Metabolomic Profiling Reveals Sex Specific Associations with Chronic Obstructive Pulmonary Disease and Emphysema
Abstract
1. Introduction
2. Results
2.1. Demographics
2.2. WGCNA Modules
2.3. Covariate Adjusted Module-Phenotype Associations in COPDGene
2.3.1. Sex
2.3.2. COPD
2.3.3. Percent Emphysema
2.3.4. Covariates
2.4. Individual Associations
2.4.1. COPD Modules
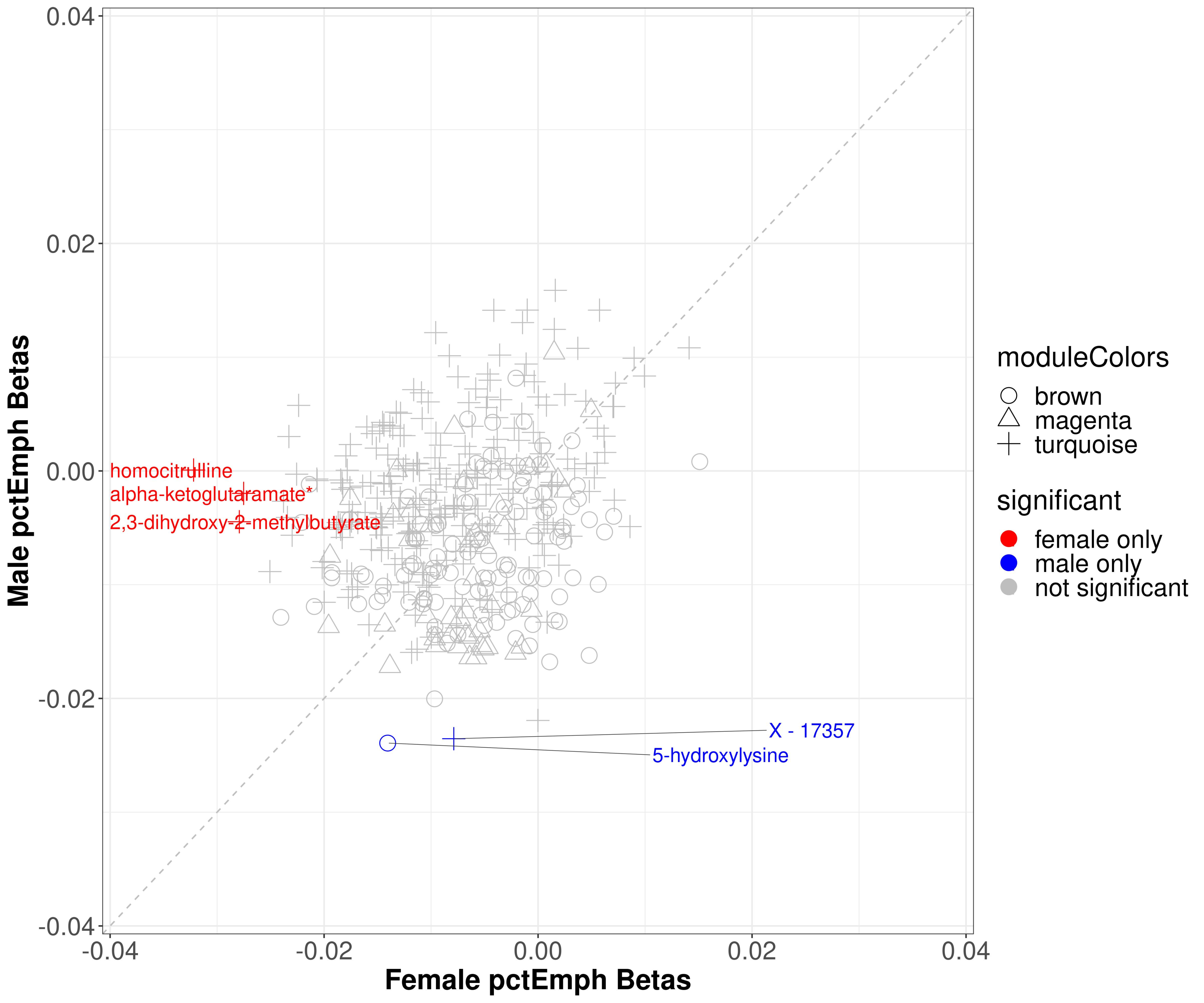
2.4.2. Percent Emphysema
3. Discussion
4. Methods
4.1. Study Populations
4.2. Clinical Data and Definitions
4.3. Statistical Analysis
4.3.1. Data Sets and Availability
4.3.2. Software
4.3.3. Individual Associations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Trabado, S.; Al-Salameh, A.; Croixmarie, V.; Masson, P.; Corruble, E.; Fève, B.; Colle, R.; Ripoll, L.; Walther, B.; Boursier-Neyret, C.; et al. The human plasma-metabolome: Reference values in 800 French healthy volunteers; impact of cholesterol, gender and age. PLoS ONE 2017, 12, e0173615. [Google Scholar] [CrossRef]
- Han, M.K. Chronic Obstructive Pulmonary Disease in Women: A Biologically Focused Review with a Systematic Search Strategy. Int. J. Chronic Obstr. Pulm. Dis. 2020, 15, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Terzikhan, N.; Verhamme, K.M.C.; Hofman, A.; Stricker, B.H.; Brusselle, G.G.; la Housse, L. Prevalence and incidence of COPD in smokers and non-smokers: The Rotterdam Study. Eur. J. Epidemiol. 2016, 31, 785–792. [Google Scholar] [CrossRef]
- Heron, M. Deaths: Leading Causes for 2017. Natl. Vital. Stat. Rep. 2019, 68, 1–77. [Google Scholar] [PubMed]
- Barnes, P.J. Sex Differences in Chronic Obstructive Pulmonary Disease Mechanisms. Am. J. Respir. Crit. Care Med. 2016, 193, 813–814. [Google Scholar] [CrossRef] [PubMed]
- Silverman, E.K.; Weiss, S.T.; Drazen, J.M.; Chapman, H.A.; Carey, V.; Campbell, E.J.; Denish, P.; Silverman, R.A.; Celedon, J.C.; Reilly, J.J.; et al. Gender-Related Differences in Severe, Early-Onset Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2000, 162, 2152–2158. [Google Scholar] [CrossRef] [PubMed]
- Foreman, M.G.; Zhang, L.; Murphy, J.; Hansel, N.N.; Make, B.; Hokanson, J.E.; Washko, G.; Regan, E.A.; Crapo, J.D.; Silverman, E.K.; et al. Early-Onset Chronic Obstructive Pulmonary Disease Is Associated with Female Sex, Maternal Factors, and African American Race in the COPDGene Study. Am. J. Respir. Crit. Care Med. 2011, 184, 414–420. [Google Scholar] [CrossRef]
- Martinez, F.J.; Curtis, J.L.; Sciurba, F.; Mumford, J.; Giardino, N.D.; Weinmann, G.; Kazerooni, E.; Murray, S.; Criner, G.J.; Sin, D.D.; et al. Sex Differences in Severe Pulmonary Emphysema. Am. J. Respir. Crit. Care Med. 2007, 176, 243–252. [Google Scholar] [CrossRef]
