Discovering the Next-Generation Plant Protection Products: A Proof-of-Concept via the Isolation and Bioactivity Assessment of the Olive Tree Endophyte Bacillus sp. PTA13 Lipopeptides
Abstract
:1. Introduction
2. Results and Discussion
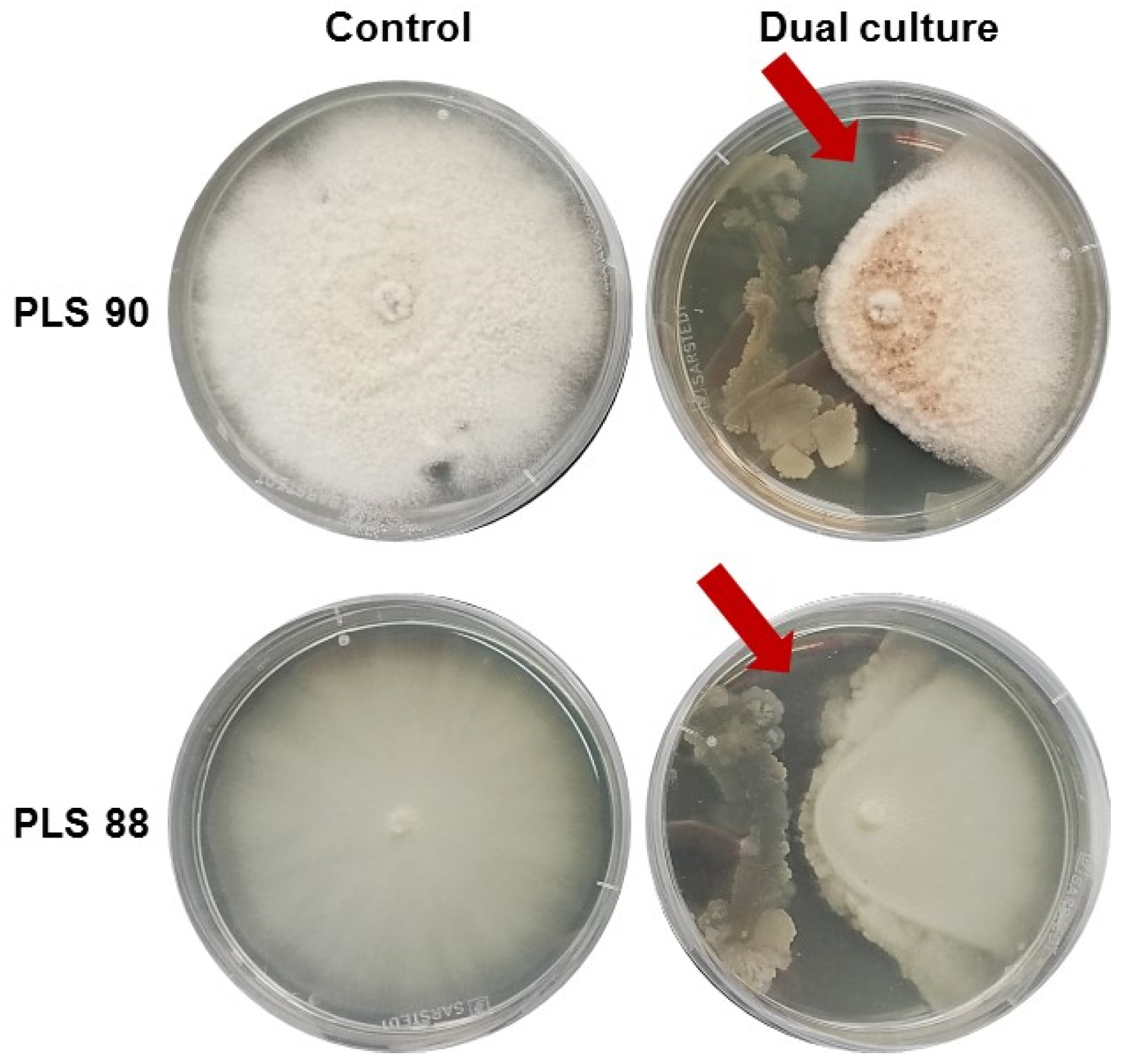
2.1. Olive Tree Endophytic Microorganisms (EMs) and Bioactivity-Driven Selection
2.2. The Bacillus sp. Is the Predominant Endophytic Microorganism (EM) of the Olive Tree Roots Studied
2.3. Isolation of the Bacillus sp. PTA13 Lipopeptides (LPs) and Deconvolution of Their Metabolite Composition Applying LC/ESI/MS/MS Analysis
2.4. Time-Course Study of the Bacillus sp. PTA13 Growth Rate and Its Lipopeptide-Producing Capacity
2.5. Isolation Protocols for the Separation of Bacillus sp. PTA13 Lipopeptide (LP) Groups When Applying Liquid–Liquid Extraction and Chromatography Techniques
2.5.1. Selection of the Optimum Biphasic Solvent System
2.5.2. Rapid Separation of the Three Major Lipopeptide (LP) Groups (Surfactins, Bacillomycins, Fengycins) Using Liquid-Liquid Extraction and Size Exclusion Chromatography
2.5.3. Application of Centrifugal Partition Chromatography (CPC) Resulted in the Detection and Separation of Low and High Abundance Lipopeptides (LPs)
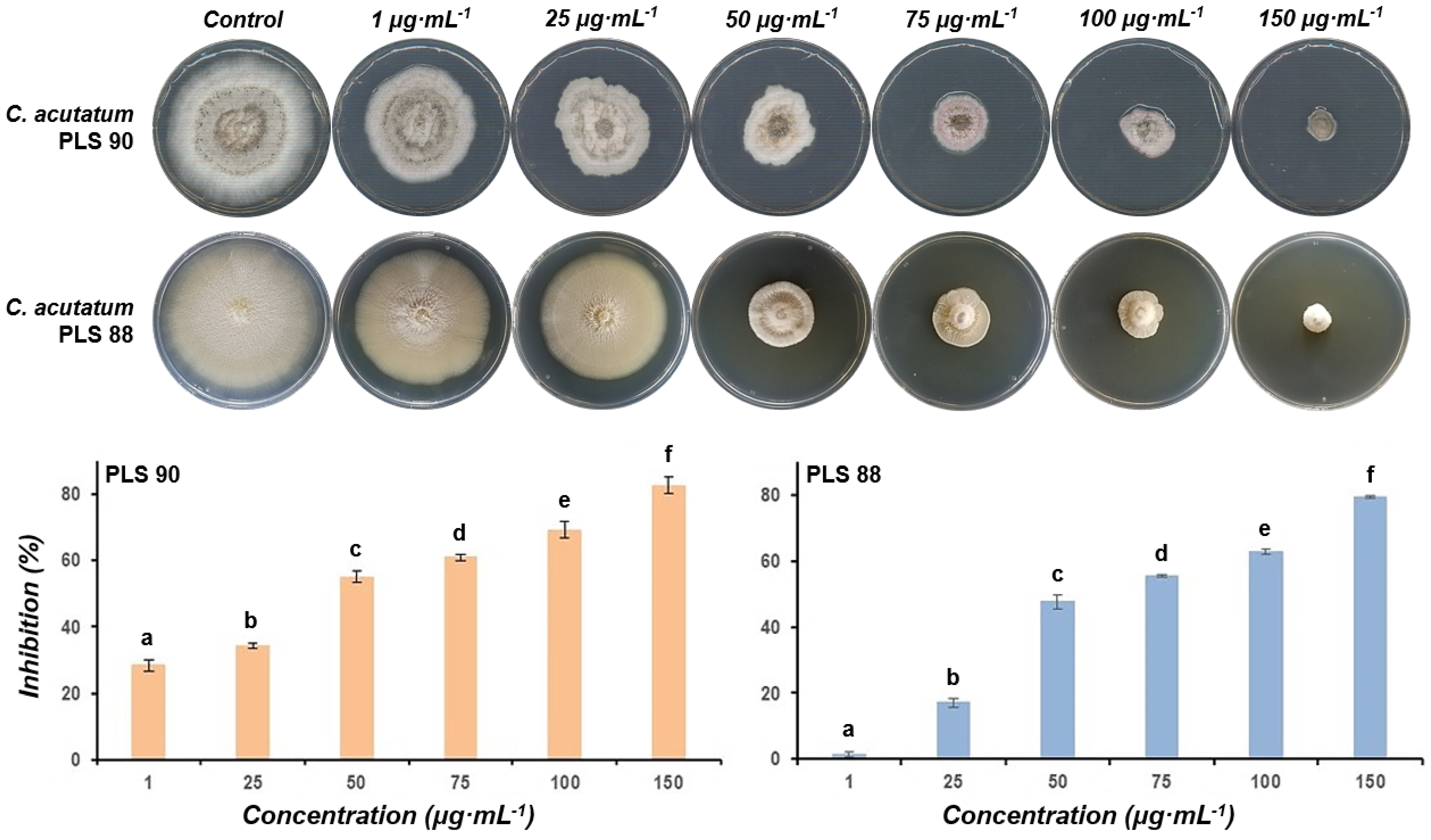
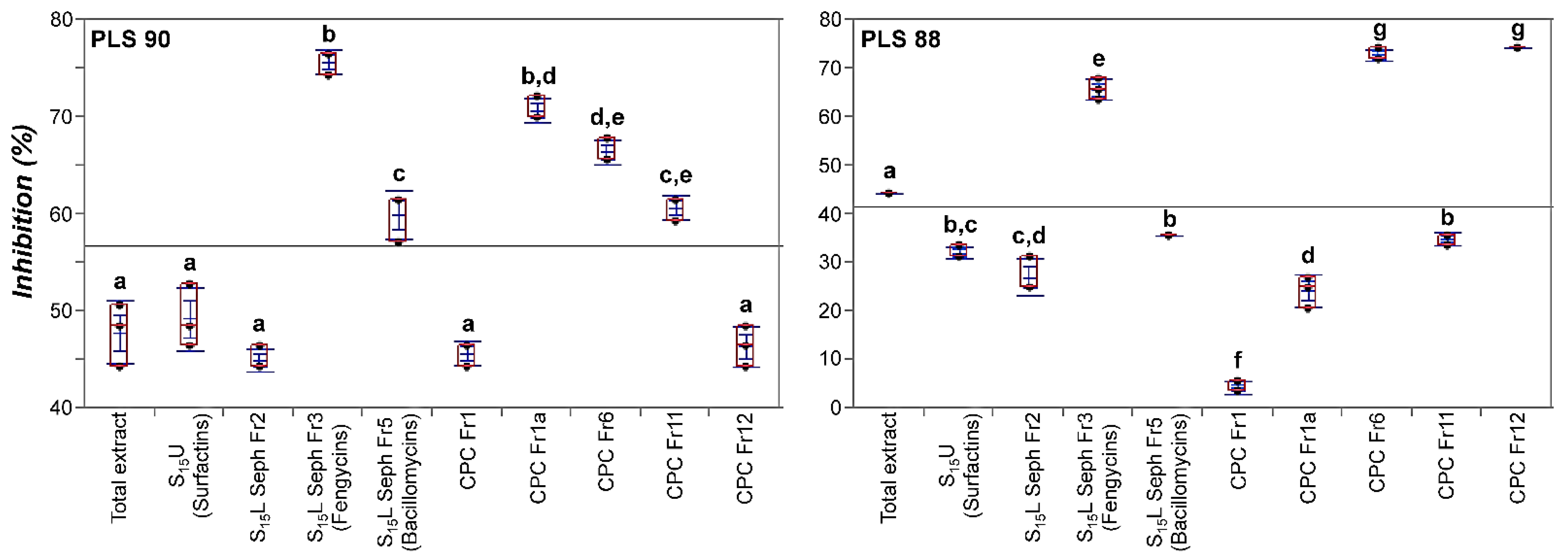
2.6. The LP Extract of Bacillus sp. PTA13 and Its Fractions Are Highly Toxic to Both the Susceptible and Resistant to Fungicides Isolates of the Colletotrichum Acutatum Species Complex
3. Materials and Methods
3.1. Plant Material and Sample Collection
3.2. Isolation and Cultivation of Olive Tree Endophytic Microorganisms (EMs)
3.3. Assessment of the Bioactivity of the Olive Tree Endophytes and That of Lipopeptide (LP) Extract/Fractions to Colletotrichum Acutatum Species Complex Isolates
3.4. Molecular Identification and Characterization of Endophytic Bacteria
3.5. Isolation of Lipopeptides (LPs) from Liquid Cultures of the Endophytic Bacillus Strain PTA13
3.6. Time-Course Study of the Bacillus sp. PTA13 Growth Rate and Its Lipopeptide-Producing Capacity
3.7. Fractionation of the Bacillus sp. PTA13 Lipopeptide (LP) Extract by Applying Liquid–Liquid Extraction and Chromatography Techniques
3.7.1. Assessment of Biphasic Solvent Systems
3.7.2. Liquid–Liquid Extraction
3.7.3. Size-Exclusion Chromatography
3.7.4. Semi-Preparative Centrifugal Partition Chromatography (CPC) Analysis
