfio, G.P.; Reid, D.M.; Olson, M.E. Characterization of Biofilm Formation by Xylella fastidiosa In Vitro. Plant Dis. 2002, 86, 633–638. [Google Scholar] [CrossRef] [Green Version]
- Abbott, A. Italy’s olive crisis intensifies as deadly tree disease spreads. Nature 2018, 563, 306–307. [Google Scholar] [CrossRef] [Green Version]
- Hopkins, D.L.; Purcell, A.H. Xylella fastidiosa: Cause of Pierce’s Disease of Grapevine and Other Emergent Diseases. Plant Dis. 2002, 86, 1056–1066. [Google Scholar] [CrossRef] [Green Version]
- Hernandez-Martinez, R.; Costa, H.S.; Dumenyo, C.K.; Cooksey, D.A. Differentiation of Strains of Xylella fastidiosa Infecting Grape, Almonds, and Oleander Using a Multiprimer PCR Assay. Plant Dis. 2006, 90, 1382–1388. [Google Scholar] [CrossRef] [Green Version]
- Janse, J.D.; Obradovic, A. Xylella fastidiosa: Its biology, diagnosis, control and risks. J. Plant Pathol. 2010, 92. [Google Scholar] [CrossRef]
- Maddox, C.E.; Laur, L.M.; Tian, L. Antibacterial Activity of Phenolic Compounds Against the Phytopathogen Xylella fastidiosa. Curr. Microbiol. 2010, 60, 53–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarrete, F.; De La Fuente, L. Response of Xylella fastidiosa to Zinc: Decreased Culturability, Increased Exopolysaccharide Production, and Formation of Resilient Biofilms under Flow Conditions. Appl. Environ. Microbiol. 2014, 80, 1097–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Lee, Y.; Igo, M.M.; Roper, M.C. Tolerance to oxidative stress is required for maximal xylem colonization by the xylem-limited bacterial phytopathogen, Xylella fastidiosa. Mol. Plant Pathol. 2017, 18, 990–1000. [Google Scholar] [CrossRef] [PubMed]
- Baldassarre, F.; De Stradis, A.; Altamura, G.; Vergaro, V.; Citti, C.; Cannazza, G.; Capodilupo, A.L.; Dini, L.; Ciccarella, G. Application of calcium carbonate nanocarriers for controlled release of phytodrugs against Xylella fastidiosa pathogen. Pure Appl. Chem. 2020, 92, 429–444. [Google Scholar] [CrossRef] [Green Version]
