The Impact of Metabolic Scion–Rootstock Interactions in Different Grapevine Tissues and Phloem Exudates
Abstract
:1. Introduction
2. Results
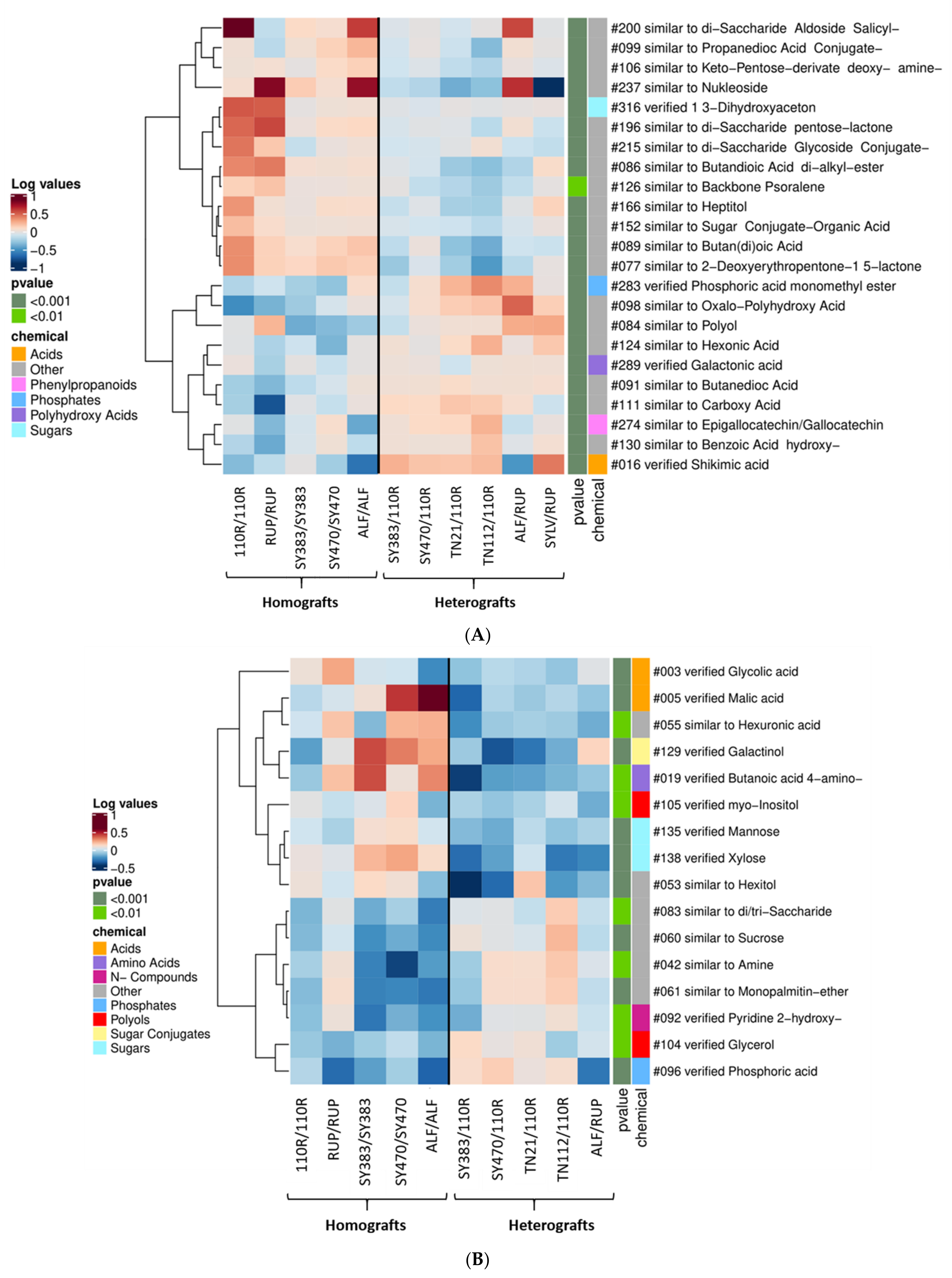
2.1. Metabolic Profile of Homografted and Heterografted Grapevines in Tissues and Phloem Exudates Collected from the Scion and Rootstock
2.2. Grafting Partner Induced Changes in the Scion and Rootstock Metabolome
2.3. Metabolic Profiles of Scion and Rootstock Phloem Exudates and Stems in Grafted Grapevines
3. Discussion
3.1. Heterografting Enhances Defense-Responses in Both Scions and Rootstocks
3.2. Scions and Rootstocks Are Able to Affect Specific Tissues and Phloem Exudates within the Grafted Plant
3.3. Phloem Exudate Composition Appears Significantly Altered between Scion and Rootstock
4. Materials and Methods
4.1. Experimental Design and Plant Material
4.2. Sample Collection
4.3. GC-MS Metabolite Profiling of Leaves, Dtems, and Phloem Exudate
4.4. Comprehensive Non-Targeted and Targeted Data Analysis of GC-MS Profiles
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Juniper, B.E. The Mysterious Origin of the Sweet Apple. Am. Sci. 2007, 95, 44–51. [Google Scholar] [CrossRef]
- Assunção, M.; Tedesco, S.; Fevereiro, P. Molecular Aspects of Grafting in Woody Plants. In Annual Plant Reviews Online, 1st ed.; Wiley Online Library: New York, NY, USA, 2021; Volume 4, pp. 87–126. [Google Scholar]
- Tandonnet, J.P.; Cookson, S.J.; Vivin, P.; Ollat, N. Scion genotype controls biomass allocation and root development in grafted grapevine. Aust. J. Grape Wine Res. 2010, 16, 290–300. [Google Scholar] [CrossRef]
- Ahsan, M.U.; Hayward, A.; Alam, M.; Hiti-Bandaralage, J.; Topp, B.; Beveridge, C.A.; Mitter, N. Scion control of miRNA abundance and tree maturity in grafted avocado. BMC Plant Biol. 2019, 19, 382. [Google Scholar] [CrossRef] [Green Version]
- Foster, T.M.; McAtee, P.A.; Waite, C.N.; Boldingh, H.L.; McGhie, T.K. Apple dwarfing rootstocks exhibit an imbalance in carbohydrate allocation and reduced cell growth and metabolism. Hortic. Res. 2017, 4, 17009. [Google Scholar] [CrossRef] [Green Version]
- Han, Q.; Guo, Q.; Korpelainen, H.; Niinemets, Ü.; Li, C. Rootstock determines the drought resistance of poplar grafting combinations. Tree Physiol. 2019, 39, 1855–1866. [Google Scholar] [CrossRef]
- Zhang, Z.; Cao, B.; Gao, S.; Xu, K. Grafting improves tomato drought tolerance through enhancing photosynthetic capacity and reducing ROS accumulation. Protoplasma 2019, 256, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, S.; Wei, M.; Gong, B.; Shi, Q. Effect of Different Rootstocks on the Salt Stress Tolerance in Watermelon Seedlings. Hortic. Plant J. 2018, 4, 239–249. [Google Scholar] [CrossRef]
- Zhang, H.; Li, X.; Zhang, S.; Yin, Z.; Zhu, W.; Li, J.; Meng, L.; Zhong, H.; Xu, N.; Wu, Y.; et al. Rootstock alleviates salt stress in grafted mulberry seedlings: Physiological and PSII function responses. Front. Plant Sci. 2018, 9, 1806. [Google Scholar] [CrossRef] [PubMed]