3.8. Thin-Layer Chromatography (TLC) Analysis of the Total Lipopeptide (LP) Extract and the Obtained Lipopeptide Fractions
3.9. Ultra-High-Performance Liquid Chromatography–High-Resolution MS/MS (LC-HRMS/MS) Analysis of the Total Lipopeptide (LP) Extract and the Obtained Lipopeptide Fractions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cooper, J.; Dobson, H. The benefits of pesticides to mankind and the environment. Crop Protect. 2007, 26, 1337–1348. [Google Scholar] [CrossRef]
- Umetsu, N.; Shirai, Y. Development of novel pesticides in the 21st century. J. Pestic. Sci. 2020, 45, 54–74. [Google Scholar] [CrossRef] [Green Version]
- FAO. Available online: http://www.fao.org/faostat/en/#data/RP (accessed on 8 October 2021).
- Aliferis, K.A.; Jabaji, S. Metabolomics—A robust bioanalytical approach for the discovery of the modes-of-action of pesticides: A review. Pestic. Biochem. Physiol. 2011, 100, 105–117. [Google Scholar] [CrossRef]
- Duke, S.O. Why have no new herbicide modes of action appeared in recent years? Pest Manag. Sci. 2012, 68, 505–512. [Google Scholar] [CrossRef] [Green Version]
- Nisa, H.; Kamili, A.N.; Nawchoo, I.A.; Shafi, S.; Shameem, N.; Bandh, S.A. Fungal endophytes as prolific source of phytochemicals and other bioactive natural products: A review. Microb. Pathog. 2015, 82, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Uzma, F.; Mohan, C.D.; Hashem, A.; Konappa, N.M.; Rangappa, S.; Kamath, P.V.; Singh, B.P.; Mudili, V.; Gupta, V.K.; Siddaiah, C.N. Endophytic fungi-alternative sources of cytotoxic compounds: A review. Front. Pharmacol. 2018, 9, 309. [Google Scholar] [CrossRef]
- Khare, E.; Mishra, J.; Arora, N.K. Multifaceted interactions between endophytes and plant: Developments and prospects. Front. Microbiol. 2018, 9, 2732. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Kumar, N.; Shandilya, C.; Varma, A. Unravelling the role of endophytes in micronutrient uptake and enhanced crop productivity. In Symbiotic Soil Microorganisms; Shrivastava, N., Mahajan, S., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2020; pp. 63–85. [Google Scholar]
- Lata, R.; Chowdhury, S.; Gond, S.K.; White, J.F., Jr. Induction of abiotic stress tolerance in plants by endophytic microbes. Lett. Appl. Microbiol. 2018, 66, 268–276. [Google Scholar] [CrossRef] [Green Version]