- Bextine, B.; Tuan, S.-J.; Shaikh, H.; Blua, M.; Miller, T.A. Evaluation of Methods for Extracting Xylella fastidiosa DNA from the Glassy-Winged Sharpshooter. J. Econ. Entomol. 2004, 97, 757–763. [Google Scholar] [CrossRef]
- Loconsole, G.; Potere, O.; Boscia, D.; Altamura, G.; Djelouah, K.; Elbeaino, T.; Frasheri, D.; Lorusso, D.; Palmisano, F.; Pollastro, P.; et al. Detection of Xylella fastidiosa in olive trees by molecular and serological methods. J. Plant Pathol. 2014, 96, 7–14. [Google Scholar]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-based metabolomics. Mol. BioSyst. 2012, 8, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, X.; Wang, C.; Zhang, H.; Cai, Z. Non-targeted and targeted metabolomics approaches to diagnosing lung cancer and predicting patient prognosis. Oncotarget 2016, 7, 63437–63448. [Google Scholar] [CrossRef] [Green Version]
- Di Donna, L.; Mazzotti, F.; Naccarato, A.; Salerno, R.; Tagarelli, A.; Taverna, D.; Sindona, G. Secondary metabolites of Olea europaea leaves as markers for the discrimination of cultivars and cultivation zones by multivariate analysis. Food Chem. 2010, 121, 492–496. [Google Scholar] [CrossRef]
- Kritikou, E.; Kalogiouri, N.P.; Kolyvira, L.; Thomaidis, N.S. Target and Suspect HRMS Metabolomics for the Determination of Functional Ingredients in 13 Varieties of Olive Leaves and Drupes from Greece. Molecules 2020, 25, 4889. [Google Scholar] [CrossRef] [PubMed]
- Guodong, R.; Xiaoxia, L.; Weiwei, Z.; Wenjun, W.; Jianguo, Z. Metabolomics reveals variation and correlation among different tissues of olive (Olea europaea L.). Biol. Open 2017, 6, 1317–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stilo, F.; Liberto, E.; Reichenbach, S.E.; Tao, Q.; Bicchi, C.; Cordero, C. Untargeted and Targeted Fingerprinting of Extra Virgin Olive Oil Volatiles by Comprehensive Two-Dimensional Gas Chromatography with Mass Spectrometry: Challenges in Long-Term Studies. J. Agric. Food Chem. 2019, 67, 5289–5302. [Google Scholar] [CrossRef] [PubMed]
