Are Fecal Metabolome and Microbiota Profiles Correlated with Autism Severity? A Cross-Sectional Study on ASD Preschoolers
Abstract
:1. Introduction
2. Results
2.1. Fecal Metabolome
2.2. Fecal Microbiota and Intestinal Inflammation
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Clinical Assessments
4.3. Metabolomics Analysis by 1H-NMR
4.4. Microbiota Analysis
4.5. Calprotectin Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix A.1. Metabolomics Analysis by 1H-NMR
Appendix A.2. Microbiota Analysis
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-5®); American Psychiatric Association: Washington, DC, USA, 2013. [Google Scholar]
- Goodwin, M.S.; Cowen, M.A.; Goodwin, T.C. Malabsorption and cerebral dysfunction: A multivariate and comparative study of autistic children. J. Autism Child. Schizophr. 1971, 1, 48–62. [Google Scholar] [CrossRef]
- Bresnahan, M.; Hornig, M.; Schultz, A.F.; Gunnes, N.; Hirtz, D.; Lie, K.K.; Magnus, P.; Reichborn-Kjennerud, T.; Roth, C.; Schjølberg, S.; et al. Association of maternal report of infant and toddler gastrointestinal symptoms with autism: Evidence from a prospective birth cohort. JAMA Psychiatry 2015, 72, 466–474. [Google Scholar] [CrossRef] [Green Version]
- Fulceri, F.; Morelli, M.; Santocchi, E.; Cena, H.; Del Bianco, T.; Narzisi, A.; Calderoni, S.; Muratori, F. Gastrointestinal symptoms and behavioral problems in preschoolers with Autism Spectrum Disorder. Dig. Liver Dis. 2016, 48, 248–254. [Google Scholar] [CrossRef]
- Klukowski, M.; Wasilewska, J.; Lebensztejn, D. Sleep and gastrointestinal disturbances in autism spectrum disorder in children. Dev. Period Med. 2015, 19, 157–161. [Google Scholar] [PubMed]
- Prosperi, M.; Santocchi, E.; Balboni, G.; Narzisi, A.; Bozza, M.; Fulceri, F.; Apicella, F.; Igliozzi, R.; Cosenza, A.; Tancredi, R.; et al. Behavioral phenotype of ASD preschoolers with gastrointestinal symptoms or food selectivity. J. Autism Dev. Disord. 2017, 47, 3574–3588. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.; Rubin, R.A. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011, 11, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coretti, L.; Paparo, L.; Riccio, M.P.; Amato, F.; Cuomo, M.; Natale, A.; Borrelli, L.; Corrado, G.; Comegna, M.; Buommino, E.; et al. Gut microbiota features in young children with autism spectrum disorders. Front. Microbiol. 2018, 9, 3146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, H.T.; Taur, Y.; Walkup, J.T. Gut microbiota and autism: Key concepts and findings. J. Autism Dev. Disord. 2016, 47, 480–489. [Google Scholar] [CrossRef]
- Laghi, L.; Picone, G.; Capozzi, F. Nuclear magnetic resonance for foodomics beyond food analysis. TrAC Trends Anal. Chem. 2014, 59, 93–102. [Google Scholar] [CrossRef]
- Ursell, L.K.; Haiser, H.J.; Van Treuren, W.; Garg, N.; Reddivari, L.; Vanamala, J.; Dorrestein, P.C.; Turnbaugh, P.; Knight, R. The intestinal metabolome: An intersection between microbiota and host. Gastroenterology 2014, 146, 1470–1476. [Google Scholar] [CrossRef] [Green Version]
- Tursi, A.; Mastromarino, P.; Capobianco, D.; Elisei, W.; Miccheli, A.; Capuani, G.; Tomassini, A.; Campagna, G.; Picchio, M.; Giorgetti, G.; et al. Assessment of fecal microbiota and fecal metabolome in symptomatic uncomplicated diverticular disease of the colon. J. Clin. Gastroenterol. 2016, 50, S9–S12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, D.-W.; Ilhan, Z.E.; Isern, N.G.; Hoyt, D.W.; Howsmon, D.P.; Shaffer, M.; Lozupone, C.A.; Hahn, J.; Adams, J.B.; Krajmalnik-Brown, R. Differences in fecal microbial metabolites and microbiota of children with autism spectrum disorders. Anaerobe 2018, 49, 121–131. [Google Scholar] [CrossRef]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Dig. Dis. Sci. 2012, 57, 2096–2102. [Google Scholar] [CrossRef] [PubMed]
