Characterization of Flavonoids and Transcripts Involved in Their Biosynthesis in Different Organs of Cissus rotundifolia Lam
Abstract
:1. Introduction
2. Results
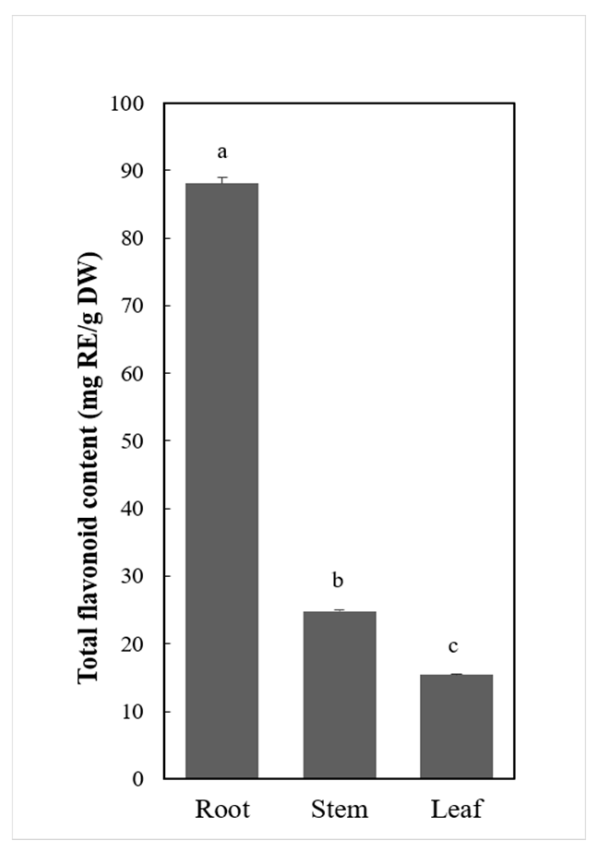
2.1. Total Flavonoids Content Estimation
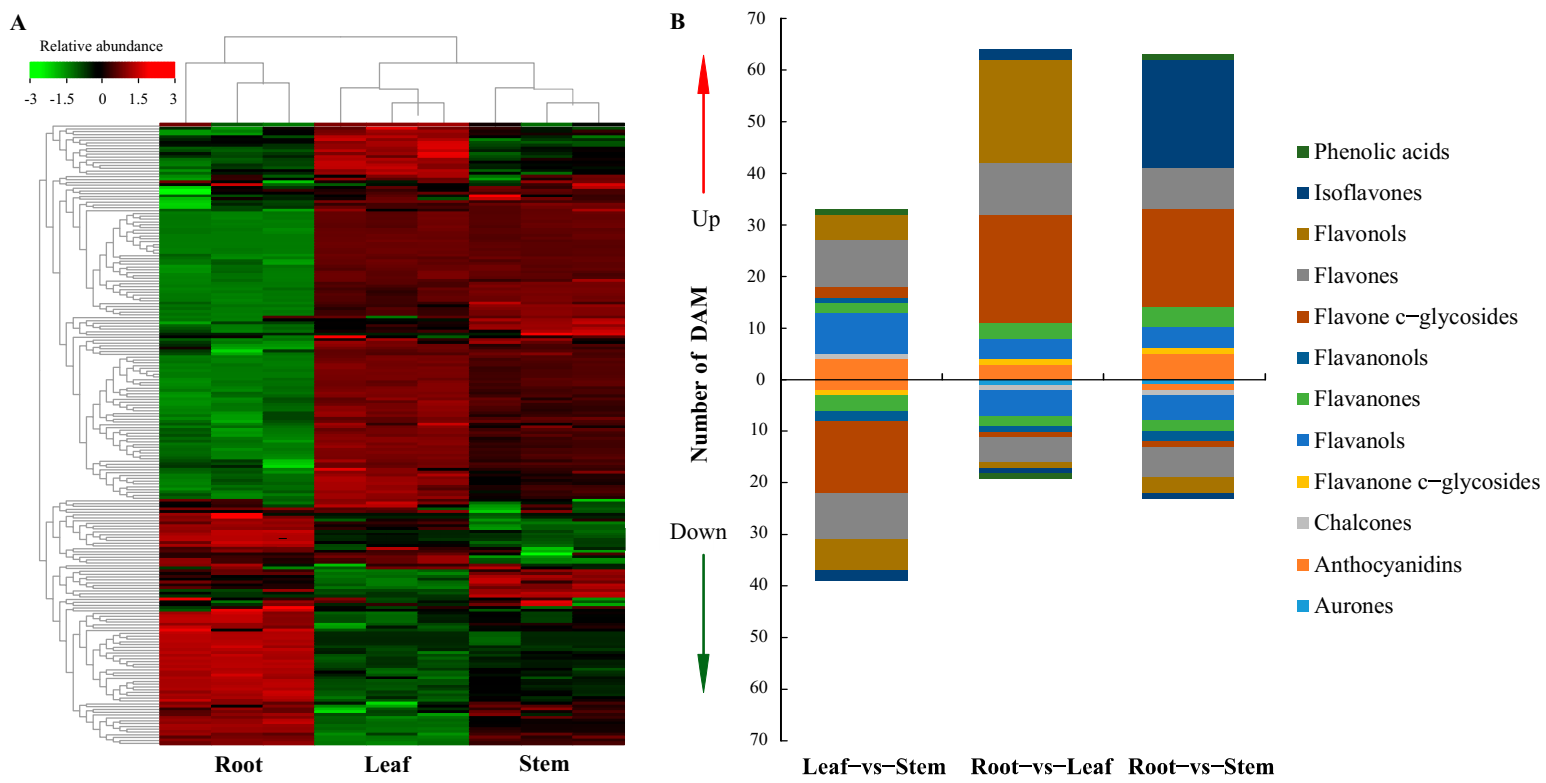
2.2. Flavonoids Profiling in C. rotundifolia
2.3. Identification of Differentially Accumulated Metabolites (DAMs)
2.4. Transcript Sequencing and Mapping
2.5. Functional Annotation of Identified Genes
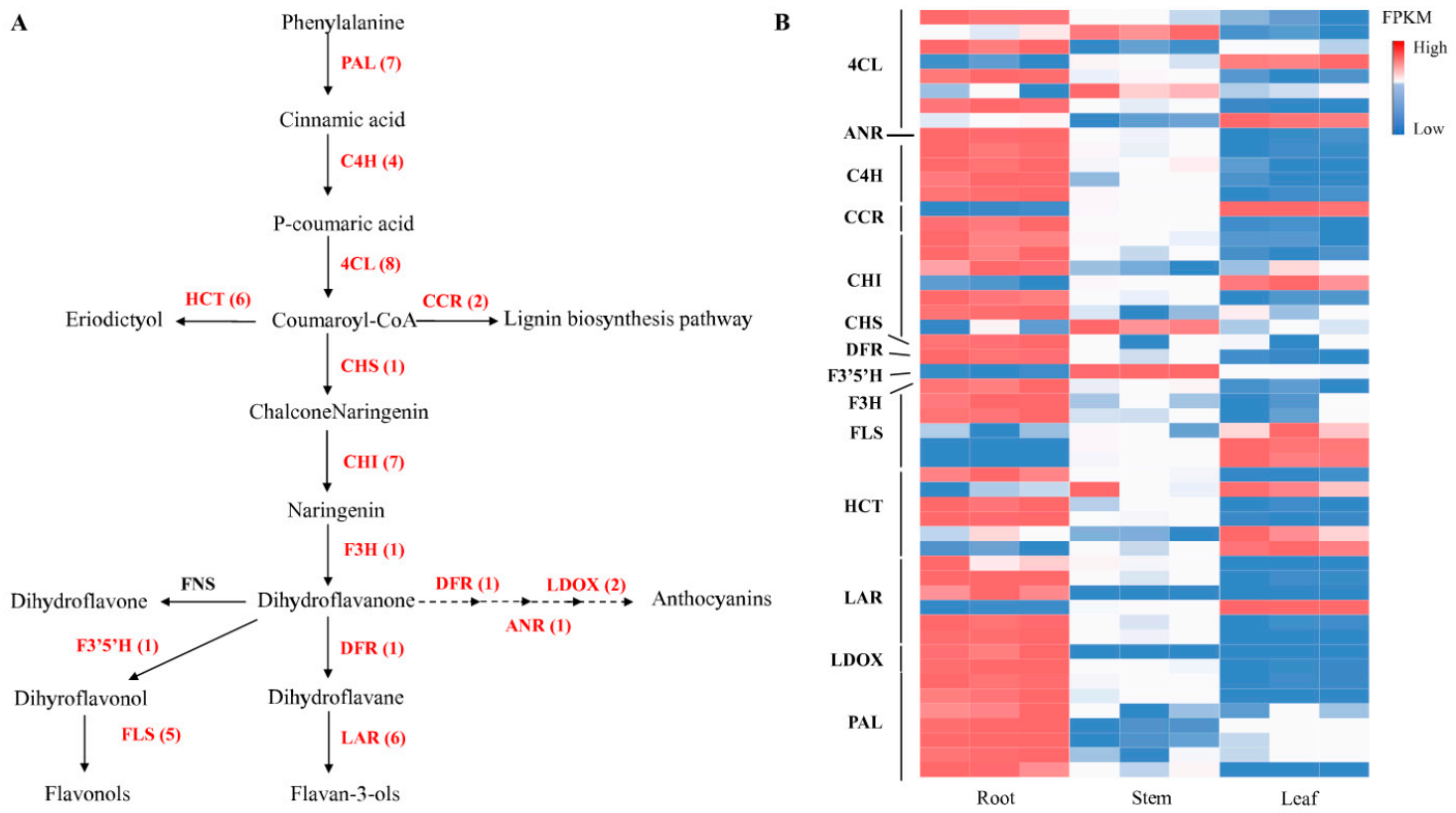
2.6. Candidate Genes Involved in Flavonoids Biosynthesis
2.7. Validation of RNA-Seq Data Using qPCR
2.8. Candidate Transcription Factors Related to Flavonoids Biosynthesis
2.9. Differential Gene Expression between Tissues of C. rotundifolia
2.10. GO Enrichment and KEGG Pathway Analysis for Differentially Expressed Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Metabolome
4.2.1. Sample Preparation and Extraction
4.2.2. HPLC Conditions
4.2.3. ESI-Q TRAP-MS/MS
4.2.4. Qualitative and Quantitative Analysis of Metabolites
4.3. Determination of Total Flavonoids Content (TFC)
4.3.1. TFC Extraction
4.3.2. Determination of TFC
4.4. Transcriptomics
4.4.1. Total RNA Extraction, RNA Library Construction, and Sequencing
4.4.2. RNA-Seq Data Analysis and Functional Annotation
4.4.3. RNA-Seq Data Validation Using qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alzoreky, N.S.; Nakahara, K. Antibacterial activity of extracts from some edible plants commonly consumed in Asia. Int. J. Food Microbiol. 2003, 80, 223–230. [Google Scholar] [CrossRef]
- Onyechi, U.A.; Judd, P.A.; Ellis, P.R. African plant foods rich in non-starch polysaccharides reduce postprandial blood glucose and insulin concentrations in healthy human subjects. Br. J. Nutr. 1998, 80, 419–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alshehri, S.A. Antidiabetic Activity of Cissus Rotundifolia Plant Growing in Saudi Arabia; South Dakota State University: Brookings, SD, USA, 2020. [Google Scholar]
- Fernandes, G. Medicinal properties of plants from the genus Cissus: A Review. J. Med. Plants Res. 2012, 30, 6. [Google Scholar] [CrossRef]
- Korish, M. Nutritional evaluation of wild plant Cissus rotundifolia. Ital. J. Food Sci. 2016, 28, 43–49. [Google Scholar]
- Onyechi, U.A.; Ibeanu, V.N. Effects of diets containing Cissus rotundifolia flour on lipid profile of rats and postprandial glucose levels of normoglycemic human adults. Afr. J. Biotechnol. 2016, 15, 557–564. [Google Scholar]
- Al-Mehdar, A.A.; Al-Battah, A.M. Evaluation of hypoglycemic activity of Boswellia carterii and Cissus rotundifolia in streptozotocin/nicotinamide-induced diabetic rats. Yemeni J. Med. Sci. 2016, 10, 30–38. [Google Scholar] [CrossRef]