- Sales, C.; Portolés, T.; Johnsen, L.G.; Danielsen, M.; Beltran, J. Olive oil quality classification and measurement of its organoleptic attributes by untargeted GC–MS and multivariate statistical-based approach. Food Chem. 2019, 271, 488–496. [Google Scholar] [CrossRef]
- Liigand, P.; Kaupmees, K.; Haav, K.; Liigand, J.; Leito, I.; Girod, M.; Antoine, R.; Kruve, A. Think Negative: Finding the Best Electrospray Ionization/MS Mode for Your Analyte. Anal. Chem. 2017, 89, 5665–5668. [Google Scholar] [CrossRef] [Green Version]
- Gidda, S.K.; Miersch, O.; Levitin, A.; Schmidt, J.; Wasternack, C.; Varin, L. Biochemical and Molecular Characterization of a Hydroxyjasmonate Sulfotransferase from Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 17895–17900. [Google Scholar] [CrossRef] [Green Version]
- Link, K.P. The Discovery of Dicumarol and Its Sequels. Circulation 1959, 19, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Matsuura, T.; Kawai, M.; Nakashima, R.; Butsugan, Y. Structures of physalin A and physalin B, 13,14-seco-16,24-cyclo-steroids from Physalis alkekengi var. Francheti. J. Chem. Soc. C Org. 1970, 664. [Google Scholar] [CrossRef]
- Januário, A.H.; Filho, E.R.; Pietro, R.C.L.R.; Kashima, S.; Sato, D.N.; França, S.C. Antimycobacterial physalins from Physalis angulata L. (Solanaceae). Phytother. Res. 2002, 16, 445–448. [Google Scholar] [CrossRef]
- Jensen, S.; Omarsdottir, S.; Bwalya, A.G.; Nielsen, M.A.; Tasdemir, D.; Olafsdottir, E.S. Marchantin A, a macrocyclic bisbibenzyl ether, isolated from the liverwort Marchantia polymorpha, inhibits protozoal growth in vitro. Phytomedicine 2012, 19, 1191–1195. [Google Scholar] [CrossRef]