- Bjarnason, I. The use of fecal calprotectin in inflammatory bowel disease. Gastroenterol. Hepatol. 2017, 13, 53–56. [Google Scholar]
- Mussap, M.; Siracusano, M.; Noto, A.; Fattuoni, C.; Riccioni, A.; Rajula, H.; Fanos, V.; Curatolo, P.; Barberini, L.; Mazzone, L. The urine metabolome of young autistic children correlates with their clinical profile severity. Metabolites 2020, 10, 476. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Francavilla, R.; Piccolo, M.; De Giacomo, A.; Gobbetti, M. Autism spectrum disorders and intestinal microbiota. Gut Microbes 2015, 6, 207–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burket, J.A.; Deutsch, S.I. Metabotropic functions of the NMDA receptor and an evolving rationale for exploring NR2A-selective positive allosteric modulators for the treatment of autism spectrum disorder. Prog. Neuro Psychopharmacol. Biol. Psychiatry 2019, 90, 142–160. [Google Scholar] [CrossRef]
- Crespi, B.J. Comparative psychopharmacology of autism and psychotic-affective disorders suggests new targets for treatment. Evol. Med. Public Health 2019, 2019, 149–168. [Google Scholar] [CrossRef]
- Schneider, M.; Al-Shareffi, E.; Haltiwanger, R.S. Biological functions of fucose in mammals. Glycobiology 2017, 27, 601–618. [Google Scholar] [CrossRef] [Green Version]
- Ke, J.; Li, Y.; Han, C.; He, R.; Lin, R.; Qian, W.; Hou, X. Fucose ameliorate intestinal inflammation through modulating the crosstalk between bile acids and gut microbiota in a chronic colitis murine model. Inflamm. Bowel Dis. 2020, 26, 863–873. [Google Scholar] [CrossRef]
- Pickard, J.M.; Maurice, C.F.; Kinnebrew, M.A.; Abt, M.C.; Schenten, D.; Golovkina, T.V.; Bogatyrev, S.R.; Ismagilov, R.F.; Pamer, E.G.; Turnbaugh, P.J. Rapid fucosylation of intestinal epithelium sustains host–commensal symbiosis in sickness. Nature 2014, 514, 638–641. [Google Scholar] [CrossRef] [Green Version]
- Rossignol, D.A.; Frye, R.E. Mitochondrial dysfunction in autism spectrum disorders: A systematic review and meta-analysis. Mol. Psychiatry 2011, 17, 290–314. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, S.; Kim, J.-M.; Yamada, H. Microbial enzymes for creatinine assay: A review. Clin. Chim. Acta 1989, 185, 241–252. [Google Scholar] [CrossRef]
- Boczonadi, V.; Jennings, M.J.; Horvath, R. The role of tRNA synthetases in neurological and neuromuscular disorders. FEBS Lett. 2018, 592, 703–717. [Google Scholar] [CrossRef] [Green Version]
- Auerbach, B.D.; Osterweil, E.; Bear, M.F. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature 2011, 480, 63–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, S.A.; Wijetunge, L.S.; Jackson, A.; Katsanevaki, D.; Osterweil, E.; Komiyama, N.H.; Grant, S.G.; Bear, M.F.; Nägerl, U.V.; Kind, P.C.; et al. Convergence of Hippocampal Pathophysiology in Syngap+/− and Fmr1−/y Mice. J. Neurosci. 2015, 35, 15073–15081. [Google Scholar] [CrossRef] [Green Version]
- Bhakar, A.L.; Dölen, G.; Bear, M.F. The pathophysiology of fragile X (and what it teaches us about synapses). Annu. Rev. Neurosci. 2012, 35, 417–443. [Google Scholar] [CrossRef] [Green Version]
- Finegold, S.M.; Dowd, S.; Gontcharova, V.; Liu, C.; Henley, K.E.; Wolcott, R.D.; Youn, E.; Summanen, P.H.; Granpeesheh, D.; Dixon, D.; et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 2010, 16, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Averina, O.V.; Kovtun, A.S.; Polyakova, S.I.; Savilova, A.M.; Rebrikov, D.V.; Danilenko, V.N. The bacterial neurometabolic signature of the gut microbiota of young children with autism spectrum disorders. J. Med. Microbiol. 2020, 69, 558–571. [Google Scholar] [CrossRef] [PubMed]