- Rho, H.; Hsieh, M.; Kandel, S.L.; Cantillo, J.; Doty, S.L.; Kim, S.-H. Do endophytes promote growth of host plants under stress? A meta-analysis on plant stress mitigation by endophytes. Microb. Ecol. 2018, 75, 407–418. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [Green Version]
- Fadiji, A.E.; Babalola, O.O. Elucidating Mechanisms of Endophytes Used in Plant Protection and Other Bioactivities with Multifunctional Prospects. Front. Bioeng. Biotechnol. 2020, 8, 467. [Google Scholar] [CrossRef] [PubMed]
- Aliferis, K.A. Metabolomics in plant protection product research and development: Discovering the mode(s)-of-action and mechanisms of toxicity. In Environmental Metabolomics; Álvarez-Muñoz, D., Farré, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 163–194. [Google Scholar]
- Bucar, F.; Wube, A.; Schmid, M. Natural product isolation—How to get from biological material to pure compounds. Nat. Prod. Rep. 2013, 30, 525–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sticher, O. Natural product isolation. Nat. Prod. Rep. 2008, 25, 517–554. [Google Scholar] [CrossRef]
- Scott, M.; Rani, M.; Samsatly, J.; Charron, J.-B.; Jabaji, S. Endophytes of industrial hemp (Cannabis sativa L.) cultivars: Identification of culturable bacteria and fungi in leaves, petioles, and seeds. Can. J. Microbiol. 2018, 64, 664–680. [Google Scholar] [CrossRef] [Green Version]
- Frank, A.C.; Saldierna Guzmán, J.P.; Shay, J.E. Transmission of bacterial endophytes. Microorganisms 2017, 5, 70. [Google Scholar] [CrossRef] [Green Version]
- Gouda, S.; Das, G.; Sen, S.K.; Shin, H.-S.; Patra, J.K. Endophytes: A treasure house of bioactive compounds of medicinal importance. Front. Microbiol. 2016, 7, 1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branda, S.S.; González-Pastor, J.E.; Ben-Yehuda, S.; Losick, R.; Kolter, R. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2001, 98, 11621–11626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef] [Green Version]
- Medeot, D.B.; Fernandez, M.; Morales, G.M.; Jofré, E. Fengycins from Bacillus amyloliquefaciens MEP218 exhibit antibacterial activity by producing alterations on the cell surface of the pathogens Xanthomonas axonopodis pv. vesicatoria and Pseudomonas aeruginosa PA01. Front. Microbiol. 2020, 10, 3107. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; De Bruijn, I.; Nybroe, O.; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062. [Google Scholar] [CrossRef] [Green Version]
