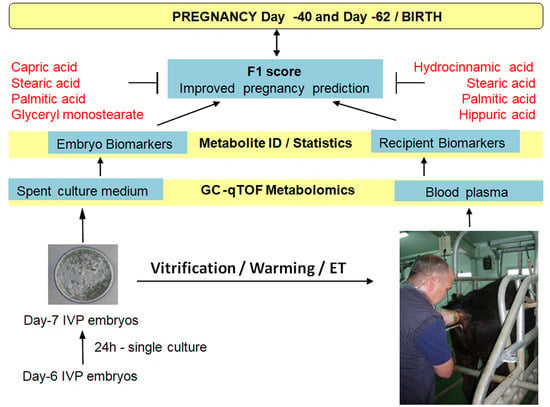
Metabolites Secreted by Bovine Embryos In Vitro Predict Pregnancies That the Recipient Plasma Metabolome Cannot, and Vice Versa
Abstract
1. Introduction
2. Results
2.1. Breed Effects on Embryo Culture Medium and Recipient Plasma
2.2. Single Candidate Biomarker Discovery
2.2.1. Culture Medium
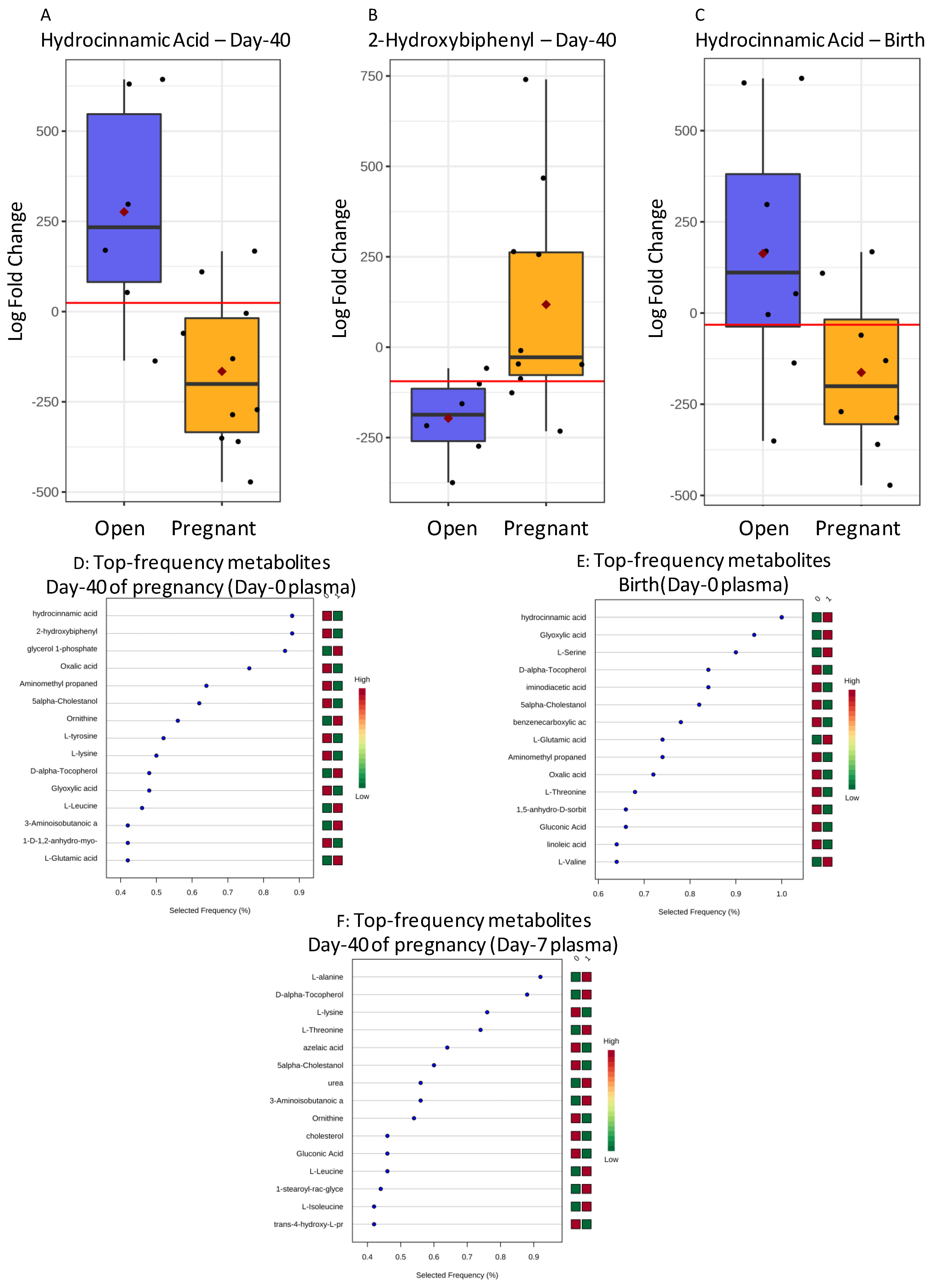
2.2.2. Plasma from Holstein Recipients
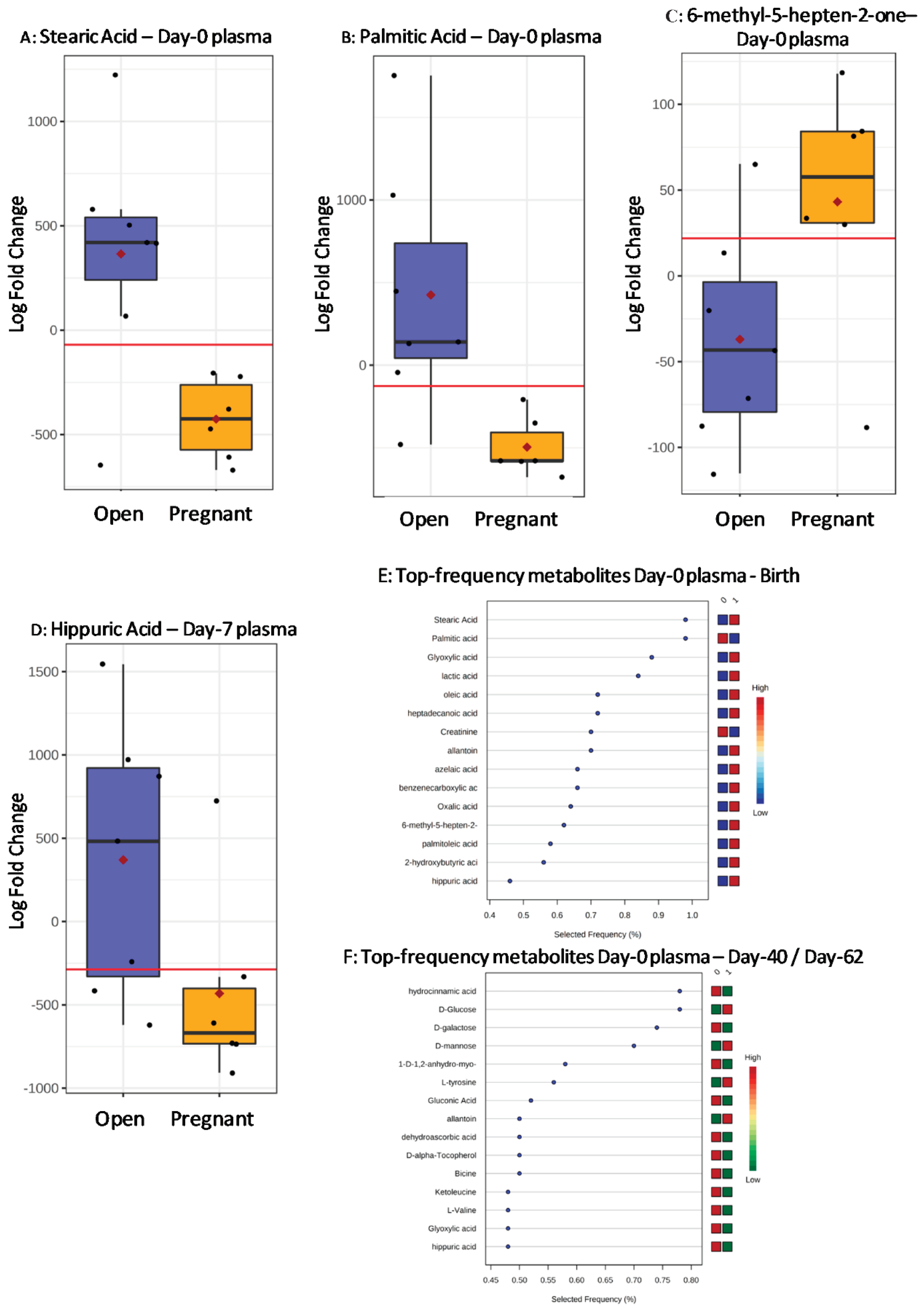
2.2.3. Plasma from AV Recipients
2.3. Combined Biomarker Discovery
2.4. Candidate Biomarker Validation
3. Discussion
3.1. Embryo Metabolites
3.2. Recipient Plasma Metabolites
3.3. Pregnancy Prediction: Single Biomarkers
3.4. New Biomarker Strategies
4. Materials and Methods
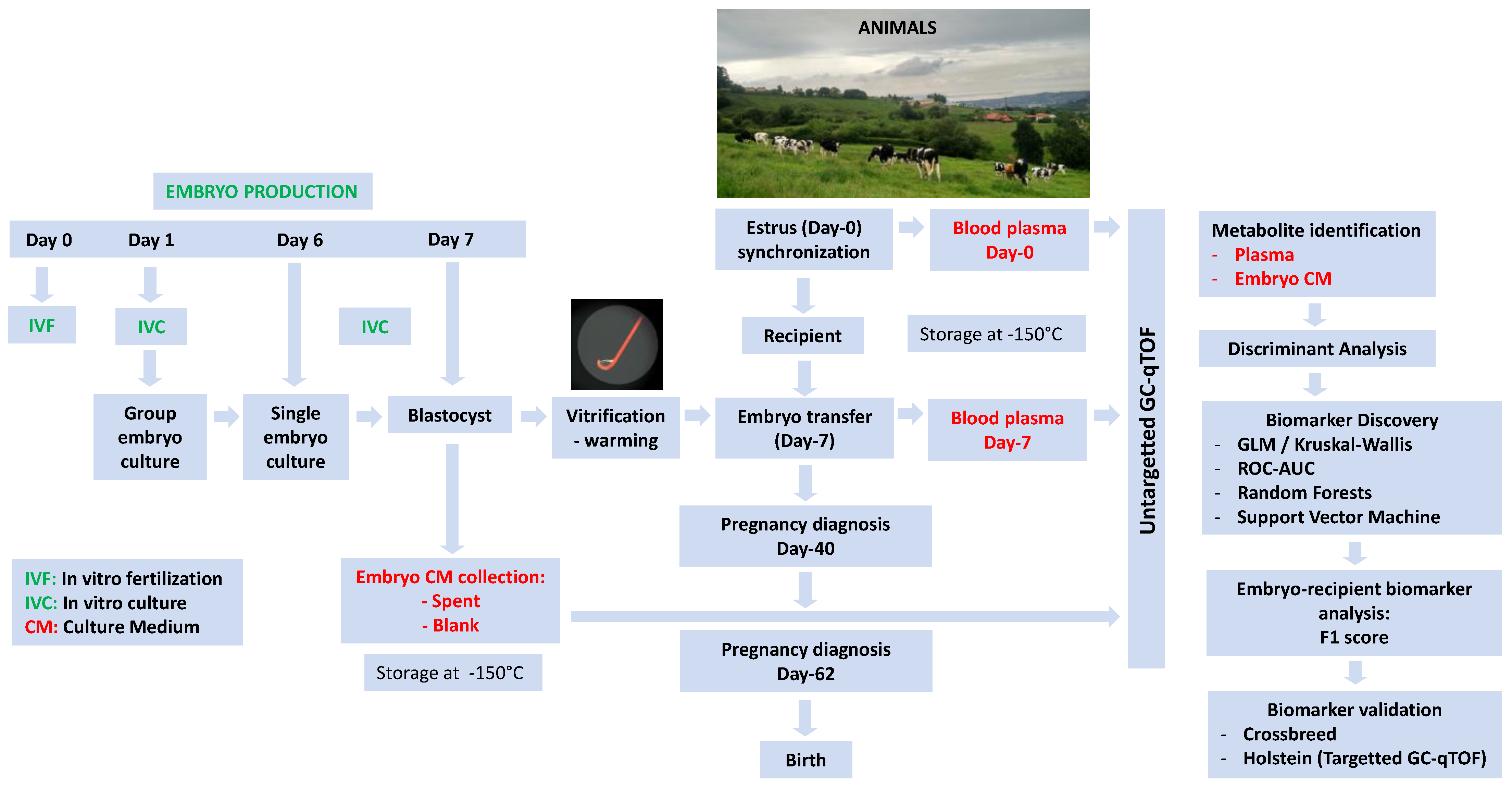
4.1. Rationale
4.2. In Vitro Embryo Production
4.3. Embryo Vitrification and Warming
4.4. Embryo Transfer and Pregnancy Diagnosis
4.5. Sample Collection for Metabolomics and Progesterone Analysis
4.6. Metabolomic Analysis of Plasma and Embryo Culture Media
4.7. Experimental Design and Statistics
4.7.1. Analyzing Breed Effects in Embryos and Recipients
4.7.2. Biomarker Studies
Identification of Candidate Metabolites
Identification of Biomarker Candidates
- Single biomarker identification: which included a univariate study with a Receiver Operator Characteristic—area under curve (ROC-AUC) (T-Test, p < 0.05, ROC-AUC > 0.650) within selected metabolites at each pregnancy endpoint in each sample subset analyzed, i.e., embryo CM and plasmas (Metaboanalyst 4.0). The effects of embryonic stages on Day-6 (i.e., morula and early blastocysts) as controllable variation sources were included in this study. Subsequently, a multivariate study with ROC curve-based exploratory analysis was carried out. The classification and feature ranking methods that showed best agreement with univariate studies were RF (Mean Decrease Accuracy –MDA- for CM metabolites) and SVM (Selected Frequency—SF—for plasma metabolites).