- Ali, A.A.N.; Al-Rahwi, K.; Lindequist, U. Some medicinal plants used in Yemeni Herbal Medicine to treat malaria. Afr. J. Tradit. Complement Altern. Med. 2004, 1, 72–76. [Google Scholar] [CrossRef]
- AL-Bukhaiti, W.Q.; Noman, A.; Mahdi, A.A.; Ali, A.H.; Abed, S.M.; Rashed, M.M.A.; Wang, H. Profiling of phenolic compounds and antioxidant activities of Cissus rotundifolia (Forssk.) as influenced by ultrasonic-assisted extraction conditions. J. Food Meas. Charact. 2019, 13, 634–647. [Google Scholar] [CrossRef]
- Said, A.A.; Aboutabl, E.A.; El Awdan, S.A.; Raslan, M.A. Proximate analysis, phytochemical screening, and bioactivities evaluation of Cissus rotundifolia (Forssk.) Vahl. (Fam. Vitaceae) and Sansevieria cylindrica Bojer ex Hook. (Fam. Dracaenaceae) growing in Egypt. Egypt Pharm. J. 2015, 14, 180. [Google Scholar] [CrossRef]
- Stavric, B. Antimutagens and anticarcinogens in foods. Food Chem. Toxicol. 1994, 32, 79–90. [Google Scholar] [CrossRef]
- Barreca, D.; Bellocco, E.; Caristi, C.; Leuzzi, U.; Gattuso, G. Elucidation of the flavonoid and furocoumarin composition and radical-scavenging activity of green and ripe chinotto (Citrus myrtifolia Raf.) fruit tissues, leaves and seeds. Food Chem. 2011, 129, 1504–1512. [Google Scholar] [CrossRef]
- Cushnie, T.P.T.; Lamb, A.J. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 2005, 26, 343–356. [Google Scholar] [CrossRef]
- García-Salas, P.; Gómez-Caravaca, A.M.; Arráez-Román, D.; Segura-Carretero, A.; Guerra-Hernández, E.; García-Villanova, B.; Fernández-Gutiérrez, A. Influence of technological processes on phenolic compounds, organic acids, furanic derivatives, and antioxidant activity of whole-lemon powder. Food Chem. 2013, 141, 869–878. [Google Scholar] [CrossRef]
- Heim, K.E.; Tagliaferro, A.R.; Bobilya, D.J. Flavonoid antioxidants: Chemistry, metabolism and structure-activity relationships. J. Nutr. Biochem. 2002, 13, 572–584. [Google Scholar] [CrossRef]
- Middleton, E.; Kandaswami, C. Effects of flavonoids on immune and inflammatory cell functions. Biochem. Pharmacol. 1992, 43, 1167–1179. [Google Scholar] [CrossRef]
- Chen, H.Q.; Zuo, W.J.; Wang, H.; Shen, H.Y.; Luo, Y.; Dai, H.F.; Mei, W.L. Two new antimicrobial flavanes from dragon’s blood of Dracaena cambodiana. J. Asian Nat. Prod. Res. 2012, 14, 436–440. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, H.; Zhao, Y.X.; Zeng, Y.-B.; Shen, H.Y.; Dai, H.F.; Mei, W.L. Cytotoxic and antibacterial flavonoids from dragon’s blood of Dracaena cambodiana. Planta Med. 2011, 77, 2053–2056. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Q.A.; Xu, M.; Yang, C.R.; Wang, D.; Li, H.Z.; Zhu, H.T.; Zhang, Y.J. Flavonoid oligomers from Chinese dragon’s blood, the red resins of Dracaena cochinchinensis. Nat. Prod. Bioprospect. 2012, 2, 111–116. [Google Scholar] [CrossRef] [Green Version]