- Shafi, J.; Tian, H.; Ji, M. Bacillus species as versatile weapons for plant pathogens: A review. Biotechnol. Biotechnol. Equip. 2017, 31, 446–459. [Google Scholar] [CrossRef] [Green Version]
- Cochrane, S.A.; Vederas, J.C. Lipopeptides from Bacillus and Paenibacillus spp.: A gold mine of antibiotic candidates. Med. Res. Rev. 2016, 36, 4–31. [Google Scholar] [CrossRef]
- Falardeau, J.; Wise, C.; Novitsky, L.; Avis, T.J. Ecological and mechanistic insights into the direct and indirect antimicrobial properties of Bacillus subtilis lipopeptides on plant pathogens. J. Chem. Ecol. 2013, 39, 869–878. [Google Scholar] [CrossRef]
- Patel, S.; Ahmed, S.; Eswari, J.S. Therapeutic cyclic lipopeptides mining from microbes: Latest strides and hurdles. World J. Microbiol. Biotechnol. 2015, 31, 1177–1193. [Google Scholar] [CrossRef]
- Santos, V.S.V.; Silveira, E.; Pereira, B.B. Toxicity and applications of surfactin for health and environmental biotechnology. J. Toxicol. Environ. Health Part B 2018, 21, 382–399. [Google Scholar] [CrossRef]
- Olishevska, S.; Nickzad, A.; Déziel, E. Bacillus and Paenibacillus secreted polyketides and peptides involved in controlling human and plant pathogens. Appl. Microbiol. Biotechnol. 2019, 103, 1189–1215. [Google Scholar] [CrossRef]
- Penha, R.O.; Vandenberghe, L.P.; Faulds, C.; Soccol, V.T.; Soccol, C.R. Bacillus lipopeptides as powerful pest control agents for a more sustainable and healthy agriculture: Recent studies and innovations. Planta 2020, 251, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, D.K.; Johri, B.N. Interactions of Bacillus spp. and plants–with special reference to induced systemic resistance (ISR). Microbiol. Res. 2009, 164, 493–513. [Google Scholar] [CrossRef]
- Wu, G.; Liu, Y.; Xu, Y.; Zhang, G.; Shen, Q.; Zhang, R. Exploring elicitors of the beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 to induce plant systemic resistance and their interactions with plant signaling pathways. Mol. Plant-Microbe Interact. 2018, 31, 560–567. [Google Scholar] [CrossRef] [Green Version]
- Kolainis, S.; Koletti, A.; Lykogianni, M.; Karamanou, D.; Gkizi, D.; Tjamos, S.E.; Paraskeuopoulos, A.; Aliferis, K.A. An integrated approach to improve plant protection against olive anthracnose caused by the Colletotrichum acutatum species complex. PLoS ONE 2020, 15, e0233916. [Google Scholar] [CrossRef]
- Eurostat. Available online: https://ec.europa.eu/eurostat/web/products-eurostat-news/-/DDN-20190301-1 (accessed on 8 October 2021).