- Rasool, A.; Mansoor, S.; Bhat, K.M.; Hassan, G.I.; Rehman Baba, T.; Nasser Alyemeni, M.; Alsahli, A.A.; El-Serehy, H.A.; Paray, B.A.; Ahmad, P. Mechanisms Underlying Graft Union Formation and Rootstock Scion Interaction in Horticultural Plants. Front. Plant Sci. 2020, 11, 590847. [Google Scholar] [CrossRef] [PubMed]
- Albacete, A.; Martínez-Andújar, C.; Martínez-Pérez, A.; Thompson, A.J.; Dodd, I.C.; Pérez-Alfocea, F. Unravelling rootstock × scion interactions to improve food security. J. Exp. Bot. 2015, 66, 2211–2226. [Google Scholar] [CrossRef] [Green Version]
- Goldschmidt, E.E. Plant grafting: New mechanisms, evolutionary implications. Front. Plant Sci. 2014, 5, 727. [Google Scholar] [CrossRef] [Green Version]
- Haywood, V.; Kragler, F.; Lucas, W.J. Plasmodesmata: Pathways for protein and ribonucleoprotein signaling. Plant Cell 2002, 14, 303–326. [Google Scholar] [CrossRef] [Green Version]
- Melnyk, C.W.; Meyerowitz, E.M. Plant grafting. Curr. Biol. 2015, 25, R183–R188. [Google Scholar] [CrossRef] [Green Version]
- Lough, T.J.; Lucas, W.J. Integrative plant biology: Role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 2006, 57, 203–232. [Google Scholar] [CrossRef] [Green Version]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT Protein Movement Contributes to Long-Distance Signaling in Floral Induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [Green Version]
- Tietel, Z.; Srivastava, S.; Fait, A.; Tel-Zur, N.; Carmi, N.; Raveh, E. Impact of scion/rootstock reciprocal effects on metabolomics of fruit juice and phloem sap in grafted Citrus reticulata. PLoS ONE 2020, 15, e0227192. [Google Scholar] [CrossRef]
- Tetyuk, O.; Benning, U.F.; Hoffmann-Benning, S. Collection and analysis of Arabidopsis phloem exudates using the EDTA-facilitated Method. J. Vis. Exp. 2013, 80, e51111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Notaguchi, M.; Okamoto, S. Dynamics of long-distance signaling via plant vascular tissues. Front. Plant Sci. 2015, 6, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melnyk, C.W.; Gabel, A.; Hardcastle, T.J.; Robinson, S.; Miyashima, S.; Grosse, I.; Meyerowitz, E.M. Transcriptome dynamics at Arabidopsis graft junctions reveal an intertissue recognition mechanism that activates vascular regeneration. Proc. Natl. Acad. Sci. USA 2018, 115, E2447–E2456. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Zhao, J.; Hu, F.; Qin, Y.; Wang, X.; Hu, G. Transcriptome changes between compatible and incompatible graft combination of Litchi chinensis by digital gene expression profile. Sci. Rep. 2017, 7, 3954. [Google Scholar] [CrossRef] [Green Version]
- He, W.; Wang, Y.; Chen, Q.; Sun, B.; Tang, H.R.; Pan, D.M.; Wang, X.R. Dissection of the Mechanism for Compatible and Incompatible Graft Combinations of Citrus grandis (L.) Osbeck (‘Hongmian Miyou’). Int. J. Mol. Sci. 2018, 19, 505. [Google Scholar] [CrossRef] [Green Version]