- Combined biomarker identification: we analyzed the PA of multiple features with the largest ROC-AUC values within the embryo CM and recipient plasma. The “Multivariate ROC curve based exploratory analysis (Explorer)” tool from Metaboanalyst was used to test the algorithms SVM, PLS-DA, and RF. Subsequently, ROC-AUC values for multiple metabolite combinations were calculated using the “ROC curve-based model evaluation (Tester)” tool.
- F1 score: The procedure was intended to collect information from the recipient and the embryo to potentially increase predictability. Each single biomarker identified in CM was combined with each metabolite identified in plasma to obtain an index (F1 score) that represents the aggregate predictive power of CM and plasma for each pregnancy specific endpoint. A Boolean matrix was performed for each metabolite in terms of allocation (1 = true) or not (0 = false) within the range of pregnant animals. F1 calculations were performed on the Boolean product vector of pairs of metabolites (i.e., 1 metabolite from CM and 1 metabolite from plasma/day/pregnancy endpoint) (False*False = False, True*False = True, False*True= True, True*True = True). For a visual representation, see a real example in Supplemental Table S4 (i.e., CM, its corresponding plasma relative concentration, and the combined F1 score). The probability value of the F1 score was calculated by a Chi-square test.
4.7.3. Biomarker Validation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leese, H.J.; Guerif, F.; Allgar, V.; Brison, D.R.; Lundin, K.; Sturmey, R.G. Biological optimization, the Goldilocks principle, and how much is lagom in the preimplantation embryo. Mol. Reprod. Dev. 2016, 83, 748–754. [Google Scholar] [CrossRef]
- Khatib, H.; Gross, N. Symposium review: Embryo survival—A genomic perspective of the other side of fertility. J. Dairy Sci. 2019, 102, 3744–3753. [Google Scholar] [CrossRef]
- Gómez, E.; Carrocera, S.; Martín, D.; Pérez-Jánez, J.J.; Prendes, J.; Prendes, J.M.; Vázquez, A.; Murillo, A.; Gimeno, I.; Muñoz, M. Efficient one-step direct transfer to recipients of thawed bovine embryos cultured in vitro and frozen in chemically defined medium. Theriogenology 2020, 146, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Mogas, T. Update on the vitrification of bovine oocytes and in vitro-produced embryos. Reprod. Fertil. Dev. 2018, 31, 105–117. [Google Scholar] [CrossRef]
- Sanches, B.V.; Lunardelli, P.A.; Tannura, J.H.; Cardoso, B.L.; Pereira, M.H.C.; Gaitkoski, D.; Basso, A.C.; Arnold, D.R.; Seneda, M.M. A new direct transfer protocol for cryopreserved IVF embryos. Theriogenology 2016, 85, 1147–1151. [Google Scholar] [CrossRef]
- Muñoz, M.; Uyar, A.; Correia, E.; Diez, C.; Fernández-González, A.; Caamaño, J.N.; Martínez-Bello, D.; Trigal, B.; Humblot, P.; Ponsart, C.; et al. Prediction of pregnancy viability in bovine in vitro-produced embryos and recipient plasma with Fourier transform infrared spectroscopy. J. Dairy Sci. 2014, 97, 5497–5507. [Google Scholar] [CrossRef]
- Xu, J.; Guo, Z.; Su, L.; Nedambale, T.; Zhang, J.; Schenk, J.; Moreno, J.; Dinnyes, A.; Ji, W.; Tian, X.; et al. Developmental Potential of Vitrified Holstein Cattle Embryos Fertilized In Vitro with Sex-Sorted Sperm. J. Dairy Sci. 2006, 89, 2510–2518. [Google Scholar] [CrossRef]
- Peterson, A.J.; Lee, R.S. Improving successful pregnancies after embryo transfer. Theriogenology 2003, 59, 687–697. [Google Scholar] [CrossRef]
- Jakobsen, A.S.; Thomsen, P.D.; Avery, B. Few polyploid blastomeres in morphologically superior bovine embryos produced in vitro. Theriogenology 2006, 65, 870–881. [Google Scholar] [CrossRef]
- Salilew-Wondim, D.; Hölker, M.; Rings, F.; Ghanem, N.; Ulas-Cinar, M.; Peippo, J.; Tholen, E.; Looft, C.; Schellander, K.; Tesfaye, D. Bovine pretransfer endometrium and embryo transcriptome fingerprints as predictors of pregnancy success after embryo transfer. Physiol. Genom. 2010, 42, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, N.; Salilew-Wondim, D.; Gad, A.; Tesfaye, D.; Phatsara, C.; Tholen, E.; Looft, C.; Schellander, K.; Hoelker, M. Bovine blas-tocysts with developmental competence to term share similar expression of developmentally important genes although derived from different culture environments. Reproduction 2011, 142, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Zolini, A.M.; Block, J.; Rabaglino, M.B.; Tríbulo, P.; Hoelker, M.; Rincon, G.; Bromfield, J.J.; Hansen, P.J. Molecular fingerprint of female bovine embryos produced in vitro with high competence to establish and maintain pregnancy†. Biol. Reprod. 2020, 102, 292–305. [Google Scholar] [CrossRef]
- Farin, P.; Britt, J.; Shaw, D.; Slenning, B. Agreement among evaluators of bovine embryos produced in vivo or in vitro. Theriogenology 1995, 44, 339–349. [Google Scholar] [CrossRef]
- Kurosawa, H.; Utsunomiya, H.; Shiga, N.; Takahashi, A.; Ihara, M.; Ishibashi, M.; Nishimoto, M.; Watanabe, Z.; Abe, H.; Kumagai, J.; et al. Development of a new clinically applicable device for embryo evaluation which measures embryo oxygen consumption. Hum. Reprod. 2016, 31, 2321–2330. [Google Scholar] [CrossRef][Green Version]
- Lopes, A.; Madsen, S.; Ramsing, N.; Løvendahl, P.; Greve, T.; Callesen, H. Investigation of respiration of individual bovine embryos produced in vivo and in vitro and correlation with viability following transfer. Hum. Reprod. 2007, 22, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, S.; Akai, T.; Imai, K. Selection of viable in vitro-fertilized bovine embryos using time-lapse monitoring in microwell culture dishes. J. Reprod. Dev. 2017, 63, 353–357. [Google Scholar] [CrossRef]