- Hardin, M.; Foreman, M.; Dransfield, M.T.; Hansel, N.; Han, M.K.; Cho, M.H.; Bhatt, S.P.; Ramsdell, J.; Lynch, D.; Curtis, J.L.; et al. Sex-specific features of emphysema among current and former smokers with COPD. Eur. Respir. J. 2016, 47, 104–112. [Google Scholar] [CrossRef]
- Trigueros, J.A.; Riesco, J.A.; Alcázar-Navarrete, B.; Campuzano, A.; Pérez, J. Clinical Features of Women With COPD: Sex Differences in A Cross-Sectional Study in Spain (“The ESPIRAL-ES Study”). Int. J. Chronic Obstr. Pulm. Disease. 2019, 14, 2469–2478. Available online: https://www.dovepress.com/clinical-features-of-women-with-copd-sex-differences-in-a-cross-sectio-peer-reviewed-fulltext-article-COPD (accessed on 16 July 2020). [CrossRef]
- Mittelstrass, K.; Ried, J.S.; Yu, Z.; Krumsiek, J.; Gieger, C.; Prehn, C.; Roemisch-Margl, W.; Polonikov, A.; Peters, A.; Theis, F.J.; et al. Discovery of Sexual Dimorphisms in Metabolic and Genetic Biomarkers. PLoS Genet. 2011, 7, e1002215. [Google Scholar] [CrossRef] [PubMed]
- Darst, B.F.; Koscik, R.L.; Hogan, K.J.; Johnson, S.C.; Engelman, C.D. Longitudinal plasma metabolomics of aging and sex. Aging 2019, 11, 1262–1282. [Google Scholar] [CrossRef]
- Krumsiek, J.; Mittelstrass, K.; Do, K.T.; Stückler, F.; Ried, J.S.; Adamski, J.; Peters, A.; Illig, T.; Kronenberg, F.; Friedrich, N.; et al. Gender-specific pathway differences in the human serum metabolome. Metabolomics 2015, 11, 1815–1833. [Google Scholar] [CrossRef]
- Bowler, R.P.; Jacobson, S.; Cruickshank, C.; Hughes, G.J.; Siska, C.; Ory, D.S.; Petrache, I.; Schaffer, J.E.; Reisdorph, N.; Kechris, K. Plasma Sphingolipids Associated with Chronic Obstructive Pulmonary Disease Phenotypes. Am. J. Respir. Crit. Care Med. 2015, 191, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Koike, K.; Berdyshev, E.V.; Bowler, R.P.; Scruggs, A.K.; Cao, D.; Schweitzer, K.S.; Serban, K.A.; Petrache, I. Bioactive Sphingolipids in the Pathogenesis of Chronic Obstructive Pulmonary Disease. Ann. Am. Thorac. Soc. 2018, 15, S249–S252. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Plata, V.; Casanova, C.; Divo, M.; Tesfaigzi, Y.; Calhoun, V.; Sui, J.; Polverino, F.; Priolo, C.; Petersen, H.; de Torres, J.P.; et al. Plasma metabolomics and clinical predictors of survival differences in COPD patients. Respir. Res. 2019, 20, 219. [Google Scholar] [CrossRef]
- Wang, L.; Tang, Y.; Liu, S.; Mao, S.; Ling, Y.; Liu, D.; He, X.; Wang, X. Metabonomic Profiling of Serum and Urine by 1H NMR-Based Spectroscopy Discriminates Patients with Chronic Obstructive Pulmonary Disease and Healthy Individuals. PLoS ONE 2013, 8, e65675. [Google Scholar] [CrossRef]
- Deidda, M.; Piras, C.; Bassareo, P.P.; Dessalvi, C.C.; Mercuro, G. Metabolomics, a promising approach to translational research in cardiology. IJC Metab. Endocr. 2015, 9, 31–38. [Google Scholar] [CrossRef]