- Zhao, H.; Shao, D.; Jiang, C.; Shi, J.; Li, Q.; Huang, Q.; Rajoka, M.S.R.; Yang, H.; Jin, M. Biological activity of lipopeptides from Bacillus. Appl. Microbiol. Biotechnol. 2017, 101, 5951–5960. [Google Scholar] [CrossRef]
- Arima, K.; Kakinuma, A.; Tamura, G. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: Isolation, characterization and its inhibition of fibrin clot formation. Biochem. Biophys. Res. Commun. 1968, 31, 488–494. [Google Scholar] [CrossRef]
- Dimkić, I.; Stanković, S.; Nišavić, M.; Petković, M.; Ristivojević, P.; Fira, D.; Berić, T. The profile and antimicrobial activity of Bacillus lipopeptide extracts of five potential biocontrol strains. Front. Microbiol. 2017, 8, 925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joyce, S.A.; Lango, L.; Clarke, D.J. The regulation of secondary metabolism and mutualism in the insect pathogenic bacterium Photorhabdus luminescens. Adv. Appl. Microbiol. 2011, 76, 1–25. [Google Scholar]
- Chen, B.; Wen, J.; Zhao, X.; Ding, J.; Qi, G. Surfactin: A Quorum-Sensing Signal Molecule to Relieve CCR in Bacillus amyloliquefaciens. Front. Microbiol. 2020, 11, 631. [Google Scholar] [CrossRef]
- Aleti, G.; Lehner, S.; Bacher, M.; Compant, S.; Nikolic, B.; Plesko, M.; Schuhmacher, R.; Sessitsch, A.; Brader, G. Surfactin variants mediate species-specific biofilm formation and root colonization in Bacillus. Environ. Microbiol. 2016, 18, 2634–2645. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Mandic-Mulec, I.; Zhang, H.; Liu, Y.; Sun, X.; Feng, H.; Xun, W.; Zhang, N.; Shen, Q.; Zhang, R. Antibiotic Bacillomycin D Affects Iron Acquisition and Biofilm Formation in Bacillus velezensis through a Btr-Mediated FeuABC-Dependent Pathway. Cell Rep. 2019, 29, 1192–1202.e1195. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.-Z.; Zheng, Q.-W.; Wei, T.; Zhang, Z.-Q.; Zhao, C.-F.; Zhong, H.; Xu, Q.-Y.; Lin, J.-F.; Guo, L.-Q. Isolation and Characterization of Fengycins Produced by Bacillus amyloliquefaciens JFL21 and Its Broad-Spectrum Antimicrobial Potential Against Multidrug-Resistant Foodborne Pathogens. Front. Microbiol. 2020, 11, 3319. [Google Scholar] [CrossRef] [PubMed]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.-S.; Villaruz, A.E.; Glose, K.A.; Fisher, E.L.; Hunt, R.L.; Li, B. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 2018, 562, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Hunt, B.J.; Holding, S.R. Size Exclusion Chromatography; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Angelis, A.; Hamzaoui, M.; Aligiannis, N.; Nikou, T.; Michailidis, D.; Gerolimatos, P.; Termentzi, A.; Hubert, J.; Halabalaki, M.; Renault, J.