- Assunção, M.; Santos, C.; Brazão, J.; Eiras-Dias, J.E.; Fevereiro, P. Understanding the molecular mechanisms underlying graft success in grapevine. BMC Plant Biol. 2019, 19, 396. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhuang, W.; Tu, X.; Gao, Z.; Xiong, A.; Yu, X.; Li, X.; Li, F.; Qu, S. Transcriptomic analysis of interstock-induced dwarfism in Sweet Persimmon (Diospyros kaki Thunb.). Hortic. Res. 2019, 6, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.Y.; Li, J.; Liu, M.M.; Yao, Q.; Chen, J.Z. Transcriptome profiling to understand the effect of citrus rootstocks on the growth of ‘Shatangju’ mandarin. PLoS ONE 2017, 12, e0169897. [Google Scholar] [CrossRef] [Green Version]
- Tzarfati, R.; Ben-Dor, S.; Sela, I.; Goldschmidt, E.E. Graft-induced Changes in MicroRNA Expression Patterns in Citrus Leaf Petioles. Open Plant Sci. J. 2013, 7, 17–23. [Google Scholar] [CrossRef] [Green Version]
- Prodhomme, D.; Fonayet, J.V.; Hévin, C.; Franc, C.; Hilbert, G.; de Revel, G.; Richard, T.; Ollat, N.; Cookson, S.J. Metabolite profiling during graft union formation reveals the reprogramming of primary metabolism and the induction of stilbene synthesis at the graft interface in grapevine. BMC Plant Biol. 2019, 19, 599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assunção, M.; Pinheiro, J.; Cruz, S.; Brazão, J.; Queiroz, J.; Eiras Dias, J.E.; Canas, S. Gallic acid, sinapic acid and catechin as potential chemical markers of Vitis graft success. Sci. Hortic. 2019, 246, 129–135. [Google Scholar] [CrossRef]
- Assunção, M.; Canas, S.; Cruz, S.; Brazão, J.; Zanol, G.C.; Eiras-Dias, J.E. Graft compatibility of Vitis spp.: The role of phenolic acids and flavanols. Sci. Hortic. 2016, 207, 140–145. [Google Scholar] [CrossRef]
- Usenik, V.; Krška, B.; Vičan, M.; Štampar, F. Early detection of graft incompatibility in apricot (Prunus armeniaca L.) using phenol analyses. Sci. Hortic. 2006, 109, 332–338. [Google Scholar] [CrossRef]
- Irisarri, P.; Zhebentyayeva, T.; Errea, P.; Pina, A. Differential expression of phenylalanine ammonia lyase (PAL) genes implies distinct roles in development of graft incompatibility symptoms in Prunus. Sci. Hortic. 2016, 204, 16–24. [Google Scholar] [CrossRef]
- Cookson, S.J.; Clemente Moreno, M.J.; Hevin, C.; Nyamba Mendome, L.Z.; Delrot, S.; Magnin, N.; Trossat-Magnin, C.; Ollat, N. Heterografting with nonself rootstocks induces genes involved in stress responses at the graft interface when compared with autografted controls. J. Exp. Bot. 2014, 65, 2473–2481. [Google Scholar] [CrossRef] [Green Version]
- Tedesco, S.; Pina, A.; Fevereiro, P.; Kragler, F. A Phenotypic Search on Graft Compatibility in Grapevine. Agronomy 2020, 10, 706. [Google Scholar] [CrossRef]
- Mandal, S.M.; Chakraborty, D.; Dey, S. Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal. Behav. 2010, 5, 359–368. [Google Scholar] [CrossRef] [Green Version]