- Ząbek, A.; Stanimirova, I.; Deja, S.; Barg, W.; Kowal, A.; Korzeniewska, A.; Orczyk-Pawiłowicz, M.; Baranowski, D.; Gdaniec, Z.; Jankowska, R.; et al. Fusion of the 1H NMR data of serum, urine and exhaled breath condensate in order to discriminate chronic obstructive pulmonary disease and obstructive sleep apnea syndrome. Metabolomics 2015, 11, 1563–1574. [Google Scholar] [CrossRef]
- Vignoli, A.; Santini, G.; Tenori, L.; Macis, G.; Mores, N.; Macagno, F.; Pagano, F.; Higenbottam, T.; Luchinat, C.; Montuschi, P. NMR-Based Metabolomics for the Assessment of Inhaled Pharmacotherapy in Chronic Obstructive Pulmonary Disease Patients. J. Proteome Res. 2020, 19, 64–74. [Google Scholar] [CrossRef]
- Halper-Stromberg, E.; Gillenwater, L.; Cruickshank-Quinn, C.; O’Neal, W.K.; Reisdorph, N.; Petrache, I.; Zhuang, Y.; Labaki, W.W.; Curtis, J.L.; Wells, J.; et al. Bronchoalveolar Lavage Fluid from COPD Patients Reveals More Compounds Associated with Disease than Matched Plasma. Metabolism 2019, 9, 157. [Google Scholar] [CrossRef] [PubMed]
- Agusti, À.; Soriano, J.B. COPD as a Systemic Disease. COPD: J. Chronic Obstr. Pulm. Dis. 2008, 5, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Austin, V.; Crack, P.J.; Bozinovski, S.; Miller, A.A.; Vlahos, R. COPD and stroke: Are systemic inflammation and oxidative stress the missing links? Clin. Sci. 2016, 130, 1039–1050. [Google Scholar] [CrossRef]
- Agarwal, A.R.; Kadam, S.; Brahme, A.; Agrawal, M.; Apte, K.; Narke, G.; Kekan, K.; Madas, S.; Salvi, S. Systemic Immuno-metabolic alterations in chronic obstructive pulmonary disease (COPD). Respir. Res. 2019, 20, 171. [Google Scholar] [CrossRef]
- Horvath, S.; Dong, J. Geometric Interpretation of Gene Coexpression Network Analysis. PLoS Comput. Biol. 2008, 4, e1000117. [Google Scholar] [CrossRef]
- Dileo, M.V.; Strahan, G.D.; Bakker, M.D.; Hoekenga, O.A. Weighted Correlation Network Analysis (WGCNA) Applied to the Tomato Fruit Metabolome. PLoS ONE 2011, 6, e26683. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Hu, T.; Likhodii, S.; Sun, G.; Zhai, G.; Fan, Z.; Xuan, C.; Zhang, W. Differential metabolomics analysis allows characterization of diversity of metabolite networks between males and females. PLoS ONE 2018, 13, e0207775. [Google Scholar] [CrossRef] [PubMed]
- Nikkilä, J.; Sysi-Aho, M.; Ermolov, A.; Seppänen-Laakso, T.; Simell, O.; Kaski, S.; Orešič, M. Gender-dependent progression of systemic metabolic states in early childhood. Mol. Syst. Biol. 2008, 4, 197. [Google Scholar] [CrossRef]
- Goodman-Gruen, D.; Barrett-Connor, E. Sex differences in the assciation of endogenous sex hormone levels and glucose tolerance status in older men and women. Diabetes Care 2000, 23, 912–918. [Google Scholar] [CrossRef]