- Hughes, H.; Rose, D.; Ashwood, P. The gut microbiota and dysbiosis in autism spectrum disorders. Curr. Neurol. Neurosci. Rep. 2018, 18, 81. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; LaBaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McElhanon, B.O.; McCracken, C.; Karpen, S.; Sharp, W.G. Gastrointestinal symptoms in autism spectrum disorder: A meta-analysis. Pediatrics 2014, 133, 872–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, D.R.; Yang, H.; Serena, G.; Sturgeon, C.; Ma, B.; Careaga, M.; Hughes, H.K.; Angkustsiri, K.; Rose, M.; Hertz-Picciotto, I.; et al. Differential immune responses and microbiota profiles in children with autism spectrum disorders and co-morbid gastrointestinal symptoms. Brain Behav. Immun. 2018, 70, 354–368. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism 2013, 4, 42. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl. Environ. Microbiol. 2011, 77, 6718–6721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Li, E.; Sun, Z.; Fu, D.; Duan, G.; Jiang, M.; Yu, Y.; Mei, L.; Yang, P.; Tang, Y.; et al. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Sci. Rep. 2019, 9, 287. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Zhu, Q.; Li, F.; Wang, J.; Shen, L.; Sheng, X. Fecal calprotectin in healthy children aged 1–4 years. PLoS ONE 2016, 11, e0150725. [Google Scholar] [CrossRef] [Green Version]
- Needham, B.D.; Adame, M.D.; Serena, G.; Rose, D.R.; Preston, G.M.; Conrad, M.C.; Campbell, A.S.; Donabedian, D.H.; Fasano, A.; Ashwood, P.; et al. Plasma and fecal metabolite profiles in autism spectrum disorder. Biol. Psychiatry 2021, 89, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Newberry, S.J.; Hempel, S.; Maher, A.R.; Wang, Z.; Miles, J.N.V.; Shanman, R.; Johnsen, B.; Shekelle, P.G. Probiotics for the prevention and treatment of antibiotic-associated diarrhea: A systematic review and meta-analysis. JAMA 2012, 307, 1959–1969. [Google Scholar] [CrossRef] [Green Version]
- Williams, B.L.; Hornig, M.; Parekh, T.; Lipkin, W.I. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. mBio 2012, 3, e00261-11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santocchi, E.; Guiducci, L.; Fulceri, F.; Billeci, L.; Buzzigoli, E.; Apicella, F.; Calderoni, S.; Grossi, E.; Morales, M.A.; Muratori, F. Gut to brain interaction in Autism Spectrum Disorders: A randomized controlled trial on the role of probiotics on clinical, biochemical and neurophysiological parameters. BMC Psychiatry 2016, 16, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santocchi, E.; Guiducci, L.; Prosperi, M.; Calderoni, S.; Gaggini, M.; Apicella, F.; Tancredi, R.; Billeci, L.; Mastromarino, P.; Grossi, E.; et al. Effects of probiotic supplementation on gastrointestinal, sensory and core symptoms in autism spectrum disorders: A randomized controlled trial. Front. Psychiatry 2020, 11, 944. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Rutter, M.; DiLavore, P.; Risi, S.; Gotham, K.; Bishop, S. Autism Diagnostic Observation Schedule, (ADOS-2) Modules 1–4; Western Psychological Services: Los Angeles, CA, USA, 2012. [Google Scholar]
- Gotham, K.; Pickles, A.; Lord, C. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. J. Autism Dev. Disord. 2008, 39, 693–705. [Google Scholar] [CrossRef] [Green Version]
- Esler, A.N.; Bal, V.; Guthrie, W.; Wetherby, A.; Weismer, S.E.; Lord, C. The autism diagnostic observation schedule, toddler module: Standardized severity scores. J. Autism Dev. Disord. 2015, 45, 2704–2720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, C.K.; Melmed, R.D.; Barstow, L.E.; Enriquez, F.J.; Ranger-Moore, J.; Ostrem, J.A. Oral human immunoglobulin for children with autism and gastrointestinal dysfunction: A prospective, open-label study. J. Autism Dev. Disord. 2006, 36, 1053–1064. [Google Scholar] [CrossRef]
- Nadkarni, M.A.; Martin, F.E.; Jacques, N.A.; Hunter, N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002, 148, 257–266. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2011; Available online: https://www.R-project.org (accessed on 14 February 2019).