- Sugimura, S.; Akai, T.; Hashiyada, Y.; Somfai, T.; Inaba, Y.; Hirayama, M.; Yamanouchi, T.; Matsuda, H.; Kobayashi, S.; Aikawa, Y.; et al. Promising System for Selecting Healthy In Vitro–Fertilized Embryos in Cattle. PLoS ONE 2012, 7, e36627. [Google Scholar] [CrossRef]
- Mellisho, E.A.; Briones, M.A.; Velasquez, A.E.; Cabezas, J.; Castro, F.O.; Rodriguez-Alvarez, L. Extracellular vesicles secreted during blastulation show viability of bovine embryos. Reproduction 2019, 158, 477–492. [Google Scholar] [CrossRef]
- Lindgren, K.E.; Gülen Yaldir, F.; Hreinsson, J.; Holte, J.; Kårehed, K.; Sundström-Poromaa, I.; Kaihola, H.; Åkerud, H. Differences in secretome in culture media when comparing blastocysts and arrested embryos using multiplex proximity assay. UPS J. Med. Sci. 2018, 123, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Mellisho, E.A.; Velasquez, A.E.; Núñez, M.J.; Cabezas, J.G.; Cueto, J.A.; Fader, C.; Castro, F.O.; Rodríguez-Alvarez, L. Identification and characteristics of extracellular vesicles from bovine blastocysts produced in vitro. PLoS ONE 2017, 12, e0178306. [Google Scholar] [CrossRef]
- Saadeldin, I.M.; Kim, S.J.; Bin Choi, Y.; Lee, B.C. Improvement of Cloned Embryos Development by Co-Culturing with Parthenotes: A Possible Role of Exosomes/Microvesicles for Embryos Paracrine Communication. Cell. Reprogr. 2014, 16, 223–234. [Google Scholar] [CrossRef]
- Abu-Halima, M.; Häusler, S.; Backes, C.; Fehlmann, T.; Staib, C.; Nestel, S.; Nazarenko, I.; Meese, E.; Keller, A. Micro-ribonucleic acids and extracellular vesicles repertoire in the spent culture media is altered in women undergoing In Vitro Fertilization. Sci. Rep. 2017, 7, 13525. [Google Scholar] [CrossRef]
- Pallinger, E.; Bognar, Z.; Bodis, J.; Csabai, T.; Farkas, N.; Godony, K.; Varnagy, A.; Buzas, E.; Szekeres-Bartho, J. A simple and rapid flow cytometry-based assay to identify a competent embryo prior to embryo transfer. Sci. Rep. 2017, 7, 39927. [Google Scholar] [CrossRef]
- Heringstad, B.; Egger-Danner, C.; Charfeddine, N.; Pryce, J.; Stock, K.; Kofler, J.; Sogstad, A.; Holzhauer, M.; Fiedler, A.; Müller, K.; et al. Invited review: Genetics and claw health: Opportunities to enhance claw health by genetic selection. J. Dairy Sci. 2018, 101, 4801–4821. [Google Scholar] [CrossRef]
- Goldansaz, S.A.; Guo, A.C.; Sajed, T.; Steele, M.A.; Plastow, G.S.; Wishart, D.S. Livestock metabolomics and the livestock metabo-lome: A systematic review. PLoS ONE 2017, 12, e0177675. [Google Scholar] [CrossRef] [PubMed]
- Rubessa, M.; Ambrosi, A.; Gonzalez-Pena, D.; Polkoff, K.M.; Wheeler, M.B. Non-invasive nuclear magnetic resonance analysis of male and female embryo metabolites during in vitro embryo culture. Metabolomics 2018, 14, 113. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, M.; Uyar, A.; Correia, E.; Diez, C.; Fernández-González, A.; Caamaño, J.N.; Trigal, B.; Carrocera, S.; Seli, E.; Gómez, E.; et al. Non-invasive assessment of embryonic sex in cattle by metabolic fingerprinting of in vitro culture medium. Metabolomics 2013, 10, 443–451. [Google Scholar] [CrossRef]
- Gomez, E.; Carrocera, S.; Martin, D.; Herrero, P.; Canela, N.; Muñoz, M. Differential release of cell-signaling metabolites by male and female bovine embryos cultured in vitro. Theriogenology 2018, 114, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Gomez, E.; Muñoz, M.; Simo, C.; Ibáñez, C.; Carrocera, S.; Martín-González, D.; Cifuentes, A. Non-invasive metabolomics for improved determination of embryonic sex markers in chemically defined culture medium. J. Chromatogr. A 2016, 1474, 138–144. [Google Scholar] [CrossRef]
- Nõmm, M.; Porosk, R.; Pärn, P.; Kilk, K.; Soomets, U.; Kõks, S.; Jaakma, Ü. In vitro culture and non-invasive metabolic profiling of single bovine embryos. Reprod. Fertil. Dev. 2019, 31, 306. [Google Scholar] [CrossRef]
- Perkel, K.J.; Madan, P. Spent culture medium analysis from individually cultured bovine embryos demonstrates metabolomic differences. Zygote 2017, 25, 662–674. [Google Scholar] [CrossRef]
- Dos Santos, É.C.; Martinho, H.; Annes, K.; da Silva, T.; Soares, C.A.; Leite, R.F.; Milazzotto, M.P. Raman-based noninvasive metabolic profile evaluation of in vitro bovine embryos. J. Biomed. Opt. 2016, 21, e75002. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, M.; Uyar, A.; Correia, E.; Ponsart, C.; Guyader-Joly, C.; Martinez-Bello, D.; Guienne, B.M.-L.; Fernández-González, A.; Diez, C.; Caamaño, J.N.; et al. Metabolomic Prediction of Pregnancy Viability in Superovulated Cattle Embryos and Recipients with Fourier Transform Infrared Spectroscopy. BioMed Res. Int. 2014, 2014, 1–8. [Google Scholar] [CrossRef]
- Kanazawa, T.; Seki, M.; Ishiyama, K.; Kubo, T.; Kaneda, Y.; Sakaguchi, M.; Izaike, Y.; Takahashi, T. Pregnancy prediction on the day of embryo transfer (Day 7) and Day 14 by measuring luteal blood flow in dairy cows. Theriogenology 2016, 86, 1436–1444. [Google Scholar] [CrossRef]
- Hidalgo, C.O.; Gómez, E.; Prieto, L.; Duque, P.; Goyache, F.; Fernández, L.; Fernandez, I.; Facal, N.; Díez, C. Pregnancy rates and metabolic profiles in cattle treated with propylene glycol prior to embryo transfer. Theriogenology 2004, 62, 664–676. [Google Scholar] [CrossRef]
- Geary, T.W.; Burns, G.W.; Moraes, J.G.; Moss, J.I.; Denicol, A.C.; Dobbs, K.B.; Ortega, M.S.; Hansen, P.J.; Wehrman, M.E.; Neibergs, H.; et al. Identification of Beef Heifers with Superior Uterine Capacity for Pregnancy. Biol. Reprod. 2016, 95, 47. [Google Scholar] [CrossRef]