- Zhao, J.; Leung, J.Y.Y.; Lin, S.L.; Schooling, C.M. Cigarette smoking and testosterone in men and women: A systematic review and meta-analysis of observational studies. Prev. Med. 2016, 85, 1–10. [Google Scholar] [CrossRef]
- Key, T.J.; Pike, M.C.; Baron, J.A.; Moore, J.W.; Wang, D.Y.; Thomas, B.S.; Bulbrook, R.D. Cigarette smoking and steroid hormones in women. J. Steroid Biochem. Mol. Biol. 1991, 39, 529–534. [Google Scholar] [CrossRef]
- Dušková, M.; Šimůnková, K.; Hill, M.; Velíková, M.; Kubátová, J.; Kancheva, L.; Kazihnitková, H.; Hruškovičová, H.; Pospíšilová, H.; Rácz, B.; et al. Chronic Cigarette Smoking Alters Circulating Sex Hormones and Neuroactive Steroids in Premenopausal Women. Physiol. Res. 2012, 61, 97–111. [Google Scholar] [CrossRef]
- Liu, G.; Lee, D.P.; Schmidt, E.; Prasad, G.L. Pathway Analysis of Global Metabolomic Profiles Identified Enrichment of Caffeine, Energy, and Arginine Metabolism in Smokers but Not Moist Snuff Consumers. Bioinform. Biol. Insights 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Derkach, A.; Freedman, N.D.; Landi, M.T.; Albanes, D.; Weinstein, S.J.; Mondul, A.M.; Matthews, C.E.; Guertin, K.A.; Xiao, Q.; et al. Cigarette smoking behaviour and blood metabolomics. Int. J. Epidemiol. 2015, 45, 1421–1432. [Google Scholar] [CrossRef] [PubMed]
- Kukharenko, A.; Brito, A.; Kozhevnikova, M.V.; Moskaleva, N.; Markin, P.A.; Bochkareva, N.; Korobkova, E.O.; Belenkov, Y.N.; Privalova, E.V.; Larcova, E.V.; et al. Relationship between the plasma acylcarnitine profile and cardiometabolic risk factors in adults diagnosed with cardiovascular diseases. Clin. Chim. Acta 2020, 507, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.E.; Evans, A.M. Carnitine and acylcarnitines: Pharmacokinetic, pharmacological and clinical aspects. Clin. Pharm. 2012, 51, 553–572. [Google Scholar] [CrossRef]
- Naz, S.; Kolmert, J.; Yang, M.; Reinke, S.N.; Kamleh, M.A.; Snowden, S.; Heyder, T.; Levänen, B.; Erle, D.J.; Sköld, C.M.; et al. Metabolomics analysis identifies sex-associated metabotypes of oxidative stress and the autotaxin–lysoPA axis in COPD. Eur. Respir. J. 2017, 49, 1602322. [Google Scholar] [CrossRef]
- Lommatzsch, M.; Cicko, S.; Müller, T.; Lucattelli, M.; Bratke, K.; Stoll, P.; Grimm, M.; Dürk, T.; Zissel, G.; Ferrari, D.; et al. Extracellular Adenosine Triphosphate and Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2010, 181, 928–934. [Google Scholar] [CrossRef]
- Balgoma, D.; Yang, M.; Sjödin, M.; Snowden, S.; Karimi, R.; Levänen, B.; Merikallio, H.; Kaarteenaho, R.; Palmberg, L.; Larsson, K.; et al. Linoleic acid-derived lipid mediators increase in a female-dominated subphenotype of COPD. Eur. Respir. J. 2016, 47, 1645–1656. [Google Scholar] [CrossRef] [PubMed]