- Rajkapoor, B.; Murugesh, N.; Rama Krishna, D. Cytotoxic activity of a flavanone from the stem of Bauhinia variegata Linn. Nat. Prod. Res. 2009, 23, 1384–1389. [Google Scholar] [CrossRef]
- Alqahtani, J.; Formisano, C.; Chianese, G.; Luciano, P.; Stornaiuolo, M.; Perveen, S.; Taglialatela-Scafati, O. Glycosylated phenols and an unprecedented diacid from the Saudi plant Cissus rotundifolia. J. Nat. Prod. 2020, 83, 3298–3304. [Google Scholar] [CrossRef]
- Nabavi, S.M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Daglia, M.; Xu, S.; Rastrelli, L.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef]
- Tohge, T.; de Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef]
- Davies, K.M.; Schwinn, K.E. Flavonoids. Chemistry, Biochemistry and Applications; Andersen, O.M., Markham, K.R., Eds.; Taylor & Francis Group: Abingdon, UK, 2006; pp. 149–178. [Google Scholar]
- Tripoli, E.; La Guardia, M.; Giammanco, S.; Majo Di, D.; Giammanco, M. Citrus flavonoids: Molecular structure, biological activity and nutritional properties: A review. Food Chem. 2007, 104, 466–479. [Google Scholar] [CrossRef]
- Wang, J.L.; Li, X.; Jiang, D.F.; Ma, P.; Yang, C.R. Chemical constituents of dragons blood resin from Dracaena cochinchinensis in Yunnan and their antifungal activity. Plant Divers 1995, 17, 1–3. [Google Scholar]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB-bHLH-WD Repeat (MBW) pigment regulatory model: Addition of a WRKY Factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef] [Green Version]
- Amato, A.; Cavallini, E.; Walker, A.R.; Pezzotti, M.; Bliek, M.; Quattrocchio, F.; Koes, R.; Ruperti, B.; Bertini, E.; Zenoni, S.; et al. The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. Plant J. 2019, 99, 1220–1241. [Google Scholar] [CrossRef]
- Winkel, B. The Biosynthesis of Flavonoids. In The Science of Flavonoids; Springer: New York, NY, USA, 2006; pp. 71–95. [Google Scholar] [CrossRef]
- Sun, Y.J.; He, J.M.; Kong, J.Q. Characterization of two flavonol synthases with iron-independent flavanone 3-hydroxylase activity from Ornithogalum caudatum Jacq. BMC Plant Biol. 2019, 19, 195. [Google Scholar] [CrossRef]
- Matsuda, F.; Hirai, M.Y.; Sasaki, E.; Akiyama, K.; Yonekura-Sakakibara, K.; Provart, N.J.; Sakurai, T.; Shimada, Y.; Saito, K. AtMetExpress development: A phytochemical atlas of Arabidopsis development. Plant Physiol. 2010, 152, 566–578. [Google Scholar] [CrossRef] [Green Version]
- Albinsky, D.; Kusano, M.; Higuchi, M.; Hayashi, N.; Kobayashi, M.; Fukushima, A.; Masaki, M.; Takanari, I.; Keiko, M.; Yoko, H.; et al. Metabolomic screening applied to rice FOX Arabidopsis lines leads to the identification of a gene-changing nitrogen metabolism. Mol. Plant. 2010, 3, 125–142. [Google Scholar] [CrossRef]