-H. An integrated process for the recovery of high added-value compounds from olive oil using solid support free liquid-liquid extraction and chromatography techniques. J. Chromatogr. A 2017, 1491, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Tareq, F.S.; Lee, M.A.; Lee, H.-S.; Lee, Y.-J.; Lee, J.S.; Hasan, C.M.; Islam, M.T.; Shin, H.J. Gageotetrins A-C, noncytotoxic antimicrobial linear lipopeptides from a marine bacterium Bacillus subtilis. Org. Lett. 2014, 16, 928–931. [Google Scholar] [CrossRef]
- Tareq, F.S.; Shin, H.J. Bacilotetrins A and B, anti-staphylococcal cyclic-lipotetrapeptides from a marine-derived Bacillus subtilis. J. Nat. Prod. 2017, 80, 2889–2892. [Google Scholar] [CrossRef]
- Aliferis, K.A.; Chrysayi-Tokousbalides, M. Metabolomics in pesticide research and development: Review and future perspectives. Metabolomics 2011, 7, 35–53. [Google Scholar] [CrossRef]
- Hamley, I.W. Lipopeptides: From self-assembly to bioactivity. Chem. Commun. 2015, 51, 8574–8583. [Google Scholar] [CrossRef] [Green Version]
- Kaspar, F.; Neubauer, P.; Gimpel, M. Bioactive secondary metabolites from Bacillus subtilis: A comprehensive review. J. Nat. Prod. 2019, 82, 2038–2053. [Google Scholar] [CrossRef] [PubMed]
- Sen, R. Surfactin: Biosynthesis, genetics and potential applications. In Biosurfactants; Springer: Berlin/Heidelberg, Germany, 2010; pp. 316–323. [Google Scholar]
- Deleu, M.; Paquot, M.; Nylander, T. Effect of fengycin, a lipopeptide produced by Bacillus subtilis, on model biomembranes. Biophys. J. 2008, 94, 2667–2679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deleu, M.; Paquot, M.; Nylander, T. Fengycin interaction with lipid monolayers at the air-aqueous interface-implications for the effect of fengycin on biological membranes. J. Colloid Interface Sci. 2005, 283, 358–365. [Google Scholar] [CrossRef]
- Zakharova, A.A.; Efimova, S.S.; Malev, V.V.; Ostroumova, O.S. Fengycin induces ion channels in lipid bilayers mimicking target fungal cell membranes. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moyne, A.L.; Shelby, R.; Cleveland, T.; Tuzun, S. Bacillomycin D: An iturin with antifungal activity against Aspergillus flavus. J. Appl. Microbiol. 2001, 90, 622–629. [Google Scholar] [CrossRef]
- Chakraborty, M.; Mahmud, N.U.; Gupta, D.R.; Tareq, F.S.; Shin, H.J.; Islam, T. Inhibitory effects of linear lipopeptides from a marine Bacillus subtilis on the wheat blast fungus Magnaporthe oryzae Triticum. Front. Microbiol. 2020, 11, 665. [Google Scholar] [CrossRef]
- Schulz, B.; Wanke, U.; Draeger, S.; Aust, H.-J. Endophytes from herbaceous plants and shrubs: Effectiveness of surface sterilization methods. Mycol. Res. 1993, 97, 1447–1450. [Google Scholar] [CrossRef]