- Kohler, M.; Sandberg, A.; Kjellqvist, S.; Thomas, A.; Karimi, R.; Nyrén, S.; Eklund, A.; Thevis, M.; Sköld, C.M.; Wheelock, Å.M. Gender differences in the bronchoalveolar lavage cell proteome of patients with chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2013, 131, 743–751.e9. [Google Scholar] [CrossRef]
- Tam, A.; Churg, A.; Wright, J.L.; Zhou, S.; Kirby, M.; Coxson, H.O.; Lam, S.; Man, S.F.P.; Sin, D.D. Sex Differences in Airway Remodeling in a Mouse Model of Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2016, 193, 825–834. [Google Scholar] [CrossRef]
- Zhang, W.Z.; Rice, M.C.; Hoffman, K.L.; Oromendia, C.; Barjaktarevic, I.Z.; Wells, J.M.; Hastie, T.A.; Labaki, W.W.; Cooper, C.B.; Comellas, A.P.; et al. Association of urine mitochondrial DNA with clinical measures of COPD in the SPIROMICS cohort. Am. Soc. Clin. Investig. 2020, 5. [Google Scholar] [CrossRef]
- Berge, M.; van den Brandsma, C.-A.; Faiz, A.; Vries, M.; de Rathnayake, S.N.H.; Parém, P.D. Differential lung tissue gene expression in males and females: Implications for the susceptibility to develop COPD. Eur. Respir. J. 2019, 154. Available online: https://erj.ersjournals.com/content/54/1/1702567 (accessed on 22 February 2021).
- Agudelo, C.W.; Kumley, B.K.; Area-Gomez, E.; Xu, Y.; Dabo, A.J.; Geraghty, P.; Campos, M.; Foronjy, R.; Garcia-Arcos, I. Decreased surfactant lipids correlate with lung function in chronic obstructive pulmonary disease (COPD). PLoS ONE 2020, 15, e0228279. [Google Scholar] [CrossRef]
- Calzada, E.; Onguka, O.; Claypool, S.M. Phosphatidylethanolamine Metabolism in Health and Disease. Int. Rev. Cell Mol. Biol. 2016, 321, 29–88. [Google Scholar] [CrossRef]
- Regan, E.A.; Hokanson, J.E.; Murphy, J.R.; Make, B.; Lynch, D.A.; Beaty, T.H. Genetic Epidemiology of COPD (COPDGene) Study Design. COPD J. Chronic Obstr. Pulm. Dis. 2011, 7, 32–43. [Google Scholar] [CrossRef]
- Pauwels, R.A.; Buist, A.S.; Calverley, P.M.A.; Jenkins, C.R.; Hurd, S.S. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2001, 163, 1256–1276. [Google Scholar] [CrossRef] [PubMed]
- Wan, E.S.; Castaldi, P.J.; Cho, M.H.; Hokanson, J.E.; Regan, E.A.; Make, B.J.; Beaty, T.H.; Han, M.K.; Curtis, J.L.; Curran-Everett, D.; et al. Epidemiology, genetics, and subtyping of preserved ratio impaired spirometry (PRISm) in COPDGene. Respir. Res. 2014, 15, 1–13. [Google Scholar] [CrossRef]
- Hansel, N.N.; Paulin, L.M.; Gassett, A.J.; Peng, R.D.; Alexis, N.; Fan, V.S.; Bleecker, E.; Bowler, R.; Comellas, A.P.; Dransfield, M.; et al. Design of the Subpopulations and Intermediate Outcome Measures in COPD (SPIROMICS) AIR Study. BMJ Open Respir. Res. 2017, 4, e000186. [Google Scholar] [CrossRef]