- Abad-García, B.; Garmón-Lobato, S.; Berrueta, L.A.; Gallo, B.; Vicente, F. Online characterization of 58 phenolic compounds in Citrus fruit juices from Spanish cultivars by high-performance liquid chromatography with photodiode-array detection coupled to electrospray ionization triple quadrupole mass spectrometry. Talanta 2012, 99, 213–224. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant. 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, X.; Chen, W.; Wang, W.; Zhang, H.; Liu, X.; Luo, J. Comprehensive profiling and natural variation of flavonoids in rice. J. Integr. Plant Biol. 2014, 56, 876–886. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Chen, W.; Gao, Y.; Liu, X.; Zhang, H.; Xu, C.; Yu, S.; Zhang, Q.; Luo, J. Genetic analysis of the metabolome exemplified using a rice population. Proc. Natl. Acad. Sci. USA 2013, 110, 20320–20325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Wang, X.; Tzin, V.; Romeis, J.; Peng, Y.; Li, Y. Combined transcriptome and metabolome analyses to understand the dynamic responses of rice plants to attack by the rice stem borer Chilo suppressalis (Lepidoptera: Crambidae). BMC Plant Biol. 2016, 16, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galperin, M.Y.; Kristensen, D.M.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Microbial genome analysis: The COG approach. Brief Bioinform. 2019, 20, 1063–1070. [Google Scholar] [CrossRef]
- Zhu, J.H.; Cao, T.J.; Dai, H.F.; Li, H.L.; Guo, D.; Mei, W.L.; Peng, S.Q. De Novo transcriptome characterization of Dracaena cambodiana and analysis of genes involved in flavonoid accumulation during formation of dragon’s blood. Sci. Rep. 2016, 6, 38315. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Fan, L.; Zeng, Q.; Chen, Z.; Freidman, N.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Al-Mamary, M.A.J. Antioxidant activity of commonly consumed vegetables in Yemen. Malays J. Nutr. 2002, 8, 179–189. [Google Scholar]
- Said, A.; Aboutabl, E.A.; Melek, F.R.; Jaleel, G.A.R.B.; Raslan, M. Phytoconstituents profiling of Cissus rotundifolia (Forssk.) Vahl. by HPLC-MS/MS, and evaluation of its free radical scavenging activity (DPPH) and cytotoxicity. Trends Phytochem. Res. 2018, 2, 65–74. [Google Scholar]
- Kaewnarin, K.; Niamsup, H.; Shank, L.; Rakariyatham, N. Antioxidant and antiglycation activities of some edible and medicinal plants. Chiang Mai J. Sci. 2014, 41, 105–116. [Google Scholar]
- Vijayalakshmi, A.; Kumar, P.R.; Sakthi Priyadarsini, S.; Meenaxshi, C. In vitro antioxidant and anticancer activity of flavonoid fraction from the aerial parts of Cissus quadrangularis Linn. against human breast carcinoma cell lines. J. Chem. 2013, 2013, 150675. [Google Scholar] [CrossRef] [Green Version]