- Huang, Y.-L.; Oppong, M.B.; Guo, Y.; Wang, L.-Z.; Fang, S.-M.; Deng, Y.-R.; Gao, X.-M. The Oleaceae family: A source of secoiridoids with multiple biological activities. Fitoterapia 2019, 136, 104155. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Qiu, S.; Lindenmaier, M.; He, X.; Featherstone, T.; Cordell, G.A. Patuletin-3-O-Rutinoside from the Aerial Parts of Echinacea angustifolia. Pharm. Biol. 2002, 40, 92–95. [Google Scholar] [CrossRef]
- Machida, K.; Sasaki, H.; Iijima, T.; Kikuchi, M. Studies on the Constituents of Lonicera Species. XVII. New Iridoid Glycosides of the Stems and Leaves of Lonicera japonica THUNB. Chem. Pharm. Bull. 2002, 50, 1041–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azizah, M.; Pripdeevech, P.; Thongkongkaew, T.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. UHPLC-ESI-QTOF-MS/MS-Based Molecular Networking Guided Isolation and Dereplication of Antibacterial and Antifungal Constituents of Ventilago denticulata. Antibiotics 2020, 9, 606. [Google Scholar] [CrossRef]
- Molnár, J.; Gunics, G.; Mucsi, I.; Koltai, M.; Petri, I.; Shoyama, Y.; Matsumoto, M.; Nishioka, I. Antimicrobial and immunomodulating effects of some phenolic glycosides. Acta Microbiol. Hung. 1989, 36, 425–432. [Google Scholar]
- Tian-Shung, W.; Chang-Sheng, K.; Furukawa, H. Acridone alkaloids and a coumarin from Citrus grandis. Phytochemistry 1983, 22, 1493–1497. [Google Scholar] [CrossRef]
- Bhattacharya, L.; Roy, S.K.; Chakraborty, D.P. Structure of the carbazole alkaloid isomurrayazoline from Murraya koenigii. Phytochemistry 1982, 21, 2432–2433. [Google Scholar] [CrossRef]
- Kamleh, M.A.; Ebbels, T.M.D.; Spagou, K.; Masson, P.; Want, E.J. Optimizing the Use of Quality Control Samples for Signal Drift Correction in Large-Scale Urine Metabolic Profiling Studies. Anal. Chem. 2012, 84, 2670–2677. [Google Scholar] [CrossRef]