- Box, G.E.; Cox, D.R. An analysis of transformations. J. R. Stat. Soc. Ser. B Methodol. 1964, 26, 211–243. [Google Scholar] [CrossRef]
- Hubert, M.; Rousseeuw, P.J.; Branden, K.V. ROBPCA: A new approach to robust principal component analysis. Technometrics 2005, 47, 64–79. [Google Scholar] [CrossRef]
- Laghi, L.; Zhu, C.; Campagna, G.; Rossi, G.; Bazzano, M.; Laus, F. Probiotic supplementation in trained trotter horses: Effect on blood clinical pathology data and urine metabolomic assessed in field. J. Appl. Physiol. 2018, 125, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef]
- Kneen, M.A.; Annegarn, H. Algorithm for fitting XRF, SEM and PIXE X-ray spectra backgrounds. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 1996, 109, 209–213. [Google Scholar] [CrossRef]
- Liland, K.H.; Almøy, T.; Mevik, B.-H. Optimal choice of baseline correction for multivariate calibration of spectra. Appl. Spectrosc. 2010, 64, 1007–1016. [Google Scholar] [CrossRef]
- Foschi, C.; Laghi, L.; D’Antuono, A.; Gaspari, V.; Zhu, C.; Dellarosa, N.; Salvo, M.; Marangoni, A. Urine metabolome in women with Chlamydia trachomatis infection. PLoS ONE 2018, 13, e0194827. [Google Scholar] [CrossRef]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Takada, T.; Tanaka, R. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 2004, 70, 7220–7228. [Google Scholar] [CrossRef] [Green Version]
- Štšepetova, J.; Sepp, E.; Kolk, H.; Loivukene, K.; Songisepp, E.; Mikelsaar, M. Diversity and metabolic impact of intestinal Lactobacillus species in healthy adults and the elderly. Br. J. Nutr. 2011, 105, 1235–1244. [Google Scholar] [CrossRef] [Green Version]