- Shoibe, M.; Chy, M.; Uddin, N.; Alam, M.; Adnan, M.; Islam, M.; Nihar, S.W.; Rahman, N.; Suez, E. In vitro and in vivo biological activities of Cissus adnata (Roxb.). Biomedicines 2017, 5, 63. [Google Scholar] [CrossRef] [Green Version]
- Park, E.; Hong, K.; Kwon, B.-M.; Kim, Y.; Kim, J.-H. Jaceosidin ameliorates insulin resistance and kidney dysfunction by enhancing insulin receptor signaling and the antioxidant defense system in type 2 diabetic mice. J. Med. Food 2020, 23, 1083–1092. [Google Scholar] [CrossRef]
- Chiou, W.F.; Lee, C.H.; Liao, J.F.; Chen, C.C. 8-Prenylkaempferol accelerates osteoblast maturation through bone morphogenetic protein-2/p38 pathway to activate Runx2 transcription. Life Sci. 2011, 88, 335–342. [Google Scholar] [CrossRef]
- Rehman, S.U.; Choe, K.; Yoo, H.H. Review on a traditional herbal medicine, Eurycoma longifolia Jack (Tongkat Ali): Its traditional uses, chemistry, evidence-based pharmacology and toxicology. Molecules 2016, 21, 331. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Lv, Y.; Xu, L.; Zheng, Q. Quantitative efficacy of soy isoflavones on menopausal hot flashes. Br. J. Clin. Pharmacol. 2015, 79, 593–604. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Lu, W.; Yin, T.; Lu, L. Calycosin suppresses TGF-β-induced epithelial-to-mesenchymal transition and migration by upregulating BATF2 to target PAI-1 via the Wnt and PI3K/Akt signaling pathways in colorectal cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 240. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.; Choo, S.; Sim, H.; Bae, J.-S. Inhibitory activities of ononin on particulate matter-induced oxidative stress. Biotechnol. Bioprocess. Eng. 2021, 26, 208–215. [Google Scholar] [CrossRef]
- Behbahani, M. Anti-human immunodeficiency virus-1 activities of pratensein and pratensein glycoside from Alhaji maurorum and its parasite Cuscuta kotchiana. Chin. J. Integr. Med. 2017, 2017, 1–6. [Google Scholar] [CrossRef]
- Tan, L.; Wang, M.; Kang, Y.; Azeem, F.; Zhou, Z.; Tuo, D.; María Preciado Rojo, L.; Khan, I.A.; Pan, Z. Biochemical and functional characterization of anthocyanidin reductase (ANR) from Mangifera indica L. Molecules 2018, 23, 2876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, T.; Guo, K.; Liu, L.; Tian, W.; Xie, X.; Wen, S.; Wen, C. Integrated transcriptomic and metabolomic data reveal the flavonoid biosynthesis metabolic pathway in Perilla frutescens (L.) leaves. Sci. Rep. 2020, 10, 16207. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhang, J.; Liu, X.; Meng, M.; Wang, J.; Lin, J. Tissue-specific transcriptome for Dendrobium officinale reveals genes involved in flavonoid biosynthesis. Genomics 2020, 112, 1781–1794. [Google Scholar] [CrossRef] [PubMed]