- Gómez, E.; Salvetti, P.; Gatien, J.; Carrocera, S.; Martín-González, D.; Muñoz, M. Blood Plasma Metabolomics Predicts Preg-nancy in Holstein Cattle Transferred with Fresh and Vitrified/Warmed Embryos Produced in Vitro. J. Proteom. Res. 2020, 19, 1169–1182. [Google Scholar] [CrossRef] [PubMed]
- Gómez, E.; Muñoz, M.; Gatien, J.; Carrocera, S.; Martín-González, D.; Salvetti, P. Metabolomic identification of pregnancy-specific biomarkers in blood plasma of BOS TAURUS beef cattle after transfer of in vitro produced embryos. J. Proteom. 2020, 225, 103883. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, G.; Pedersen, H.S.; Rabaglino, M.B.; Hyttel, P.; Callesen, H.; Kadarmideen, H.N. Characterization of the endometrial transcriptome in early diestrus influencing pregnancy status in dairy cattle after transfer of in vitro-produced embryos. Physiol. Genomics. 2020, 52, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Guijas, C.; Montenegro-Burke, J.R.; Warth, B.; Spilker, M.E.; Siuzdak, G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 2018, 36, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Gómez, E.; Salvetti, P.; Gatien, J.; Muñoz, M.; Martín-González, D.; Carrocera, S.; Goyache, F. Metabolomic Profiling of Bos taurus Beef, Dairy, and Crossbred Cattle: A Between-Breeds Meta-Analysis. J. Agric. Food Chem. 2020, 68, 8732–8743. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, E.M.; Leese, H.J. A potential role for triglyceride as an energy source during bovine oocyte maturation and early embryo development. Mol. Reprod. Dev. 2006, 73, 1195–1201. [Google Scholar] [CrossRef] [PubMed]
- Homa, S.T.; Racowsky, C.; McGaughey, R.W. Lipid analysis of immature pig oocytes. Reproduction 1986, 77, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Murillo, A.; Muñoz, M.; Martín-González, D.; Carrocera, S.; Martínez-Nistal, A.; Gómez, E. Low serum concentration in bovine embryo culture enhances early blastocyst rates on Day-6 with quality traits in the expanded blastocyst stage similar to BSA-cultured embryos. Reprod. Biol. 2017, 17, 162–171. [Google Scholar] [CrossRef]
- Gómez, E.; Carrocera, S.; Uzbekova, S.; Martín, D.; Murillo, A.; Alonso-Guervós, M.; Goyache, F.; Muñoz, M. Protein in culture and endogenous lipid interact with embryonic stages in vitro to alter calf birthweight after embryo vitrification and warming. Reprod. Fertil. Dev. 2017, 29, 1932–1943. [Google Scholar] [CrossRef]
- Dunning, K.R.; Cashman, K.; Russell, D.L.; Thompson, J.G.; Norman, R.J.; Robker, R.L. Beta-Oxidation Is Essential for Mouse Oocyte Developmental Competence and Early Embryo Development1. Biol. Reprod. 2010, 83, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Sutton-McDowall, M.L.; Feil, D.; Robker, R.L.; Thompson, J.G.; Dunning, K.R. Utilization of endogenous fatty acid stores for energy production in bovine preimplantation embryos. Theriogenology 2012, 77, 1632–1641. [Google Scholar] [CrossRef] [PubMed]
- Sturmey, R.G.; Reis, A.; Leese, H.J.; McEvoy, T.G. Role of Fatty Acids in Energy Provision During Oocyte Maturation and Early Embryo Development. Reprod. Domest. Anim. 2009, 44, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Gómez, E. Acetoacetate and beta-D-hydroxybutyrate as energy substrates during early bovine embryo development in vitro. Theriogenology 1997, 48, 63–74. [Google Scholar] [CrossRef]
- Gómez, E.; Duque, P.; Díaz, E.; Facal, N.; Antolin, I.; Hidalgo, C.; Diez, C. Effects of acetoacetate and D-beta-hydroxybutyrate on bovine in vitro embryo development in serum-free medium. Theriogenology 2002, 57, 1551–1562. [Google Scholar] [CrossRef]
- Desmet, K.L.J.; Marei, W.F.A.; Richard, C.; Sprangers, K.; Beemster, G.T.S.; Meysman, P.; Laukens, K.; Declerck, K.; Berghe, W.V.; Bols, P.E.J.; et al. Oocyte maturation under lipotoxic conditions induces carryover transcriptomic and functional alterations during post-hatching development of good-quality blastocysts: Novel insights from a bovine em-bryo-transfer model. Hum. Reprod. 2020, 35, 293–307. [Google Scholar] [CrossRef]
- Marei, W.F.; De Bie, J.; Mohey-Elsaeed, O.; Wydooghe, E.; Bols, P.E.; Leroy, J.L. Alpha-linolenic acid protects the developmental capacity of bovine cumulus–oocyte complexes matured under lipotoxic conditions in vitro†. Biol. Reprod. 2017, 96, 1181–1196. [Google Scholar] [CrossRef]
- Desmet, K.L.J.; Van Hoeck, V.; Gagné, D.; Fournier, E.; Thakur, A.; O’Doherty, A.M.; Walsh, C.P.; Sirard, M.A.; Bols, P.E.J.; Leroy, J.L.M.R. Exposure of bovine oocytes and embryos to elevated non-esterified fatty acid concentrations: Integration of epigenetic and transcriptomic signatures in resultant blastocysts. BMC Genom. 2016, 17, 1–18. [Google Scholar] [CrossRef]
- Van Hoeck, V.; Sturmey, R.G.; Bermejo-Alvarez, P.; Rizos, D.; Gutierrez-Adan, A.; Leese, H.J.; Bols, P.E.; Leroy, J.L. Elevated non-esterified fatty acid concentrations during bovine oocyte maturation compromise early embryo physiology. PLoS ONE 2011, 6, e23183. [Google Scholar]
- Elis, S.; Oseikria, M.; Carvalho, A.V.; Bertevello, P.S.; Corbin, E.; Teixeira-Gomes, A.-P.; Lecardonnel, J.; Archilla, C.; Duranthon, V.; Labas, V.; et al. Docosahexaenoic acid mechanisms of action on the bovine oocyte-cumulus complex. J. Ovarian Res. 2017, 10, 1–14. [Google Scholar] [CrossRef]
- Fayezi, S.; Leroy, J.L.; Novin, M.G.; Darabi, M. Oleic acid in the modulation of oocyte and preimplantation embryo development. Zygote 2017, 26, 1–13. [Google Scholar] [CrossRef]