- Santos-Sánchez, N.F.; Salas-Coronado, R.; Hernández-Carlos, B.; Villanueva-Cañongo, C. Shikimic Acid Pathway in Biosynthesis of Phenolic Compounds. In Plant Physiological Aspects of Phenolic Compounds, 1st ed.; Soto-Hernández, M., García-Mateos, R., Palma-Tenango, M., Eds.; IntechOpen: London, UK, 2019; Volume 395, pp. 116–124. [Google Scholar]
- Bouché, N.; Fromm, H. GABA in plants: Just a metabolite? Trends Plant Sci. 2004, 9, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Sheen, J. Metabolic repression of transcription in higher plants. Plant Cell 1990, 2, 1027–1038. [Google Scholar] [CrossRef]
- Yang, M.; Soga, T.; Pollard, P.J.; Adam, J. The emerging role of fumarate as an oncometabolite. Front. Oncol. 2012, 2, 85. [Google Scholar] [CrossRef] [Green Version]
- McCammon, M.T.; Epstein, C.B.; Przybyla-Zawislak, B.; McAlister-Henn, L.; Butow, R.A. Global Transcription Analysis of Krebs Tricarboxylic Acid Cycle Mutants Reveals an Alternating Pattern of Gene Expression and Effects on Hypoxic and Oxidative Genes. Mol. Biol. Cell 2003, 14, 958–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, C.; Scheible, W.-R.; Stitt, M.; Krapp, A. Influence of malate and 2-oxoglutarate on the NIA transcript level and nitrate reductase activity in tobacco leaves. Plant. Cell Environ. 2001, 24, 191–203. [Google Scholar] [CrossRef]
- Finkemeier, I.; König, A.-C.; Heard, W.; Nunes-Nesi, A.; Pham, P.A.; Leister, D.; Fernie, A.R.; Sweetlove, L.J. Transcriptomic analysis of the role of carboxylic acids in metabolite signaling in arabidopsis leaves. Plant Physiol. 2013, 162, 239–253. [Google Scholar] [CrossRef] [Green Version]
- Maia, M.; Ferreira, A.E.N.; Nascimento, R.; Monteiro, F.; Traquete, F.; Marques, A.P.; Cunha, J.; Eiras-Dias, J.E.; Cordeiro, C.; Figueiredo, A.; et al. Integrating metabolomics and targeted gene expression to uncover potential biomarkers of fungal/oomycetes-associated disease susceptibility in grapevine. Sci. Rep. 2020, 10, 15688. [Google Scholar] [CrossRef] [PubMed]
- Filippi, A.; Petrussa, E.; Boscutti, F.; Vuerich, M.; Vrhovsek, U.; Rabiei, Z.; Braidot, E. Bioactive polyphenols modulate enzymes involved in grapevine pathogenesis and chitinase activity at increasing complexity levels. Int. J. Mol. Sci. 2019, 20, 6357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lòpez-Fernàndez, S.; Compant, S.; Vrhovsek, U. Grapevine colonization by endophytic bacteria shifts secondary metabolism and suggests activation of defense pathways. Plant Soil 2016, 405, 155–175. [Google Scholar] [CrossRef]
- Noctor, G.; Queval, G.; Gakière, B. NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J. Exp. Bot. 2006, 57, 1603–1620. [Google Scholar] [CrossRef] [Green Version]
- Martelli, G.P. Directory of virus and virus-like diseases of the grapevine and their agents. J. Plant Pathol. 2014, 96, 1–36. [Google Scholar] [CrossRef]
- Wallis, C.M.; Sudarshana, M.R. Effects of Grapevine red blotch-associated virus (GRBaV) infection on foliar metabolism of grapevines. Can. J. Plant Pathol. 2016, 38, 358–366. [Google Scholar] [CrossRef]