- Goto-Yamamoto, N.; Wan, G.H.; Masaki, K.; Kobayashi, S. Structure and transcription of three chalcone synthase genes of grapevine (Vitis vinifera). Plant Sci. 2002, 162, 867–872. [Google Scholar] [CrossRef]
- Muir, S.R.; Collins, G.J.; Robinson, S.; Hughes, S.; Bovy, A.; De Vos, C.H.R.; van Tunen, A.J.; Verhoeyen, M.E. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat. Biotechnol. 2001, 19, 470–474. [Google Scholar] [CrossRef]
- Jez, J.M.; Bowman, M.E.; Dixon, R.A.; Noel, J.P. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat. Struct. Biol. 2000, 7, 786–791. [Google Scholar]
- Forkmann, G.; Martens, S. Metabolic engineering and applications of flavonoids. Curr. Opin. Biotechnol. 2001, 12, 155–160. [Google Scholar] [CrossRef]
- Zhang, S.; Li, H.; Liang, X.; Yan, Y.; Xia, P.; Jia, Y.; Liang, Z. Enhanced production of phenolic acids in Salvia miltiorrhiza hairy root cultures by combing the RNAi-mediated silencing of chalcone synthase gene with salicylic acid treatment. Biochem. Eng. J. 2015, 103, 185–192. [Google Scholar] [CrossRef]
- Cho, K.; Cho, K.-S.; Sohn, H.-B.; Ha, I.J.; Hong, S.-Y.; Lee, H.; Kim, Y.-M.; Nam, M.H. Network analysis of the metabolome and transcriptome reveals novel regulation of potato pigmentation. J. Exp. Bot. 2016, 67, 1519–1533. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Li, J.; Zhu, L.; Chang, P.; Li, L.; Zhang, L. Identification and characterization of MYB-bHLH-WD40 regulatory complex members controlling anthocyanidin biosynthesis in blueberry fruits development. Genes 2019, 10, 496. [Google Scholar] [CrossRef] [Green Version]
- Schaart, J.G.; Dubos, C.; Romero De La Fuente, I.; van Houwelingen, A.M.M.L.; de Vos, R.C.H.; Jonker, H.H.; Bovy, A.; Xu, W.; Routaboul, J.-M.; Lepiniec, L. Identification and characterization of MYB-bHLH-WD 40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (F ragaria× ananassa) fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef]
- Chen, Y.; Pan, W.; Jin, S.; Lin, S. Combined metabolomic and transcriptomic analysis reveals key candidate genes involved in the regulation of flavonoid accumulation in Anoectochilus roxburghii. Process Biochem. 2020, 91, 339–351. [Google Scholar] [CrossRef]
- Liang, W.; Ni, L.; Carballar-Lejarazú, R.; Zou, X.; Sun, W.; Wu, L.; Yuan, X.; Mao, Y.; Huang, W.; Zou, S. Comparative transcriptome among Euscaphis konishii Hayata tissues and analysis of genes involved in flavonoid biosynthesis and accumulation. BMC Genom. 2019, 20, 1–14. [Google Scholar] [CrossRef]