- Thangavelu, G.; Colazo, M.; Ambrose, D.; Oba, M.; Okine, E.; Dyck, M. Diets enriched in unsaturated fatty acids enhance early embryonic development in lactating Holstein cows. Theriogenology 2007, 68, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Lowe, J.L.; Bartolac, L.K.; Bathgate, R.; Grupen, C.G. Supplementation of culture medium with L-carnitine improves the devel-opment and cryotolerance of in vitro-produced porcine embryos. Reprod. Fertil. Dev. 2017, 29, 2357–2366. [Google Scholar] [CrossRef] [PubMed]
- Murillo-Ríos, A.; Maillo, V.; Muñoz, M.; Gutiérrez-Adán, A.; Carrocera, S.; Martín-González, D.; Fernandez-Buznego, A.; Gómez, E. Short- and long-term outcomes of the absence of protein during bovine blastocyst formation in vitro. Reprod. Fertil. Dev. 2017, 29, 1064–1073. [Google Scholar] [CrossRef]
- Takahashi, T.; Inaba, Y.; Somfai, T.; Kaneda, M.; Geshi, M.; Nagai, T.; Manabe, N. Supplementation of culture medium with L-carnitine improves development and cryotolerance of bovine embryos produced in vitro. Reprod. Fertil. Dev. 2013, 25, 589–599. [Google Scholar] [CrossRef]
- Paschoal, D.M.; Sudano, M.J.; Schwarz, K.R.L.; Maziero, R.R.D.; Guastali, M.D.; Crocomo, L.F.; Magalhães, L.C.O.; Martins, A.; Leal, C.L.V.; Landim-Alvarenga, F.D.C. Cell apoptosis and lipid content of in vitro –produced, vitrified bovine embryos treated with forskolin. Theriogenology 2017, 87, 108–114. [Google Scholar] [CrossRef]
- Held-Hoelker, E.; Klein, S.; Rings, F.; Salilew-Wondim, D.; Saeed-Zidane, M.; Neuhoff, C.; Tesfaye, D.; Schellander, K.; Hoelker, M. Cryosurvival of in vitro produced bovine embryos supplemented with l -Carnitine and concurrent reduction of fatty acids. Theriogenology 2017, 96, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Cremin, J.D., Jr.; McLeod, K.R.; Harmon, D.L.; Goetsch, A.L.; Bourquin, L.D.; Fahey, G.C., Jr. Portal and hepatic fluxes in sheep and con-centrations in cattle ruminal fluid of 3-(4-hydroxyphenyl)propionic, benzoic, 3-phenylpropionic, and trans-cinnamic acids. J. Anim. Sci. 1995, 73, 1766–1775. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sondossi, M.; Sylvestre, M.; Ahmad, D.; Masse, R. Metabolism of hydroxybiphenyl and cholorohydroxybiphenyl by bi-phenyl/cholorobiphenyl degradign Pseudomonas testosteroni, strain B-356. J. Ind. Microbiol. 1991, 6, 77–88. [Google Scholar] [CrossRef]
- Prajapati, V.S.; Purohit, H.J.; Raj, D.V.; Parmar, N.; Pate, A.B.; Jone, O.; Joshi, C.G. The effect of a high-roughage diet on the metabolism of aromatic compounds by rumen microbes: A metagenomic study using Mehsani buffalo (Bubalus bubalis). Appl. Microbiol. Biotechnol. 2016, 100, 1319–1331. [Google Scholar] [CrossRef] [PubMed]
- Geppert, T.C.; Meyer, A.M.; Perry, G.A.; Gunn, P.J. Effects of excess metabolizable protein on ovarian function and circulating amino acids of beef cows: 2. Excessive supply in varying concentrations from corn gluten meal. Animal 2017, 11, 634–642. [Google Scholar] [CrossRef][Green Version]
- Webb, L.; Sadri, H.; Von Soosten, D.; Dänicke, S.; Egert, S.; Stehle, P.; Sauerwein, H. Changes in tissue abundance and activity of enzymes related to branched-chain amino acid catabolism in dairy cows during early lactation. J. Dairy Sci. 2019, 102, 3556–3568. [Google Scholar] [CrossRef]
- Löest, C.A.; Gilliam, G.G.; Waggoner, J.W.; Turner, J.L. Post-ruminal branched-chain amino acid supplementation and intravenous lipopolysaccharide infusion alter blood metabolites, rumen fermentation, and nitrogen balance of beef steers1. J. Anim. Sci. 2018, 96, 2886–2906. [Google Scholar] [CrossRef] [PubMed]
- McNamara, J.; Huber, K.; Kenéz, A. A dynamic, mechanistic model of metabolism in adipose tissue of lactating dairy cattle. J. Dairy Sci. 2016, 99, 5649–5661. [Google Scholar] [CrossRef]
- Drackley, J.; Donkin, S.; Reynolds, C. Major Advances in Fundamental Dairy Cattle Nutrition. J. Dairy Sci. 2006, 89, 1324–1336. [Google Scholar] [CrossRef]
- Esposito, G.; Raffrenato, E.; Lukamba, S.D.; Adnane, M.; Irons, P.C.; Cormican, P.; Tasara, T.; Chapwanya, A. Characterization of metabolic and inflammatory profiles of transition dairy cows fed an energy-restricted diet. J. Anim. Sci. 2020, 98, 391. [Google Scholar] [CrossRef] [PubMed]
- Beam, S.W.; Butler, W.R. Effects of energy balance on follicular development and first ovulation in postpartum dairy cows. Biosci. Proc. 2019, 54, 411–424. [Google Scholar] [CrossRef]
- Butler, W. Nutritional interactions with reproductive performance in dairy cattle. Anim. Reprod. Sci. 2000, 60-61, 449–457. [Google Scholar] [CrossRef]
- Li, P.; Li, L.; Zhang, C.; Cheng, X.; Zhang, Y.; Guo, Y.; Long, M.; Yang, S.; He, J. Palmitic Acid and β-Hydroxybutyrate Induce Inflammatory Responses in Bovine Endometrial Cells by Activating Oxidative Stress-Mediated NF-κB Signaling. Molecules 2019, 24, 2421. [Google Scholar] [CrossRef] [PubMed]
- Chankeaw, W.; Guo, Y.Z.; Båge, R.; Svensson, A.; Andersson, G.; Humblot, P. Elevated non-esterified fatty acids impair survival and promote lipid accumulation and pro-inflammatory cytokine production in bovine endometrial epithelial cells. Reprod. Fertil. Dev. 2018, 30, 1770–1784. [Google Scholar] [CrossRef]
- Waters, S.M.; Coyne, G.S.; Kenny, D.A.; MacHugh, D.E.; Morris, D.G. Dietary n-3 polyunsaturated fatty acid supplemen-tation alters the expression of genes involved in the control of fertility in the bovine uterine endometrium. Physiol. Genom. 2012, 44, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Kępka, A.; Chojnowska, S.; Okungbowa, O.E.; Zwierz, K. The role of carnitine in the perinatal period. Dev. Period Med. 2015, 18, 417–425. [Google Scholar]