- Bartosch, S.; Fite, A.; Macfarlane, G.T.; McMurdo, M.E.T. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 2004, 70, 3575–3581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, N.; Vogensen, F.; Berg, F.V.D.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Derrien, M.; Isolauri, E.; De Vos, W.M.; Salminen, S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 2007, 73, 7767–7770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
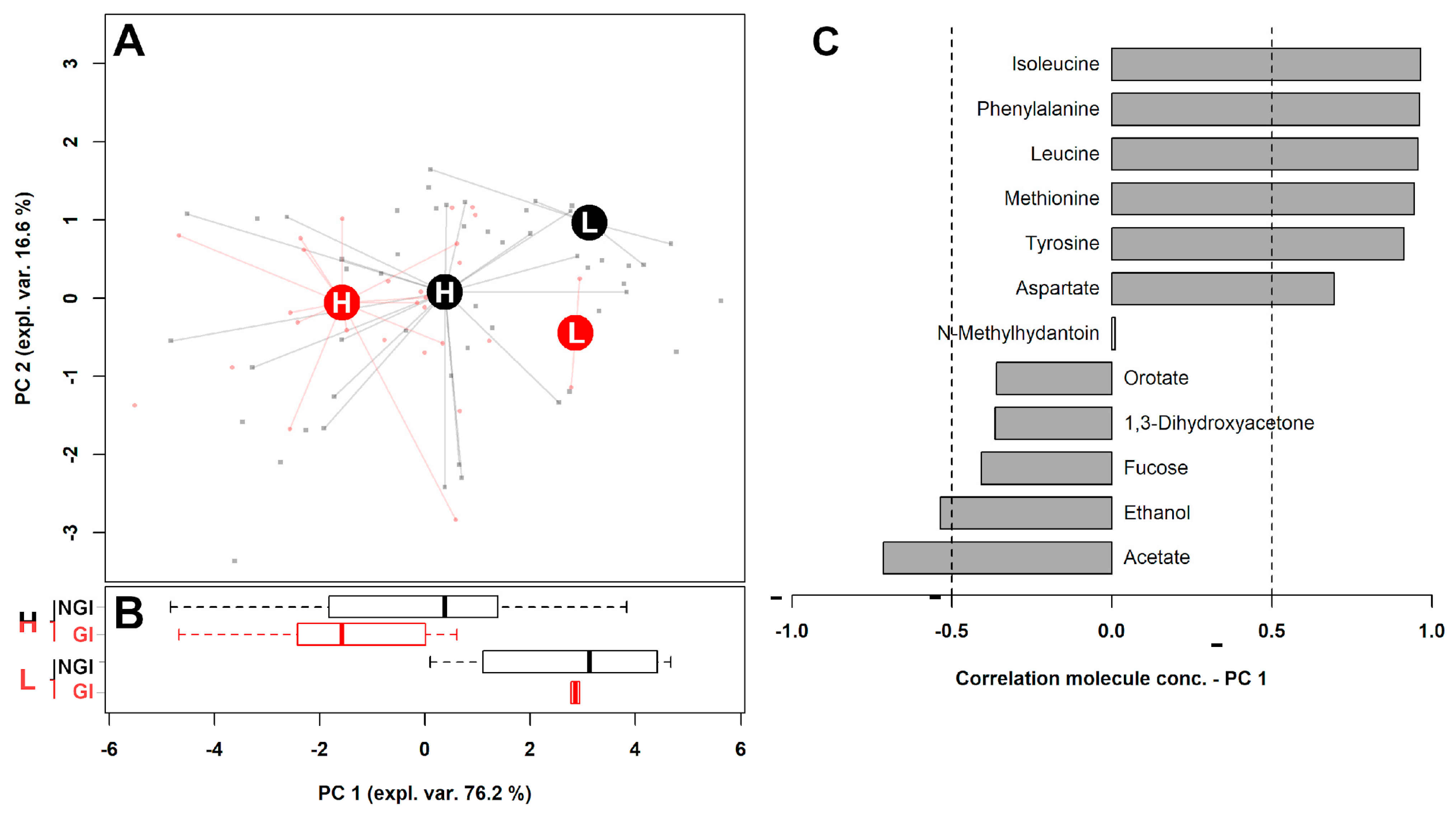
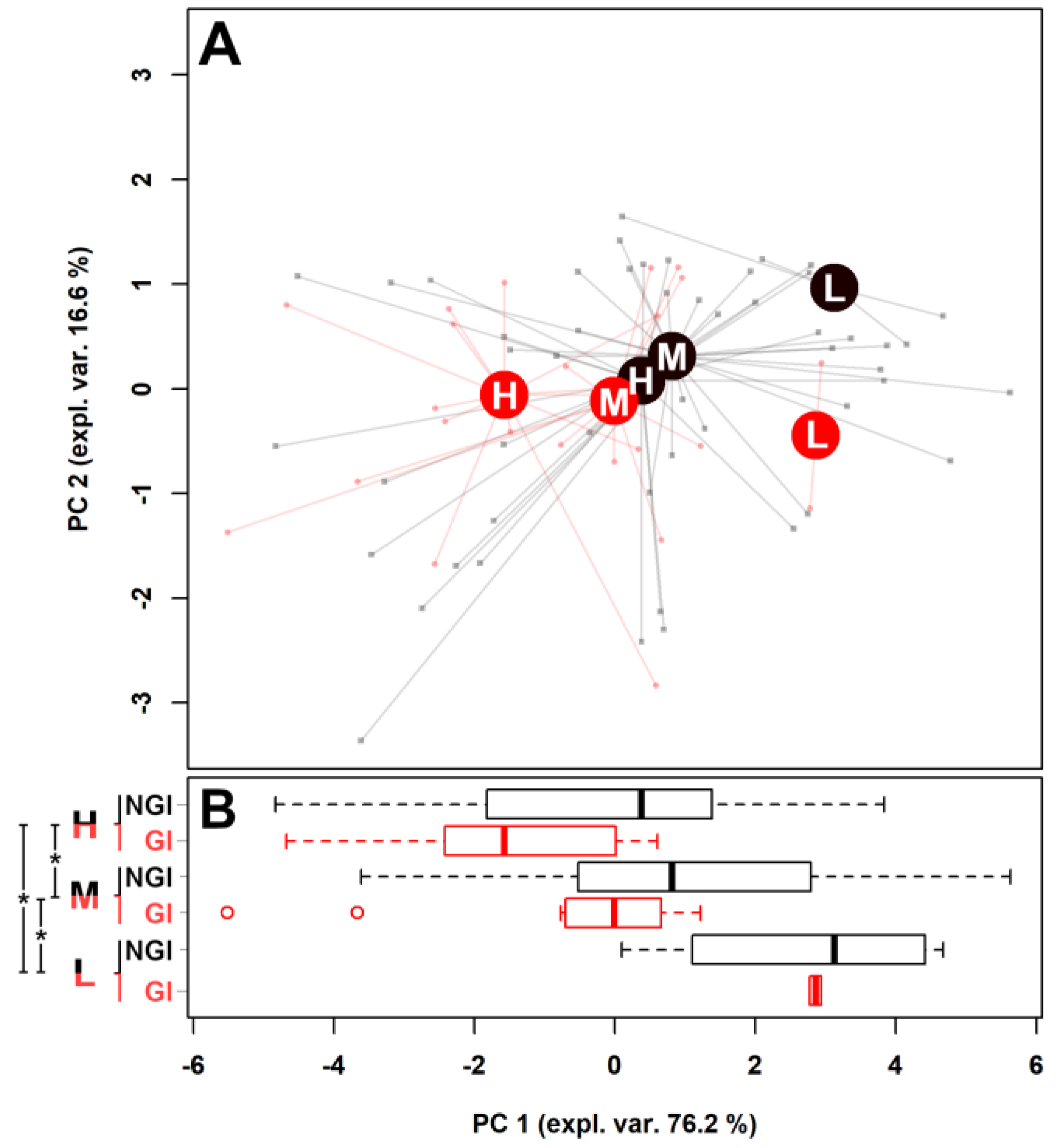
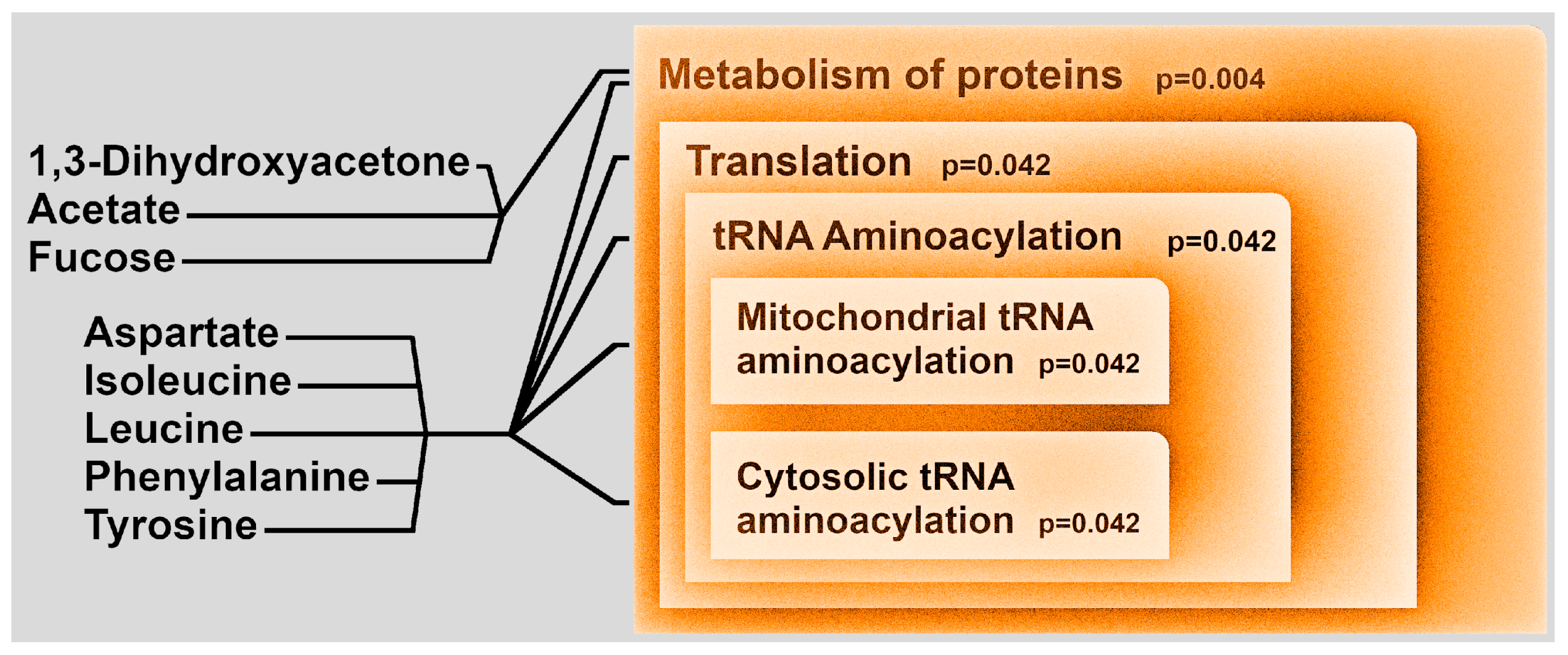
| (a) | ||||
| NGI | GI | p-Value | GI vs. NGI | |
| Acetate | 6.32 × 10−2 (4.06 × 10−2) | 7.58 × 10−2 (4.99 × 10−2) | 0.042 | ↑ |
| Alanine | 6.12 × 10−3 (2.10 × 10−3) | 4.80 × 10−3 (1.62 × 10−3) | 0.012 | ↓ |
| Ethanol | 4.62 × 10−4 (8.20 × 10−4) | 5.78 × 10−4 (9.06 × 10−4) | 0.048 | ↓ |
| Formate | 1.20 × 10−4 (5.59 × 10−5) | 1.42 × 10−4 (9.45 × 10−5) | 0.006 | ↑ |
| Isoleucine | 1.68 × 10−3 (8.23 × 10−4) | 1.43 × 10−3 (5.31 × 10−4) | 0.039 | ↓ |
| Leucine | 4.34 × 10−3 (2.31 × 10−3) | 3.61 × 10−3 (1.16 × 10−3) | 0.034 | ↓ |
| Methionine | 9.03 × 10−4 (3.75 × 10−4) | 7.87 × 10−4 (3.02 × 10−4) | 0.014 | ↓ |
| Orotate | 5.89 × 10−5 (4.18 × 10−5) | 7.70 × 10−5 (6.42 × 10−5) | 0.015 | ↑ |
| Phenylalanine | 1.34 × 10−3 (6.20 × 10−4) | 1.19 × 10−3 (4.83 × 10−4) | 0.037 | ↓ |
| Propionate | 1.82 × 10−2 (1.20 × 10−2) | 2.23 × 10−2 (1.30 × 10−2) | 0.035 | ↑ |
| Tyrosine | 2.76 × 10−3 (1.16 × 10−3) | 2.47 × 10−3 (1.02 × 10−3) | 0.048 | ↓ |
| Uridine | 4.55 × 10−5 (3.82 × 10−5) | 6.51 × 10−5 (5.33 × 10−5) | 0.003 | ↑ |