- Saigusa, D.; Okamura, Y.; Motoike, I.N.; Katoh, Y.; Kurosawa, Y.; Saijyo, R.; Koshiba, S.; Yasuda, J.; Motohashi, H.; Sugawara, J.; et al. Establishment of Protocols for Global Metabolomics by LC-MS for Biomarker Discovery. PLoS ONE 2016, 11, e0160555. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Gu, J.; Gui, Y.; Chen, L.; Yuan, G.; Lu, H.-Z.; Xu, X. Use of Natural Products as Chemical Library for Drug Discovery and Network Pharmacology. PLoS ONE 2013, 8, e62839. [Google Scholar] [CrossRef] [Green Version]
- Ruttkies, C.; Schymanski, E.L.; Wolf, S.; Hollender, J.; Neumann, S. MetFrag relaunched: Incorporating strategies beyond in silico fragmentation. J. Cheminform. 2016, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A Web-Based Platform to Process Untargeted Metabolomic Data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef] [Green Version]
- Tautenhahn, R.; Cho, K.; Uritboonthai, W.; Zhu, Z.; Patti, G.J.; Siuzdak, G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 2012, 30, 826–828. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O.; Robertson, D.; Griffin, J.; vab der Werf, M.; Nikolau, B.; Morrison, N.; Sumner, L.W.; Goodacre, R.; Hardy, N.W.; Taylor, C.; et al. The metabolomics standards initiative (MSI). Metabolomics 2007, 3, 175–178. [Google Scholar] [CrossRef]
- Wang, S.; Alseekh, S.; Fernie, A.R.; Luo, J. The Structure and Function of Major Plant Metabolite Modifications. Mol. Plant 2019, 12, 899–919. [Google Scholar] [CrossRef] [PubMed]
- Jorge, T.F.; Rodrigues, J.A.; Caldana, C.; Schmidt, R.; van Dongen, J.T.; Thomas-Oates, J.; António, C. Mass spectrometry-based plant metabolomics: Metabolite responses to abiotic stress. Mass Spectrom. Rev. 2016, 35, 620–649. [Google Scholar] [CrossRef]

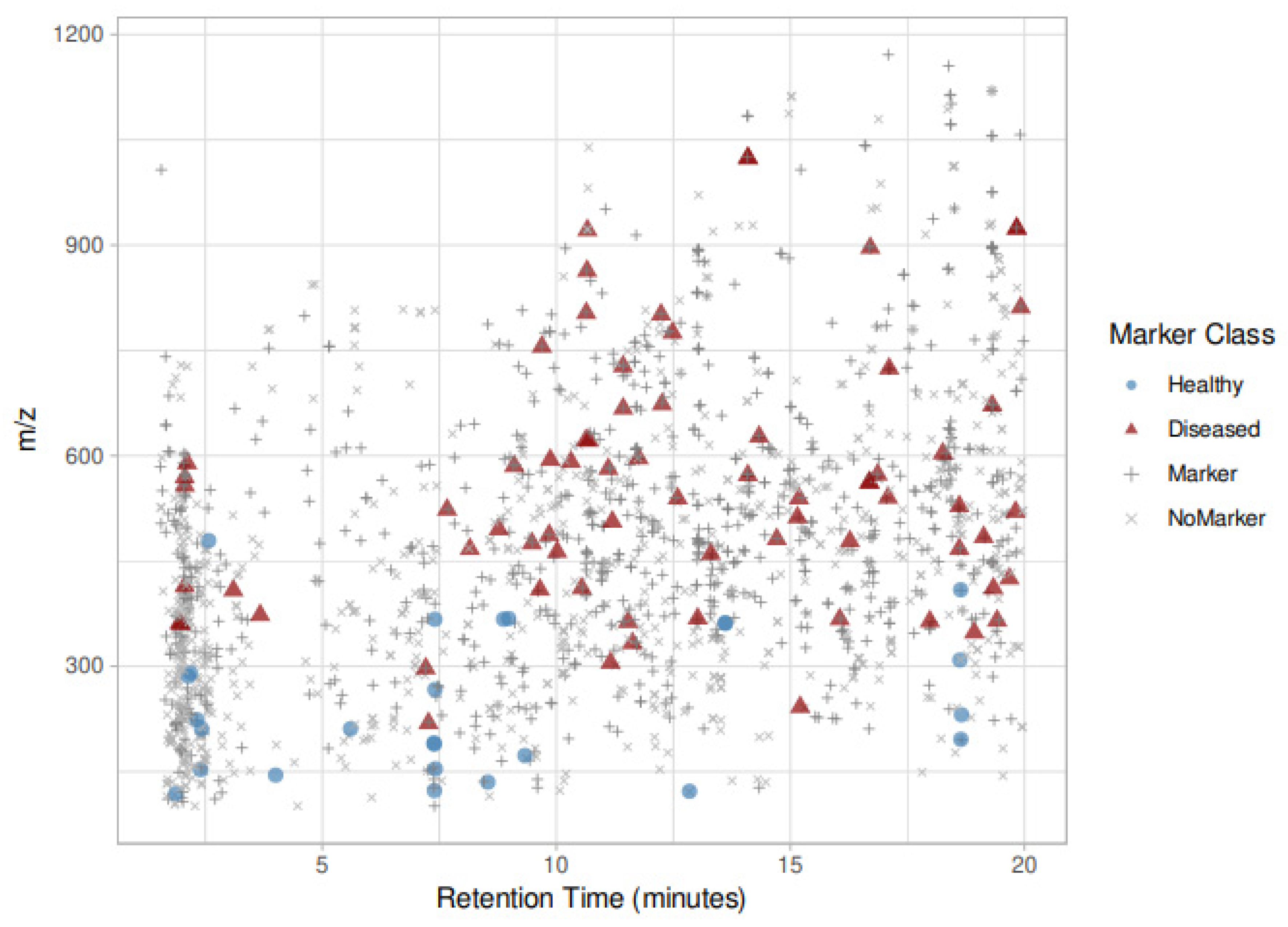
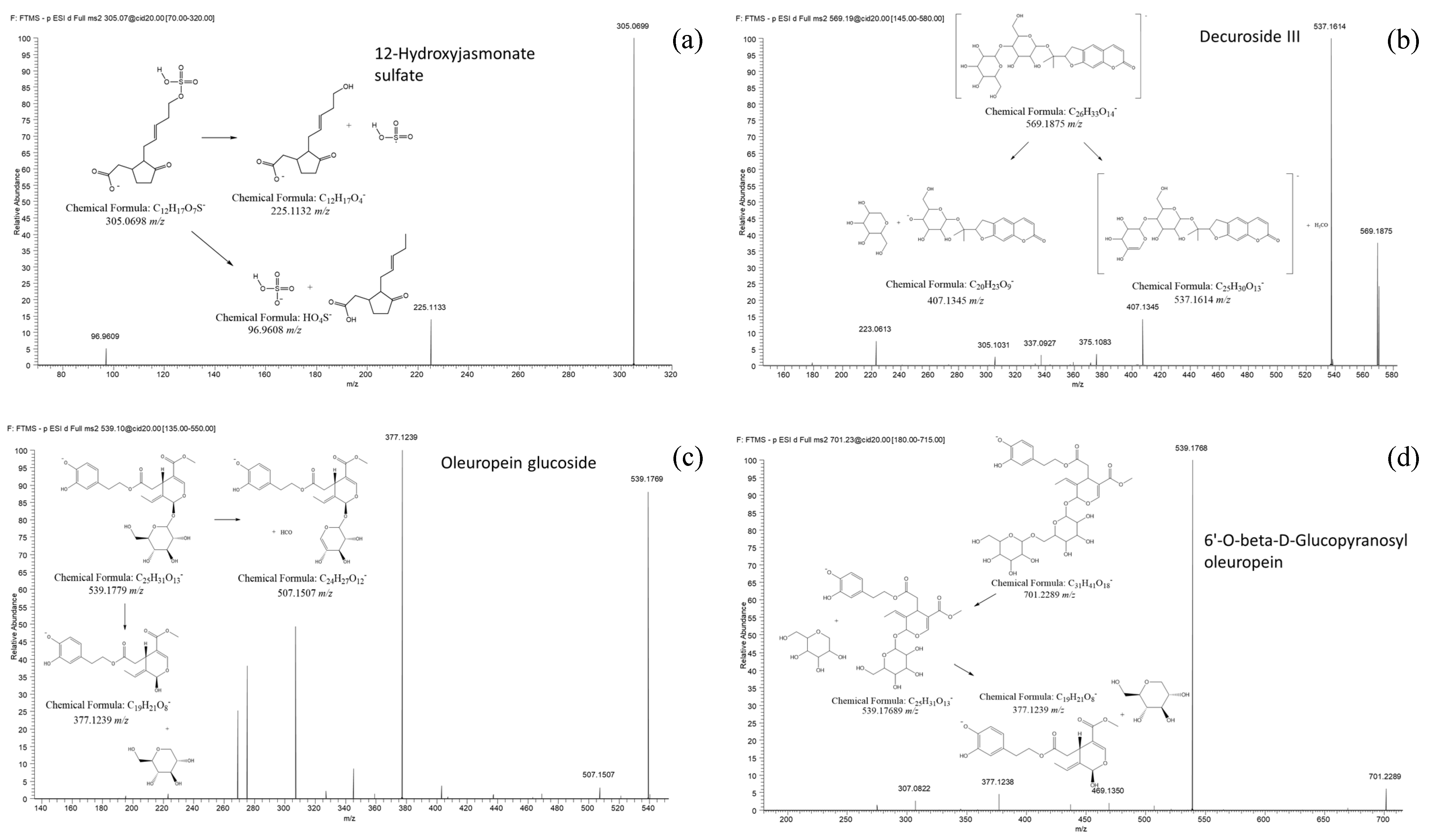

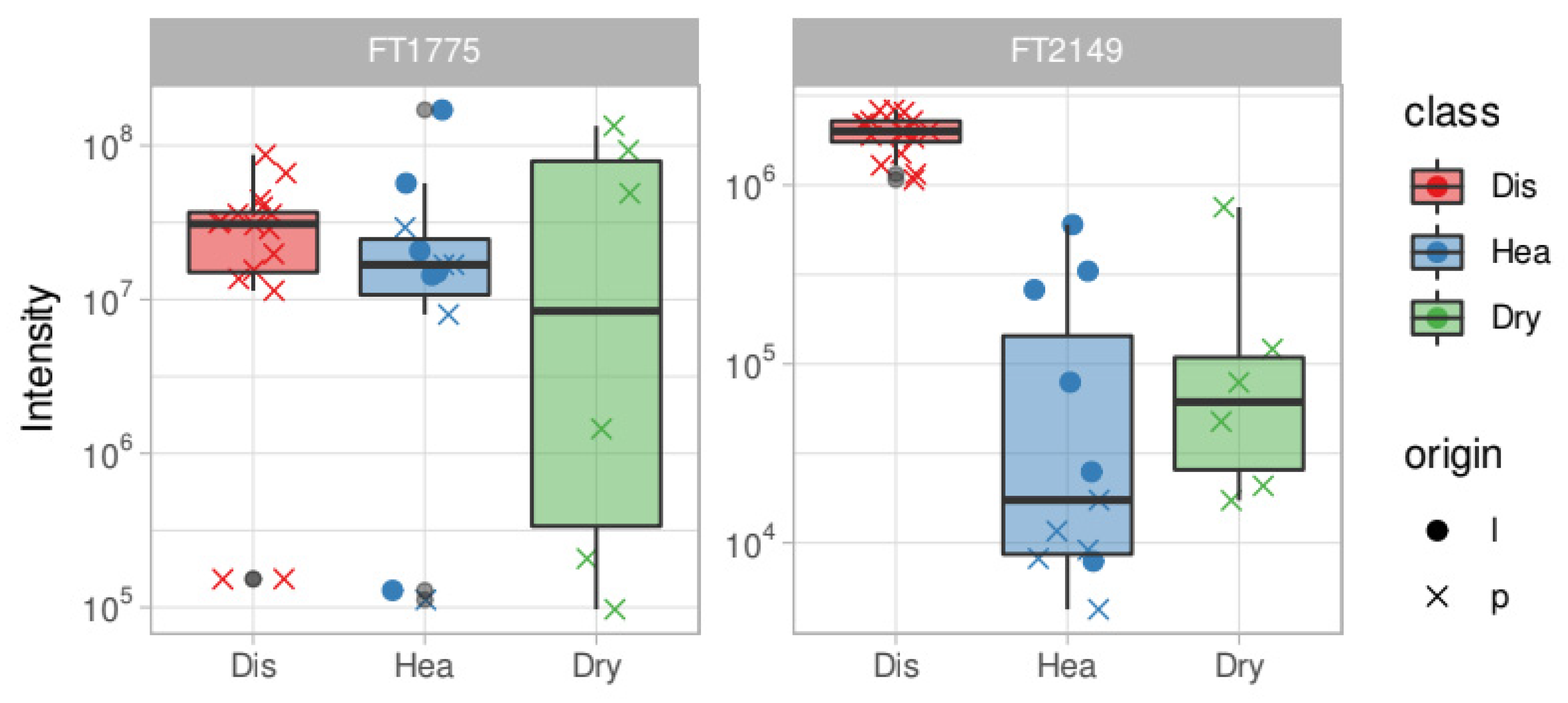
| Feature ID | Healthy–Infected | m/z [M − H]− | R.t. (min) | MSI Level | Fragments (Intensity) | Proposed Formula | Proposed Name (*) | DBE | Δ (PPM) |
|---|---|---|---|---|---|---|---|---|---|