- Golinska, P.; Wypij, M.; Agarkar, G.; Rathod, D.; Dahm, H.; Rai, M. Endophytic actinobacteria of medicinal plants: Diversity and bioactivity. Antonie Leeuwenhoek 2015, 108, 267–289. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Pi, H.; Chandrangsu, P.; Li, Y.; Wang, Y.; Zhou, H.; Xiong, H.; Helmann, J.D.; Cai, Y. Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Kim, H.-S.; Yoon, B.-D.; Lee, C.-H.; Suh, H.-H.; Oh, H.-M.; Katsuragi, T.; Tani, Y. Production and properties of a lipopeptide biosurfactant from Bacillus subtilis C9. J. Ferment. Bioeng. 1997, 84, 41–46. [Google Scholar] [CrossRef]
- Kim, P.; Bai, H.; Bai, D.; Chae, H.; Chung, S.; Kim, Y.; Park, R.; Chi, Y.T. Purification and characterization of a lipopeptide produced by Bacillus thuringiensis CMB26. J. Appl. Microbiol. 2004, 97, 942–949. [Google Scholar] [CrossRef]
- Michailidis, D.; Angelis, A.; Aligiannis, N.; Mitakou, S.; Skaltsounis, L. Recovery of sesamin, sesamolin, and minor lignans from sesame oil using solid support-free liquid–liquid extraction and chromatography techniques and evaluation of their enzymatic inhibition properties. Front. Pharmacol. 2019, 10, 723. [Google Scholar] [CrossRef]
- Friedman, M. Applications of the ninhydrin reaction for analysis of amino acids, peptides, and proteins to agricultural and biomedical sciences. J. Agric. Food Chem. 2004, 52, 385–406. [Google Scholar] [CrossRef] [PubMed]

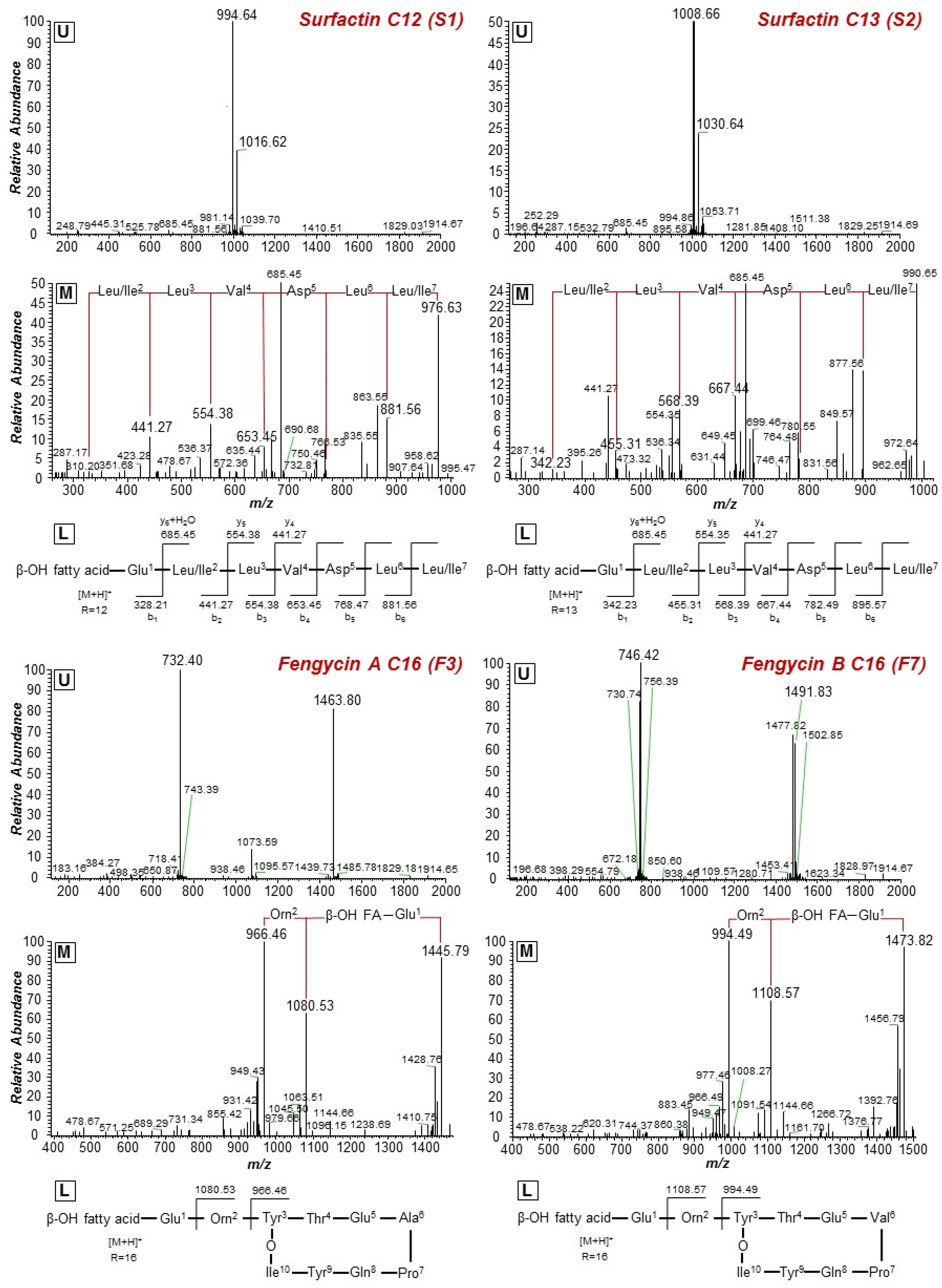
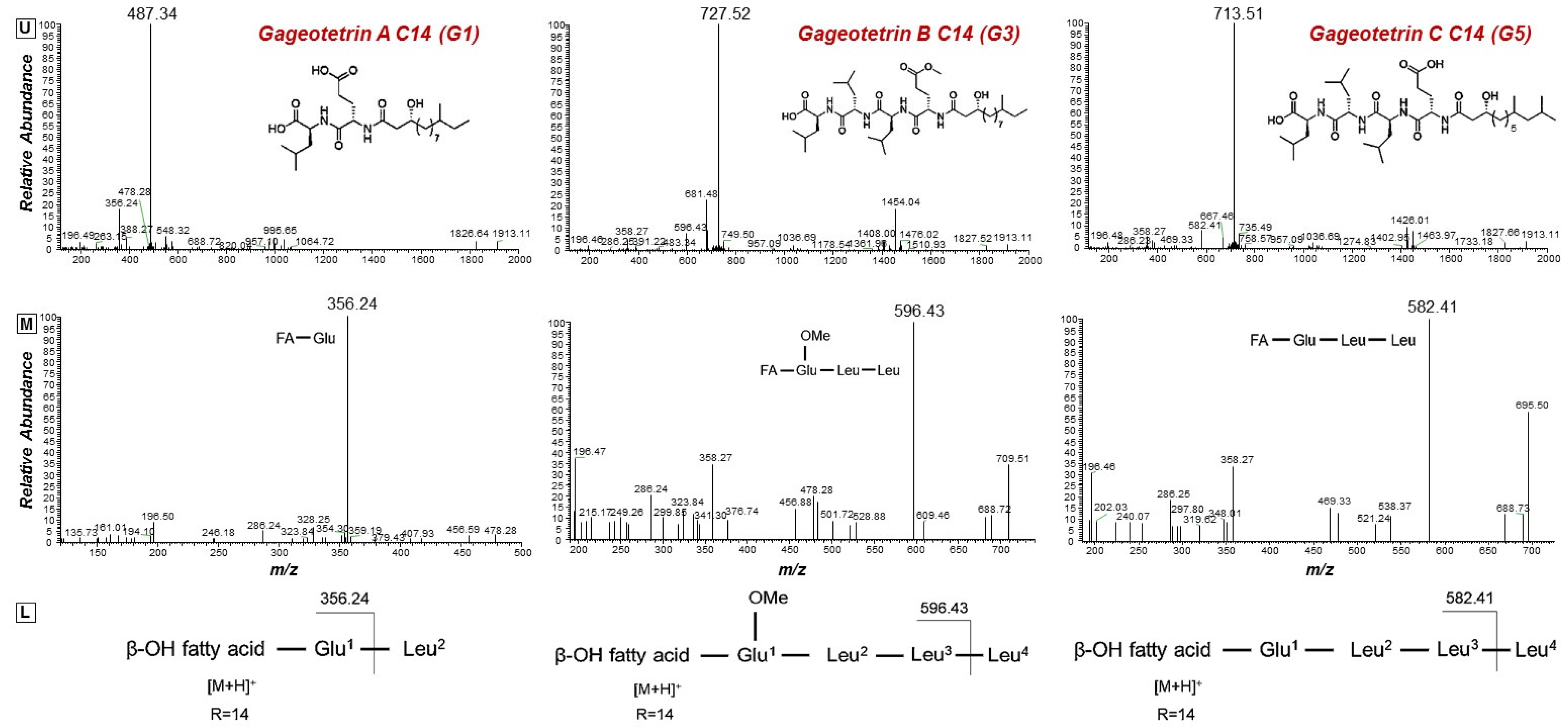

| Peak | Mass (Da) | Rt (min) | Assignment | Molecular Formula | Sequence |
|---|---|---|---|---|---|
| Surfactins | |||||
| S1 | 994.6423 | 16.18 | C12[M+H]+ | C50H87N7O13 | 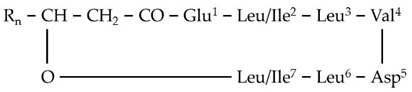 |
| 1016.6233 | C12[M+Na]+ | ||||
| S2 | 1008.6575 | 16.63 | C13[M+H]+ | C51H89N7O13 | |
| 1030.6389 | C13[M+Na]+ | ||||
| S3 | 1022.6728 | 17.32 | C14[M+H]+ | C52H91N7O13 | |
| 1044.6539 | C14[M+Na]+ | ||||
| S4 | 1036.6890 | 17.67 | C15[M+H]+ | C53H93N7O13 | |
| 1058.6700 | C15[M+Na]+ | ||||
| Bacillomycins D | |||||
| B1 | 1003.5117 | 9.23 | C12[M+H]+ | C46H70N10O15 | 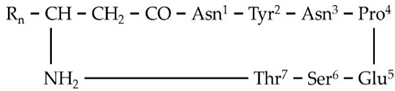 |
| 1025.4904 | C12[M+Na]+ | ||||
| B2 | 1017.5267 | 9.69 | C13[M+H]+ | C47H72N10O15 | |
| 1039.5086 | C13[M+Na]+ | ||||
| B3 | 1031.5400 | 10.40 | C14[M+H]+ | C48H74N10O15 | |
| 1053.5211 | C14[M+Na]+ | ||||
| B4 | 1045.5540 | 10.89 | C15[M+H]+ | C49H76N10O15 | |
| 1067.5359 | C15[M+Na]+ | ||||
| B5 | 1059.5710 | 11.63 | C16[M+H]+ | C50H78N10O15 | |
| 1081.5527 | C16[M+Na]+ | ||||
| B6 | 1073.5866 | 12.05 | C17[M+H]+ | C51H80N10O15 | |