- Zou, Y.; Chang, S.K.C.; Gu, Y.; Qian, S.Y. Antioxidant activity and phenolic compositions of lentil (Lens culinaris var. Morton) extract and its fractions. J. Agric. Food Chem. 2011, 59, 2268–2276. [Google Scholar] [CrossRef] [Green Version]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]


| Metabolite Name | Precursor Ion (Q1) (Da) | Product Ion (Q3) (Da) | Retention Time (min) | Main Fragments (Da) |
|---|---|---|---|---|
| Diosmetin (5,7,3′-Trihydroxy-4′-methoxyflavone) | 299.06 | 256.04 | 6.1 | 256.04, 284.03, 299.05, 299.13, 299.06 |
| Quercetin-3-o-galactoside (Hyperin) | 463.1 | 300.03 | 4.2 | 300.03, 301.03, 463.09 |
| Catechin gallate | 441.3 | 289.08 | 4.2 | 124.02, 125.03, 169.02, 193.01, 203.07, 245.08, 289.08, 331.05, 441.08 |
| Kaempferol-3-o-rhamnoside (Afzelin)(Kaempferin) | 431 | 284.04 | 5 | 229.05, 227.04, 255.03, 284.04, 285.05, 431.11 |
| Epiafzelechin | 275.1 | 139.04 | 4.2 | 107.05, 111.04, 121.06, 139.04, 149.06, 145.06, 173.06, 191.07, 275.09 |
| Delphinidin | 303.05 | 137.02 | 4.8 | 137.02, 153.02, 165.02, 129.05, 257.04 |
| Isoorientin-7-o-(6″-p-coumaroyl)glucoside | 757.2 | 637.16 | 4.2 | 147.04, 291.08, 309.09, 329.07, 353.07, 431.1, 449.11, 577.13, 637.15, 757.20 |
| Kaempferol-3-o-rutinoside(Nicotiflorin) | 593.16 | 285 | 4.3 | 285.04, 593.15 |
| Kaempferol-3-arabinopyranoside | 419.1 | 133.05 | 5 | 133.05, 287.06 |
| Epicatechin | 291 | 123.05 | 3.8 | 119.05, 123.05, 139.04, 147.04, 165.06, 161.06, 179.07, 207.07, 291.09 |
| Isohemiphloin | 433.12 | 313.07 | 4.2 | 125.02, 152.99, 211.06, 271.06, 331.07, 343.08, 359.15, 433.23, 433.11, 433.2 |
| Calycosin | 285 | 225.06 | 5.5 | 225.09, 242.06, 269.04, 270.05, 285.08 |
| 5-Hydroxy-6,7,8,3′,4′-pentamethoxyflavone | 389.1 | 359.08 | 7.2 | 341.08, 359.07, 389.12 |
| Pratensein | 301.07 | 269.04 | 6.4 | 167.04, 181.07, 258.05, 269.08, 286.05, 301.07 |
| Aureusidin | 287.05 | 153.02 | 5.6 | 153.02, 287.06 |
| 5-Hydroxyauranetin | 389.1 | 359.07 | 7 | 341.06, 359.07, 389.12 |
| Epigallocatechin | 305 | 219.07 | 3 | 125.02, 137.02, 167.03, 165.02, 179.03, 219.07, 221.05, 305.07 |
| Gallocatechin | 307 | 163.04 | 2.8 | 123.04, 139.04, 163.04, 177.05, 195.06, 233.06 |
| Kaempferol (3,5,7,4′-Tetrahydroxyflavone) | 285.05 | 229.05 | 6.2 | 151.00, 185.06, 211.04, 229.05, |
| Naringenin-7-o-glucoside (Prunin) | 433 | 151 | 4.8 | 119.05, 151.00, 177.02, 255.03, 271.06, 284.03, 301.03, 417.08, 433.11 |
| Luteolin-8-c-glucoside (Orientin) | 449.1 | 329.07 | 4 | 287.06, 299.06, 329.07, 353.07, 383.08, 413.09, 431.10, 449.11 |
| Avicularin(Quercetin-3-o-α-L-arabinofuranoside) | 435.08 | 303 | 4.5 | 303.05, 257.04, 229.05 |
| Quercetin-3-o-arabinoside (Guaijaverin) | 433.08 | 255.03 | 4.7 | 151.00, 179.00, 255.03, 271.02, 300.03, 301.04, 433.08 |