| FT0449 | Healthy | 286.0732 | 2.1 | 3 | C15H13NO5 | Grandisine III (b) CAS: 53421-39-9 | 10 | 5.79 | |
| FT0534 | Infected | 305.0698 | 11.2 | 2 | 305.0698 (100) 225.1133(14) 96.9608(5) | C12H18O7S | 12-Hydroxy Jasmonate sulfate (a) CID: 44815853 | 4.5 | 0.92 |
| FT0659 | Healthy | 330.1871 | 25.1 | 3 | C23H25NO | Isomurrayazoline (b) CAS: 85547-20-2 | 12 | 3.94 | |
| FT0852 | Infected | 368.1366 | 13.0 | 3 | C17H23NO8 | Niazicinin (b) CID: 101920262 | 7 | 5.64 | |
| FT1073 | Infected | 407.1346 | 3.1 | 2 | 337.0925(100) 305.1029 (90) 375.1081 (48) 407.1344 (41)151.0405 (20) | C20H24O9 | Nodakenin (a) CID: 73191 | 9.5 | 0.95 |
| FT1089 | Infected | 409.2005 | 9.6 | 3 | C25H30O5 | Vismione D (b) CID: 5281573 | 11 | 2.37 | |
| FT1133 | Infected | 419.1847 | 13.4 | 3 | C26H28O5 | Ovaliflavanone D (b) CID: 42607825 | 13 | 2.52 | |
| FT1228 | Infected | 439.1536 | 21.1 | 2 | 393.1762(100) 197.0821 (35) | C28H24O5 | Marchantin A (a) CID: 88418-46-6 | 17.5 | 0.39 |
| FT1775 | O.e Marker | 539.1769 | 20.9 | 2 | 377.1239 (100) 507.1507 (5) | C25H32O13 | Oleuropein glucoside (c) CID: 5281544 | 10 | 0.79 |
| FT1879 | Infected | 557.2002 | 2.1 | 2 | 513.2335 (100) 345.1185 (21) 227.1288 (8) | C29H34O11 | Physalin (a) Metlin ID: 89909 | 13.5 | 0.46 |
| FT1947 | Infected | 569.1844 | 20.1 | 2 | 537.1612 (100) 403.1242 (95) 569.1873 (46) 407.1344 (13) | C26H34O14 | Decuroside III (a) CAS: 96638-81-2 | 10.5 | 1.69 |
| FT2071 | Infected | 593.1404 | 18.2 | 2 | 593.1507 (100) 285.0403 (98) | C27H30O15 | Isoorientin rhamnoside (a) CID: 16126794 | 13 | 3.79 |
| FT2149 | O.e Marker | 609.1458 | 14.5 | 3 | C27H30O16 | Rutin (c) CID: 5280805 | 13 | 0.38 | |
| FT2403 | Infected | 701.2271 | 11.7 | 2 | 539.1768 (100) 377.1238 (5) | C31H42O18 | 6’-O-beta-D-Glucopyranosyl-oleuropein (a) CID: 102078602 | 11 | 3.13 |
| FT2416 | Infected | 707.1863 | 24.7 | 3 | C32H36O18 | Patuletin 3-(4’’-acetylrhamnosid)7-(2’’’-acetylrhamnoside) (b) CID: 44259840 | 15 | 5.73 | |