- Wang, Y.; Tang, C.; Wu, M.; Pan, Y.; Ruan, H.; Chen, L.; Yao, H.; Zhu, H.; Wu, X. Dehydroascorbic acid taken up by glucose trans-porters stimulates estradiol production through inhibition of JNK/c-Jun/AP1 signaling in JAR cells. Mol. Hum. Reprod. 2014, 20, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Rejeb, M.; Najar, T. A survey on the effect of plasma vitamin C on white blood constituents under heat stress condition for dairy cows. J. Anim. Plant. Sci. 2018, 38, 6206–6216. [Google Scholar]
- Obeidat, Y.; Catandi, G.; Carnevale, E.; Chicco, A.J.; Demann, A.; Field, S.; Chen, T. A multi-sensor system for measuring bovine embryo metabolism. Biosens. Bioelectron. 2019, 126, 615–623. [Google Scholar] [CrossRef] [PubMed]
- García-Herreros, M.; Simintiras, C.A.; Lonergan, P. Temporally differential protein expression of glycolytic and glycogenic en-zymes during in vitro preimplantation bovine embryo development. Reprod. Fertil. Dev. 2018, 30, 1245–1252. [Google Scholar] [CrossRef]
- Sudano, M.J.; Rascado, T.D.; Tata, A.; Belaz, K.R.; Santos, V.G.; Valente, R.S.; Mesquita, F.S.; Ferreira, C.R.; Araújo, J.P.; Eberlin, M.N.; et al. Lipidome signatures in early bovine embryo development. Theriogenology 2016, 86, 472–484.e1. [Google Scholar] [CrossRef] [PubMed]
- Donnay, I.; Feugang, J.; Bernard, S.; Marchandise, J.; Pampfer, S.; Moens, A.; Dessy, F. Impact of adding 5.5 mM glucose to SOF medium on the development, metabolism and quality of in vitro produced bovine embryos from the morula to the blastocyst stage. Zygote 2002, 10, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Pepe, M.S.; Li, C.I.; Feng, Z. Improving the Quality of Biomarker Discovery Research: The Right Samples and Enough of Them. Cancer Epidemiol. Biomark. Prev. 2015, 24, 944–950. [Google Scholar] [CrossRef] [PubMed]
- Gómez, E.; Rodríguez, A.; Muñoz, M.; Caamaño, J.N.; Carrocera, S.; Martín, D.; Facal, N.; Díez, C. Development and quality of bovine morulae cultured in serum-free medium with specific retinoid receptor agonists. Reprod. Fertil. Dev. 2008, 20, 884–891. [Google Scholar] [CrossRef]
- Caamaño, J.; Gómez, E.; Trigal, B.; Muñoz, M.; Carrocera, S.; Martín, D.; Díez, C. Survival of vitrified in vitro–produced bovine embryos after a one-step warming in-straw cryoprotectant dilution procedure. Theriogenology 2015, 83, 881–890. [Google Scholar] [CrossRef]
- Grobet, L.; Royo Martin, L.J.; Poncelet, D.; Pirottin, D.; Brouwers, B.; Riquet, J.; Schoeberlein, A.; Dunner, S.; Meńissier, F.; Massabanda, J.; et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 1997, 17, 71–74. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lee, S.-J. Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. USA 1997, 94, 12457–12461. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]

| p Value | Pregnancy | Univariate Study | Biomarker Discovery Study | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite | BB 1 | Day-6-E 2 | Endpoint | p-Value | Open | Pregnant | p-Value | FCh | ROC-AUC | Open | Pregnant |
| Glyceryl monostearate | 0.406 | 0.077 | Birth | 0.023 | 2454 ± 698 | 940 ± 468 | 0.060 | 2.829 | 0.670 | 11/17 | 11/18 |
| Capric acid | 0.364 | 0.742 | Birth | 0.048 | 329 ± 68 | 188 ± 45 | 0.021 | 4.472 | 0.728 | 10/17 | 15/19 |
| Palmitic acid | 0.507 | 0.341 | Birth | 0.089 | 17,719 ± 2992 | 12,460 ± 1978 | 0.051 | 5.381 | 0.669 | 12/17 | 12/19 |
| Stearic acid | 0.350 | 0.188 | Birth | 0.019 | 75,827 ± 9612 | 51,951 ± 6356 | 0.016 | 5.899 | 0.743 | 13/17 | 14/19 |
| Stearic acid | 0.350 | 0.188 | D-62 | 0.023 | 75,692 ± 11,580 | 52,453 ± 6092 | 0.022 | 5.612 | 0.733 | 11/15 | 14/21 |
| Day | Pregnancy | Univariate Study | Biomarker Discovery Study | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite | Plasma | Endpoint | p | Open | Pregnant | p | FCh | ROC-AUC | Open | Pregnant |
| Hydrocinnamic acid | 0 | Birth | 0.059 | 362 ± 34 | 255 ± 44 | 0.044 | 0.495 | 0.797 | 6/8 | 6/8 |
| Hydrocinnamic acid | 0 | Day-40 | 0.012 | 400 ± 34 | 254 ± 26 | 0.005 | 0.666 | 0.900 | 5/6 | 8/10 |
| 2-hydroxybiphenyl | 0 | Day-40 | 0.013 | 271 ± 30 | 363 ± 23 | 0.031 | −0.395 | 0.900 | 5/6 | 8/10 |
| Glycerol-phosphate | 0 | Day-40 | 0.017 | 1315 ± 307 | 1401 ± 238 | 0.062 | −0.586 | 0.883 | 6/6 | 8/10 |
| Alanine | 7 | Birth | 0.065 | 1662 ± 182 | 2163 ± 172 | 0.065 | −0.357 | 0.792 | 6/8 | 6/9 |
| Lysine | 7 | Birth | 0.067 | 148 ± 25 | 223 ± 24 | 0.049 | −0.497 | 0.778 | 6/8 | 7/9 |
| Threonine | 7 | Day-62 | 0.051 | 93 ± 18 | 141 ± 15 | 0.052 | −0.669 | 0.800 | 5/7 | 8/10 |
| Day | Pregnancy | Univariate Study | Biomarker Discovery Study | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Plasma | Endpoint | p | Open | Pregnant | p | FCh | ROC-AUC | Open | Pregnant | |
| Stearic acid | 0 | Birth | 0.022 | 3668 ± 96 | 3204 ± 104 | 0.007 | 0.191 | 0.881 | 6/7 | 6/6 |
| Palmitic acid | 0 | Birth | 0.007 | 2901 ± 215 | 2136 ± 221 | 0.013 | 0.367 | 0.976 | 6/7 | 6/6 |
| 6-methyl-5-hepten-2-one | 0 | Birth | 0.055 | 108 ± 1.9 | 115 ± 2.0 | 0.055 | −0.078 | 0.833 | 6/7 | 5/6 |
| Palmitic acid | 0 | D40–D62 | 0.015 | 286 ± 17 | 223 ± 16 | 0.020 | 0.342 | 0.929 | 5/6 | 6/7 |
| Hippuric acid | 7 | Birth | 0.021 | 2385 ± 144 | 1663 ± 148 | 0.071 | 0.430 | 0.857 | 5/7 | 5/6 |
| Hippuric acid | 7 | D40–D62 | 0.045 | 2458 ± 230 | 1839 ± 213 | 0.045 | 0.454 | 0.833 | 4/6 | 6/7 |
| L-Valine | 7 | D40–D62 | 0.056 | 4254 ± 443 | 3607 ± 410 | 0.056 | 0.596 | 0.857 | 5/6 | 6/7 |