| Isoorientin-7-o-glucoside | 611.1 | 329.07 | 3.5 | 299.06, 319.04, 329.07, 383.08, 431.10, 449.11, 465.10, 611.15 |
| Vitexin-7-o-(6″-p-coumaroyl)glucoside | 741.2 | 415.1 | 4.4 | 147.04, 309.11, 313.07, 337.07, 415.10, 433.11, 741.20 |
| Luteolin-7-o-rutinoside | 595.16 | 287.05 | 4.3 | 287.06, 449.11 |
| Phloretin | 273.08 | 123.04 | 6 | 119.05, 123.04, 167.03, 273.08 |
| Kaempferol-3,7-o-dirhamnoside (Kaempferitrin) | 579.2 | 433.11 | 4.3 | 287.05, 433.11 |
| Quercetin | 303.04 | 229.05 | 5.6 | 153.02, 165.02, 229.05, 257.04, 285.04 |
| Kaempferol-3-o-arabinoside (Juglanin) | 417.1 | 284.03 | 4.9 | 227.04, 255.03, 284.04, 285.04, 417.09 |
| Enzyme | Gene Designation | No. of Annotated Sequences (C. rotundifolia) | No. of Annotated Sequences (D. cambodiana) |
|---|---|---|---|
| Phenylalanine ammonia-lyase | PAL | 7 | 6 |
| Cinnamic acid 4-hydroxylase | C4H | 4 | 1 |
| 4-Coumaric acid: CoA ligase | 4CL | 8 | 18 |
| Chalcone synthase | CHS | 1 | 10 |
| Chalcone isomerase | CHI | 7 | 6 |
| Cinnamoyl-CoA | CCR | 2 | - |
| Flavanone 3-hydroxylase | F3H | 1 | 7 |
| Flavonoid 3′,5′-hydroxylase | F3′5′H | 1 | - |
| Shikimate-o-hydroxycinnamoyltransferase | HCT | 6 | - |
| Flavonol synthase | FLS | 5 | 10 |
| Dihydroflavanol 4-reductase | DFR | 1 | 16 |
| Leucoanthocyanidin dioxygenase/anthocyanidin synthase | LDOX/ANS | 2 | - |
| Anthocyanidin reductase | ANR | 1 | - |
| Leucoanthocyanidin reductase | LAR | 6 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gichuki, D.K.; Li, Q.; Hou, Y.; Liu, Y.; Ma, M.; Zhou, H.; Xu, C.; Zhu, Z.; Wang, L.; Musila, F.M.; et al. Characterization of Flavonoids and Transcripts Involved in Their Biosynthesis in Different Organs of Cissus rotundifolia Lam. Metabolites 2021, 11, 741. https://doi.org/10.3390/metabo11110741
Gichuki DK, Li Q, Hou Y, Liu Y, Ma M, Zhou H, Xu C, Zhu Z, Wang L, Musila FM, et al. Characterization of Flavonoids and Transcripts Involved in Their Biosynthesis in Different Organs of Cissus rotundifolia Lam. Metabolites. 2021; 11(11):741. https://doi.org/10.3390/metabo11110741
Chicago/Turabian StyleGichuki, Duncan Kiragu, Qingyun Li, Yujun Hou, Yuanshuang Liu, Mengxue Ma, Huimin Zhou, Chen Xu, Zhenfei Zhu, Lina Wang, Fredrick Mutie Musila, and et al. 2021. "Characterization of Flavonoids and Transcripts Involved in Their Biosynthesis in Different Organs of Cissus rotundifolia Lam" Metabolites 11, no. 11: 741. https://doi.org/10.3390/metabo11110741
APA StyleGichuki, D. K., Li, Q., Hou, Y., Liu, Y., Ma, M., Zhou, H., Xu, C., Zhu, Z., Wang, L., Musila, F. M., Wang, Q., & Xin, H. (2021). Characterization of Flavonoids and Transcripts Involved in Their Biosynthesis in Different Organs of Cissus rotundifolia Lam. Metabolites, 11(11), 741. https://doi.org/10.3390/metabo11110741