| FT2574 | Infected | 757.2582 | 21.9 | 3 | C34H46O19 | Aldosecologanin (b) CID: 10908841 | 12 | 3.58 | |
| FT2646 | Infected | 783.2375 | 24.8 | 3 | C35H44O20 | Rhamnazin 3-rhamninoside (b) CID: 44259609 | 14 | 3.51 | |
| FT2654 | Infected | 785.2533 | 24.8 | 3 | C35H46O20 | Purpureaside (b) CID: 11953944 | 13 | 3.76 |
| Healthy | Infected | |
|---|---|---|
| Normal (HP) | Desiccated (DHP) | Desiccated (OP) |
| 11 | 6 | 16 |
| 5 Puglia, 6 Liguria | 6 Puglia | 16 Puglia |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asteggiano, A.; Franceschi, P.; Zorzi, M.; Aigotti, R.; Dal Bello, F.; Baldassarre, F.; Lops, F.; Carlucci, A.; Medana, C.; Ciccarella, G. HPLC-HRMS Global Metabolomics Approach for the Diagnosis of “Olive Quick Decline Syndrome” Markers in Olive Trees Leaves. Metabolites 2021, 11, 40. https://doi.org/10.3390/metabo11010040
Asteggiano A, Franceschi P, Zorzi M, Aigotti R, Dal Bello F, Baldassarre F, Lops F, Carlucci A, Medana C, Ciccarella G. HPLC-HRMS Global Metabolomics Approach for the Diagnosis of “Olive Quick Decline Syndrome” Markers in Olive Trees Leaves. Metabolites. 2021; 11(1):40. https://doi.org/10.3390/metabo11010040
Chicago/Turabian StyleAsteggiano, Alberto, Pietro Franceschi, Michael Zorzi, Riccardo Aigotti, Federica Dal Bello, Francesca Baldassarre, Francesco Lops, Antonia Carlucci, Claudio Medana, and Giuseppe Ciccarella. 2021. "HPLC-HRMS Global Metabolomics Approach for the Diagnosis of “Olive Quick Decline Syndrome” Markers in Olive Trees Leaves" Metabolites 11, no. 1: 40. https://doi.org/10.3390/metabo11010040
APA StyleAsteggiano, A., Franceschi, P., Zorzi, M., Aigotti, R., Dal Bello, F., Baldassarre, F., Lops, F., Carlucci, A., Medana, C., & Ciccarella, G. (2021). HPLC-HRMS Global Metabolomics Approach for the Diagnosis of “Olive Quick Decline Syndrome” Markers in Olive Trees Leaves. Metabolites, 11(1), 40. https://doi.org/10.3390/metabo11010040