| (b) | ||||
| Low-ADOS | High-ADOS | p-Value Low vs. High-ADOS | ||
| 1,3-Dihydroxyacetone | 8.08 × 10−5 (6.53 × 10−5) | 1.67 × 10−4 (1.65 × 10−4) | 0.037 | |
| Acetate | 4.17 × 10−2 (1.14 × 10−2) | 7.87 × 10−2 (3.72 × 10−2) | 0.011 | |
| Aspartate | 2.16 × 10−3 (4.78 × 10−4) | 1.18 × 10−3 (6.75 × 10−4) | 1.78 × 10−4 | |
| Ethanol | 1.97 × 10−4 (2.91 × 10−4) | 1.02 × 10−3 (1.10 × 10−3) | 0.007 | |
| Fucose | 7.40 × 10−5 (3.47 × 10−5) | 1.37 × 10−4 (8.47 × 10−5) | 0.021 | |
| Isoleucine | 2.15 × 10−3 (4.07 × 10−4) | 1.34 × 10−3 (7.11 × 10−4) | 0.006 | |
| Leucine | 5.40 × 10−3 (9.46 × 10−4) | 3.39 × 10−3 (2.04 × 10−3) | 0.007 | |
| Methionine | 1.19 × 10−3 (2.78 × 10−4) | 8.04 × 10−4 (3.35 × 10−4) | 0.004 | |
| N-Methylhydantoin | 1.89 × 10−5 (8.73 × 10−6) | 3.93 × 10−5 (4.35 × 10−5) | 0.026 | |
| Orotate | 3.12 × 10−5 (1.55 × 10−5) | 6.34 × 10−5 (6.89 × 10−5) | 0.011 | |
| Phenylalanine | 1.57 × 10−3 (2.81 × 10−4) | 1.16 × 10−3 (6.09 × 10−4) | 0.005 | |
| Tyrosine | 3.24 × 10−3 (6.49 × 10−4) | 2.42 × 10−3 (1.13 × 10−3) | 0.011 | |
| NGI | GI | p Value | |
|---|---|---|---|
| 1,3-Dihydroxyacetone | 1.50 × 10−4 (1.51 × 10−4) | 2.59 × 10−4 (1.90 × 10−4) | NS |
| Acetate | 6.52 × 10−2 (3.96 × 10−2) | 5.49 × 10−2 (2.03 × 10−2) | NS |
| Aspartate | 1.26 × 10−3 (6.79 × 10−4) | 1.22 × 10−3 (6.84 × 10−4) | NS |
| Ethanol | 3.59 × 10−4 (3.45 × 10−4) | 5.42 × 10−4 (9.63 × 10−4) | NS |
| Fucose | 1.01 × 10−4 (7.41 × 10−5) | 1.15 × 10−4 (4.43 × 10−5) | NS |
| Isoleucine | 1.68 × 10−3 (9.22 × 10−4) | 1.55 × 10−3 (3.83 × 10−4) | NS |