| 1095.5682 | C17[M+Na]+ | ||||
| Fengycins | |||||
| Fengycins A | |||||
| F1 | 1435.7688 | 11.20 | C14[M+H]+ | C70H106N12O20 | 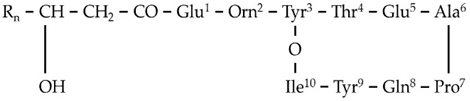 |
| 718.3880 | C14[M+H]2+ | ||||
| F2 | 1449.7848 | 11.54 | C15[M+H]+ | C71H108N12O20 | |
| 725.3958 | C15[M+H]2+ | ||||
| F3 | 1463.8005 | 12.02 | C16[M+H]+ | C72H110N12O20 | |
| 732.4037 | C16[M+H]2+ | ||||
| F4 | 1477.8176 | 12.27 | C17[M+H]+ | C73H112N12O20 | |
| 739.4125 | C17[M+H]2+ | ||||
| Fengycins B | |||||
| F5 | 1463.8009 | 11.54 | C14[M+H]+ | C72H110N12O20 | 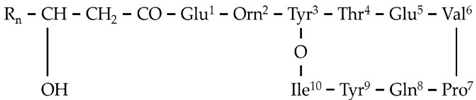 |
| 732.4033 | C14[M+H]2+ | ||||
| F6 | 1477.8173 | 11.84 | C15[M+H]+ | C73H112N12O20 | |
| 739.4122 | C15[M+H]2+ | ||||
| F7 | 1491.8318 | 12.32 | C16[M+H]+ | C74H114N12O20 | |
| 746.4196 | C16[M+H]2+ | ||||
| F8 | 1505.8480 | 12.64 | C17[M+H]+ | C75H116N12O20 | |
| 753.4275 | C17[M+H]2+ | ||||
| Gageotetrins | |||||
| Gageotetrins A | |||||
| G1 | 487.3378 | 13.95 | C14[M+H]+ | C25H46N2O7 | 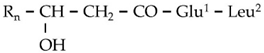 |
| G2 | 501.3536 | 14.49 | C15[M+H]+ | C26H48N2O7 | |
| Gageotetrins B | |||||
| G3 | 727.5231 | 16.14 | C14[M+H]+ | C38H70N4O9 | 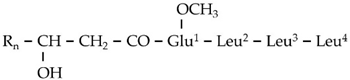 |
| Gageotetrins C | |||||
| G4 | 699.4916 | 14.69 | C13[M+H]+ | C36H66N4O9 | 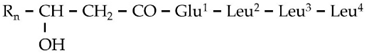 |
| G5 | 713.5078 | 15.61 | C14[M+H]+ | C37H68N4O9 | |
| Bacilotetrins |  | ||||
| Bacilotetrin A | |||||
| Bt1 | 695.4964 | 17.01 | C37H66N4O8 | ||
| Bacilotetrin B | |||||
| Bt2 | 709.5120 | 17.43 | C38H68N4O8 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadopoulou, E.-A.; Angelis, A.; Antoniadi, L.; Aliferis, K.A.; Skaltsounis, A.-L. Discovering the Next-Generation Plant Protection Products: A Proof-of-Concept via the Isolation and Bioactivity Assessment of the Olive Tree Endophyte Bacillus sp. PTA13 Lipopeptides. Metabolites 2021, 11, 833. https://doi.org/10.3390/metabo11120833
Papadopoulou E-A, Angelis A, Antoniadi L, Aliferis KA, Skaltsounis A-L. Discovering the Next-Generation Plant Protection Products: A Proof-of-Concept via the Isolation and Bioactivity Assessment of the Olive Tree Endophyte Bacillus sp. PTA13 Lipopeptides. Metabolites. 2021; 11(12):833. https://doi.org/10.3390/metabo11120833
Chicago/Turabian StylePapadopoulou, Evgenia-Anna, Apostolis Angelis, Lemonia Antoniadi, Konstantinos A. Aliferis, and Alexios-Leandros Skaltsounis. 2021. "Discovering the Next-Generation Plant Protection Products: A Proof-of-Concept via the Isolation and Bioactivity Assessment of the Olive Tree Endophyte Bacillus sp. PTA13 Lipopeptides" Metabolites 11, no. 12: 833. https://doi.org/10.3390/metabo11120833