| N-(2-hydroxyethyl) iminodiacetic acid 2 | 7 | D40–D62 | 0.022 | 10,743 ± 492 | 9150 ± 455 | 0.037 | 0.230 | 0.881 | 5/6 | 7/7 |
| Dehydroascorbic acid | 7 | D40–D62 | 0.034 | 1213 ± 66 | 1012 ± 49 | 0.034 | 0.302 | 0.857 | 5/6 | 6/7 |
| Breed | Embryo Culture Medium | Recipient Plasma | Endpoint | F1 Score | |||||
|---|---|---|---|---|---|---|---|---|---|
| Metabolite | ROC-AUC | Metabolite | Day | ROC-AUC | Aggregate | B/M | p | ||
| AV | Capric acid | 0.728 | Palmitic acid | 0 | 0.952 | Birth | 1.000 | <0.001 | |
| AV | Capric acid | 0.728 | Stearic acid | 0 | 0.881 | Birth | 1.000 | <0.001 | |
| AV | Capric acid | 0.728 | Heptadecanoic acid | 0 | 0.810 | Birth | 0.923 | 0.002 | |
| AV | Glyceryl-Monostearate | 0.721 | Palmitic acid | 0 | 0.952 | Birth | 0.923 | 0.002 | |
| AV | Glyceryl-Monostearate | 0.721 | Stearic acid | 0 | 0.881 | Birth | 0.923 | 0.002 | |
| AV | Glyceryl-Monostearate | 0.721 | Heptadecanoic acid | 0 | 0.810 | Birth | 0.923 | 0.002 | |
| AV | Capric acid | 0.728 | L-Valine | 7 | 0.762 | Birth | 0.900 | 0.008 | |
| AV | Capric acid | 0.728 | 2-hydroxybiphenyl | 7 | 0.714 | Birth | 0.923 | 0.002 | |
| AV | Capric acid | 0.728 | N-(2-hydroxyethyl) iminodiacetic acid 2 | 7 | 0.762 | Birth | 0.923 | 0.002 | |
| AV | Capric acid | 0.728 | Hippuric acid | 7 | 0.857 | Birth | 0.857 | 0.008 | |
| AV | Capric acid | 0.728 | Dehydroascorbic acid | 7 | 0.738 | Birth | 0.857 | 0.008 | |
| AV | Glyceryl-Monostearate | 0.695 | Palmitic acid | 0 | 0.904 | D62 | 0.875 | 0.008 | |
| AV | Glyceryl-Monostearate | 0.695 | Stearic acid | 0 | 0.733 | D62 | 0.875 | 0.008 | |
| AV | Glyceryl-Monostearate | 0.695 | Heptadecanoic acid | 0 | 0.810 | D62 | 0.875 | 0.008 | |
| AV | Capric acid | 0.662 | Stearic acid | 0 | 0.785 | D40 | 0.933 | 0.002 | |
| AV | Capric acid | 0.662 | Heptadecanoic acid | 0 | 0.786 | D40 | 1.000 | <0.001 | |
| AV | Glyceryl-Monostearate | 0.687 | Palmitic acid | 0 | 0.904 | D40 | 0.875 | 0.008 | |
| AV | Glyceryl-Monostearate | 0.687 | Stearic acid | 0 | 0.733 | D40 | 0.875 | 0.008 | |
| AV | Glyceryl-Monostearate | 0.687 | Heptadecanoic acid | 0 | 0.810 | D40 | 0.933 | 0.002 | |
| Holstein | Capric acid | 0.728 | Hydrocinnamic acid | 0 | 0.797 | Birth | 0.889 | 0.933 | <0.001 |
| Holstein | Capric acid | 0.728 | Glycerol-phosphate | 0 | 0.625 | Birth | 0.842 | 0.933 | <0.001 |
| Holstein | Capric acid | 0.728 | Citric acid | 0 | 0.625 | Birth | 0.889 | 0.941 | <0.001 |
| Holstein | Glyceryl-Monostearate | 0.695 | Citric acid | 0 | 0.625 | Birth | 0.842 | 0.889 | 0.002 |
| Holstein | Capric acid | 0.728 | L-Alanine | 7 | 0.792 | Birth | 0.900 | 0.900 | 0.001 |
| Holstein | Capric acid | 0.728 | L-Threonine | 7 | 0.764 | Birth | 0.900 | 0.947 | <0.001 |
| Holstein | Capric acid | 0.728 | Hydrocinnamic acid | 7 | 0.583 | Birth | 0.900 | 0.947 | <0.001 |
| Holstein | Capric acid | 0.728 | Glycerol-phosphate | 7 | 0.652 | Birth | 0.900 | 0.947 | <0.001 |
| Holstein | Glyceryl-Monostearate | 0.695 | Hydrocinnamic acid | 0 | 0.794 | D62 | 0.857 | 0.903 | 0.001 |
| Holstein | Capric acid | 0.670 | Hydrocinnamic acid | 0 | 0.900 | D40 | 0.952 | 0.952 | <0.001 |
| Holstein | Glyceryl-Monostearate | 0.687 | Hydrocinnamic acid | 0 | 0.900 | D40 | 0.909 | 0.952 | <0.001 |
| Holstein | Capric acid | 0.670 | L-Valine | 7 | 0.606 | D40 | 0.917 | 0.917 | 0.002 |
| Holstein | Capric acid | 0.670 | L-Threonine | 7 | 0.757 | D40 | 0.917 | 0.957 | <0.001 |
| Holstein | Glyceryl-Monostearate | 0.687 | L-Threonine | 7 | 0.757 | D40 | 0.917 | 0.917 | 0.002 |
| Combined Metabolites | With Day-6 Embryonic Stage | Without Day-6 Embryonic Stage | |||||
|---|---|---|---|---|---|---|---|
| Culture Medium | Plasma | True | F1 Score | p Value | True | F1 Score | p Value |
| Capric acid | Hydrocinnamic acid | 14/19 | 0.848 | 0.013 | ND | ||
| Glyceryl-monostearate | Hydrocinnamic acid | 15/19 | 0.882 | 0.005 | 14/19 | 0.848 | 0.013 |
| Capric acid | Citric acid | 16/19 | 0.889 | 0.002 | ND | ||
| Glyceryl-monostearate | Citric acid | 16/19 | 0.914 | 0.002 | ND | ||
| Capric acid | Hippuric acid | 13/19 | 0.813 | 0.033 | ND | ||
| Glyceryl-monostearate | Hippuric acid | 15/19 | 0.882 | 0.005 | 14/19 | 0.848 | 0.013 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomez, E.; Canela, N.; Herrero, P.; Cereto, A.; Gimeno, I.; Carrocera, S.; Martin-Gonzalez, D.; Murillo, A.; Muñoz, M. Metabolites Secreted by Bovine Embryos In Vitro Predict Pregnancies That the Recipient Plasma Metabolome Cannot, and Vice Versa. Metabolites 2021, 11, 162. https://doi.org/10.3390/metabo11030162
Gomez E, Canela N, Herrero P, Cereto A, Gimeno I, Carrocera S, Martin-Gonzalez D, Murillo A, Muñoz M. Metabolites Secreted by Bovine Embryos In Vitro Predict Pregnancies That the Recipient Plasma Metabolome Cannot, and Vice Versa. Metabolites. 2021; 11(3):162. https://doi.org/10.3390/metabo11030162
Chicago/Turabian StyleGomez, Enrique, Nuria Canela, Pol Herrero, Adrià Cereto, Isabel Gimeno, Susana Carrocera, David Martin-Gonzalez, Antonio Murillo, and Marta Muñoz. 2021. "Metabolites Secreted by Bovine Embryos In Vitro Predict Pregnancies That the Recipient Plasma Metabolome Cannot, and Vice Versa" Metabolites 11, no. 3: 162. https://doi.org/10.3390/metabo11030162
APA StyleGomez, E., Canela, N., Herrero, P., Cereto, A., Gimeno, I., Carrocera, S., Martin-Gonzalez, D., Murillo, A., & Muñoz, M. (2021). Metabolites Secreted by Bovine Embryos In Vitro Predict Pregnancies That the Recipient Plasma Metabolome Cannot, and Vice Versa. Metabolites, 11(3), 162. https://doi.org/10.3390/metabo11030162