| Leucine | 4.40 × 10−3 (2.03 × 10−3) | 3.94 × 10−3 (9.69 × 10−4) | NS |
| Methionine | 9.26 × 10−4 (3.31 × 10−4) | 8.07 × 10−4 (1.67 × 10−4) | NS |
| N-Methylhydantoin | 3.15 × 10−5 (2.31 × 10−5) | 3.97 × 10−5 (1.93 × 10−5) | NS |
| Orotate | 6.04 × 10−5 (3.61 × 10−5) | 7.25 × 10−5 (3.75 × 10−5) | NS |
| Phenylalanine | 1.35 × 10−3 (7.01 × 10−4) | 1.19 × 10−3 (1.97 × 10−4) | NS |
| Tyrosine | 2.73 × 10−3 (1.23 × 10−3) | 2.49 × 10−3 (4.87 × 10−4) | NS |
| Lact. | Akk. | Bifi. | Bact. | Prev. | Sutt. | Calpr. | ||
|---|---|---|---|---|---|---|---|---|
| Molecules altered only by GI | Formate | 0.32 | - | - | - | 0.28 | - | - |
| Uridine | - | - | −0.26 | 0.30 | - | - | −0.4 | |
| Alanine | 0.27 | 0.45 | 0.33 | −0.46 | - | −0.28 | - | |
| Propionate | - | - | - | - | 0.35 | - | - | |
| Molecules altered by GI and ADOS | Acetate | −0.23 | - | −0.34 | - | - | 0.22 | - |
| Ethanol | −0.23 | −0.28 | - | - | - | 0.28 | - | |
| Isoleucine | 0.37 | - | 0.51 | −0.26 | - | −0.38 | - | |
| Leucine | 0.41 | 0.27 | 0.54 | −0.32 | - | −0.41 | - | |
| Methionine | - | 0.23 | 0.33 | - | - | - | - | |
| Orotate | - | - | −0.30 | - | - | - | - | |
| Phenylalanine | 0.32 | - | 0.46 | - | - | −0.34 | - | |
| Tyrosine | - | - | 0.31 | - | - | −0.32 | - | |
| Molecules altered only by ADOS | Aspartate | - | - | - | - | - | −0.44 | - |
| N-Methylhydantoin | - | - | - | - | −0.26 | - | - | |
| 1,3-Dihydroxyacetone | - | - | −0.23 | - | - | - | - | |
| Fucose | - | - | - | 0.26 | - | 0.31 | −0.44 | |
| Metabolomics PC 1 | 0.27 | - | 0.46 | - | - | −0.35 | - | |
| Whole Sample | Low-ADOS | Moderate-ADOS | High-ADOS | |
|---|---|---|---|---|