- Vestbo, J.; Hurd, S.S.; Agustí, A.G.; Jones, P.W.; Vogelmeier, C.; Anzueto, A. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 2013, 187, 347–365. [Google Scholar] [CrossRef]
- Pompe, E.; Strand, M.; van Rikxoort, E.M.; Hoffman, E.A.; Barr, R.G.; Charbonnier, J.P.; Humphries, S.; Han, M.K.; Hokanson, J.E.; Make, B.J.; et al. Five-year Progression of Emphysema and Air Trapping at CT in Smokers with and Those without Chronic Obstructive Pulmonary Disease: Results from the COPDGene Study. Radiology 2020, 295, 218–226. [Google Scholar] [CrossRef]
- Lynch, D.A.; Moore, C.M.; Wilson, C.; Nevrekar, D.; Jennermann, T.; Humphries, S.M.; Austin, J.H.M.; Grenier, P.A.; Kauczor, H.-U.; Han, M.K.; et al. CT-based Visual Classification of Emphysema: Association with Mortality in the COPDGene Study. Radiology 2018, 288, 859–866. [Google Scholar] [CrossRef]
- Sieren, J.P.; Newell, J.D.; Barr, R.G.; Bleecker, E.R.; Burnette, N.; Carretta, E.E.; Couper, D.; Goldin, J.; Guo, J.; Han, M.K.; et al. SPIROMICS Protocol for Multicenter Quantitative Computed Tomography to Phenotype the Lungs. Am. J. Respir. Crit. Care Med. 2016, 194, 794–806. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.P.; Nath, H.P.; Kim, Y.-I.; Ramachandran, R.; Watts, J.R.; Terry, N.L.J. Centrilobular emphysema and coronary artery calcification: Mediation analysis in the SPIROMICS cohort. Respir. Res. 2018, 19, 257. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.M.; de Haven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, Nontargeted Ultrahigh Performance Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry Platform for the Identification and Relative Quantification of the Small-Molecule Complement of Biological Systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef]
- De Haven, C.D.; Evans, A.M.; Dai, H.; A Lawton, K. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J. Chemin. 2010, 2, 9. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.J.; Kennedy, A.D.; Eckhart, A.D.; Burrage, L.C.; Wulff, J.E.; Miller, L.A.; Milburn, M.V.; Ryals, J.A.; Beaudet, A.L.; Sun, Q.; et al. Untargeted metabolomic analysis for the clinical screening of inborn errors of metabolism. J. Inherit. Metab. Dis. 2015, 38, 1029–1039. [Google Scholar] [CrossRef]
- Gillenwater, L.A.; Pratte, K.A.; Hobbs, B.D.; Cho, M.H.; Zhuang, Y.; Halper-Stromberg, E.; Cruickshank-Quinn, C.; Reisdorph, N.; Petrache, I.; Labaki, W.W.; et al. Plasma Metabolomic Signatures of Chronic Obstructive Pulmonary Disease and the Impact of Genetic Variants on Phenotype-Driven Modules. Netw. Syst. Med. 2020, 3, 159–181. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. 2018. Available online: https://www.R-project.org (accessed on 30 December 2020).
- Langfelder, P.; Zhang, B.; Horvath, S. Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinformatics 2007, 24, 719–720. [Google Scholar] [CrossRef]
- Pei, G.; Chen, L.; Zhang, W. Chapter Nine—WGCNA Application to Proteomic and Metabolomic Data Analysis. In Methods in Enzymology; Shukla, A.K., Ed.; Academic Press: Cambridge, MA, USA, 2017; Volume 585, pp. 135–158. Available online: http://www.sciencedirect.com/science/article/pii/S0076687916302890 (accessed on 3 June 2020).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Langfelder, P.; Luo, R.; Oldham, M.C.; Horvath, S. Is My Network Module Preserved and Reproducible? PLoS Comput. Biol. 2011, 7, e1001057. [Google Scholar] [CrossRef] [PubMed]
- Fuller, T.F.; Ghazalpour, A.; Aten, J.E.; Drake, T.A.; Lusis, A.J.; Horvath, S. Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm. Genome 2007, 18, 463–472. [Google Scholar] [CrossRef] [PubMed]




| COPDGene | SPIROMICS | |||||
|---|---|---|---|---|---|---|
| Variable a | Males | Females | p-Value b | Males | Females | p-Value b |
| Participants | 434 | 405 | 232 | 214 | ||
| Age | 68.5 (8.4) | 66.1 (8.8) | <0.0001 | 64 (8.0) | 63 (8.8) | 0.2244 |
| NHW (%) | 399 (91.9) | 370 (91.4) | 0.8593 | 191 (82.3) | 163 (76.2) | 0.1365 |
| BMI | 29.1 (5.6) | 28.6 (6.6) | 0.1815 | 28.6 (4.9) | 28.4 (5.6) | 0.6088 |
| Current Smokers (%) | 88 (20.3) | 111 (27.4) | 0.0190 | 90 (39.1) | 70 (32.9) | 0.2030 |
| Smoking Pack-years | 50.1 (27.1) | 39.4 (20.5) | <0.0001 | 55.6 (38.1) | 45.5 (21.3) | 0.0006 |
| COPD Cases | 224 (51.6) | 167 (41.2) | 0.0033 | 140 (61.1) | 102 (47.7) | 0.0193 |
| Percent Emphysema c | 9 (11.3) | 6.3 (10.2) | 0.0005 | 5.4 (9.1) | 5 (8.8) | 0.6870 |
| COPDGene Module | Most Preserved SPIROMICS Module | Metabolite Classes * |
|---|---|---|
| blue | turquoise | Acyl Carnitines, Fatty Acids (Dicarboxylate, Monohydroxy, Long chain, Medium chain), Endocannabinoids, Nucleotides |
| red | yellow | Ceramides, Sphingomyelins |
| turquoise | blue | Xenobiotics, Amino Acids (Tryptophan metabolism, Glutamate metabolism, Histidine metabolism, Branched Chain Amino Acids, Glycine, Serine and Threonine Metabolism, Methionine, Cysteine, SAM and Taurine Metabolism, Polyamine Metabolism, Urea cycle; Arginine and Proline Metabolism), TCA cycle metabolites |
| brown | brown | Amino Acids (Gamma-glutamyl Amino Acid, Glutamate Metabolism, Branched Chain Amino Acids, Urea cycle; Arginine and Proline Metabolism, Lysine Metabolism, Methionine, Cysteine, SAM and Taurine Metabolism, Phenylalanine Metabolism), Bile Acids, Acyl Cholines, Lysophospholipids |
| yellow | black | Xenobiotics (Benzoates, Xanthines, Nutritional) |
| green | green | Lysophospholipids, Phosphatidylcholines (PC), Phosphatidylinositols (PI), Plasmalogens |
| magenta | pink | Sterioids (Androgenic, Pregnenolone, Corticosteroids, Progestin) |
| black | purple | Diacylglycerols, Phosphatidylethanolamines (PE), Acyl Carnitines |
| greenyellow | magenta | Cofactors and Vitamins |
| purple | NA | Acetylated peptides, Xenobiotics (Benzoates), Secondary Bile Acids |
| pink | NA | Xenobiotics (Chemicals), Dipeptides, Hemoglobin and Porphyrin Metabolites |