| n | 80 | 6 | 42 | 32 |
| NGI/GI | 52/28 | 4/2 | 29/13 | 19/13 |
| Females/Males | 14/66 | 2/4 | 9/33 | 3/29 |
| Age (years) | 4.14 ± 1.01 | 3.60 ± 1.01 | 4.29 ± 1.18 | 4.05 ± 0.97 |
| BMI (Kg/m2) | 16.00 ± 1.66 | 15.21 ± 1.61 | 15.81 ± 1.61 | 16.40 ± 1.69 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laghi, L.; Mastromarino, P.; Prosperi, M.; Morales, M.A.; Calderoni, S.; Santocchi, E.; Muratori, F.; Guiducci, L. Are Fecal Metabolome and Microbiota Profiles Correlated with Autism Severity? A Cross-Sectional Study on ASD Preschoolers. Metabolites 2021, 11, 654. https://doi.org/10.3390/metabo11100654
Laghi L, Mastromarino P, Prosperi M, Morales MA, Calderoni S, Santocchi E, Muratori F, Guiducci L. Are Fecal Metabolome and Microbiota Profiles Correlated with Autism Severity? A Cross-Sectional Study on ASD Preschoolers. Metabolites. 2021; 11(10):654. https://doi.org/10.3390/metabo11100654
Chicago/Turabian StyleLaghi, Luca, Paola Mastromarino, Margherita Prosperi, Maria Aurora Morales, Sara Calderoni, Elisa Santocchi, Filippo Muratori, and Letizia Guiducci. 2021. "Are Fecal Metabolome and Microbiota Profiles Correlated with Autism Severity? A Cross-Sectional Study on ASD Preschoolers" Metabolites 11, no. 10: 654. https://doi.org/10.3390/metabo11100654
APA StyleLaghi, L., Mastromarino, P., Prosperi, M., Morales, M. A., Calderoni, S., Santocchi, E., Muratori, F., & Guiducci, L. (2021). Are Fecal Metabolome and Microbiota Profiles Correlated with Autism Severity? A Cross-Sectional Study on ASD Preschoolers. Metabolites, 11(10), 654. https://doi.org/10.3390/metabo11100654