| COPDGene Module | HubMets | Kme * | SPIROMICS Module | HubMets | Kme * |
|---|---|---|---|---|---|
| Black | 1-stearoyl-2-arachidonoyl-GPE (18:0/20:4) | 0.81 | Purple | 1-stearoyl-2-arachidonoyl-GPE (18:0/20:4) | 0.87 |
| Blue | 10-nonadecenoate (19:1n9) | 0.90 | Turquoise | 10-nonadecenoate (19:1n9) | 0.86 |
| Blue | 10-heptadecenoate (17:1n7) | 0.87 | Turquoise | 10-heptadecenoate (17:1n7) | 0.87 |
| Blue | oleate/vaccenate (18:1) | 0.88 | Turquoise | oleate/vaccenate (18:1) | 0.89 |
| Brown | gamma-glutamylleucine | 0.85 | Brown | gamma-glutamylleucine | 0.85 |
| Green | 1-stearoyl-GPE (18:0) | 0.82 | Green | 1-stearoyl-GPE (18:0) | 0.81 |
| Greenyellow | oxalate (ethanedioate) | 0.88 | Magenta | oxalate (ethanedioate) | 0.87 |
| Magenta | androstenediol (3beta,17beta) disulfate (2) | 0.92 | Pink | androstenediol (3beta,17beta) disulfate (2) | 0.888 |
| Pink | X-11442 | 0.92 | None | ||
| Pink | biliverdin | 0.76 | Turquoise | biliverdin | 0.4 |
| Purple | p-cresol sulfate | 0.89 | Blue | p-cresol sulfate | 0.4 |
| Red | sphingomyelin (d17:2/16:0, d18:2/15:0) * | 0.80 | Yellow | sphingomyelin (d17:2/16:0, d18:2/15:0) * | 0.78 |
| Red | sphingomyelin (d18:2/23:0, d18:1/23:1, d17:1/24:1) * | 0.75 | Yellow | sphingomyelin (d18:2/23:0, d18:1/23:1, d17:1/24:1) * | 0.87 |
| Turquoise | 2,3-dihydroxy-5-methylthio-4-pentenoate (DMTPA) * | 0.84 | Blue | 2,3-dihydroxy-5-methylthio-4-pentenoate (DMTPA) * | 0.85 |
| Turquoise | pseudouridine | 0.84 | Blue | pseudouridine | 0.8 |
| Yellow | 3-hydroxypyridine sulfate | 0.88 | Black | 3-hydroxypyridine sulfate | 0.76 |
| Yellow | catechol sulfate | 0.87 | Black | catechol sulfate | 0.8 |
| Yellow | trigonelline (N′-methylnicotinate) | 0.79 | Black | trigonelline (N′-methylnicotinate) | 0.81 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gillenwater, L.A.; Kechris, K.J.; Pratte, K.A.; Reisdorph, N.; Petrache, I.; Labaki, W.W.; O’Neal, W.; Krishnan, J.A.; Ortega, V.E.; DeMeo, D.L.; et al. Metabolomic Profiling Reveals Sex Specific Associations with Chronic Obstructive Pulmonary Disease and Emphysema. Metabolites 2021, 11, 161. https://doi.org/10.3390/metabo11030161
Gillenwater LA, Kechris KJ, Pratte KA, Reisdorph N, Petrache I, Labaki WW, O’Neal W, Krishnan JA, Ortega VE, DeMeo DL, et al. Metabolomic Profiling Reveals Sex Specific Associations with Chronic Obstructive Pulmonary Disease and Emphysema. Metabolites. 2021; 11(3):161. https://doi.org/10.3390/metabo11030161
Chicago/Turabian StyleGillenwater, Lucas A., Katerina J. Kechris, Katherine A. Pratte, Nichole Reisdorph, Irina Petrache, Wassim W. Labaki, Wanda O’Neal, Jerry A. Krishnan, Victor E. Ortega, Dawn L. DeMeo, and et al. 2021. "Metabolomic Profiling Reveals Sex Specific Associations with Chronic Obstructive Pulmonary Disease and Emphysema" Metabolites 11, no. 3: 161. https://doi.org/10.3390/metabo11030161
APA StyleGillenwater, L. A., Kechris, K. J., Pratte, K. A., Reisdorph, N., Petrache, I., Labaki, W. W., O’Neal, W., Krishnan, J. A., Ortega, V. E., DeMeo, D. L., & Bowler, R. P. (2021). Metabolomic Profiling Reveals Sex Specific Associations with Chronic Obstructive Pulmonary Disease and Emphysema. Metabolites, 11(3), 161. https://doi.org/10.3390/metabo11030161