- Rowhani, A.; Uyemoto, J.K.; Golino, D.A.; Daubert, S.D.; Al Rwahnih, M. Viruses Involved in Graft Incompatibility and Decline. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 289–302. [Google Scholar]
- Martelli, G.P. An Overview on Grapevine Viruses, Viroids, and the Diseases They Cause. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 31–46. [Google Scholar]
- Chitarra, W.; Perrone, I.; Avanzato, C.G.; Minio, A.; Boccacci, P.; Santini, D.; Gilardi, G.; Siciliano, I.; Gullino, M.L.; Delledonne, M.; et al. Grapevine Grafting: Scion Transcript Profiling and Defense-Related Metabolites Induced by Rootstocks. Front. Plant Sci. 2017, 8, 654. [Google Scholar] [CrossRef]
- Balmer, A.; Pastor, V.; Glauser, G.; Mauch-Mani, B. Tricarboxylates induce defense priming against bacteria in arabidopsis thaliana. Front. Plant Sci. 2018, 9, 1221. [Google Scholar] [CrossRef]
- Batovska, D.I.; Todorova, I.T.; Parushev, S.P.; Nedelcheva, D.V.; Bankova, V.S.; Popov, S.S.; Ivanova, I.I.; Batovski, S.A. Biomarkers for the prediction of the resistance and susceptibility of grapevine leaves to downy mildew. J. Plant Physiol. 2009, 166, 781–785. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wu, F. Artificially applied vanillic acid changed soil microbial communities in the rhizosphere of cucumber (cucumis sativus L.). Can. J. Soil Sci. 2013, 93, 13–21. [Google Scholar] [CrossRef]
- Guo, X.W.; Wang, B.; Li, K.; Liu, Z.-D.; Han, X.; Xu, S.-J.; Guo, Y.-S.; Xie, H.-J. Effect of 4-hydroxybenzoic acid on grape (Vitis vinifera L.) soil microbial community structure and functional diversity. Biotechnol. Biotechnol. Equip. 2015, 29, 637–645. [Google Scholar] [CrossRef]
- Pate, J.; Shedley, E.; Arthur, D.; Adams, M. Spatial and temporal variations in phloem sap composition of plantation-grown Eucalyptus globulus. Oecologia 1998, 117, 312–322. [Google Scholar] [CrossRef]
- Jeandet, P.; Douillet-Breuil, A.C.; Bessis, R.; Debord, S.; Sbaghi, M.; Adrian, M. Phytoalexins from the vitaceae: Biosynthesis, phytoalexin gene expression in transgenic plants, antifungal activity, and metabolism. J. Agric. Food Chem. 2002, 50, 2731–2741. [Google Scholar] [CrossRef] [PubMed]
- Chitarrini, G.; Soini, E.; Riccadonna, S.; Franceschi, P.; Zulini, L.; Masuero, D.; Vecchione, A.; Stefanini, M.; Di Gaspero, G.; Mattivi, S.; et al. Identification of biomarkers for defense response to Plasmopara viticola in a resistant grape variety. Front. Plant Sci. 2017, 8, 1524. [Google Scholar] [CrossRef] [PubMed]
- Erban, A.; Martinez-Seidel, F.; Rajarathinam, Y.; Dethloff, F.; Orf, I.; Fehrle, I.; Alpers, J.; Beine-Golovchuk, O.; Kopka, J. Multiplexed Profiling and Data Processing Methods to Identify Temperature-Regulated Using Gas Chromatography Coupled to Mass Spectrometry. In Plant Cold Acclimation, 1st ed.; Methods in Molecular Biology; Hincha, D., Zuther, E., Eds.; Humana: New York, NY, USA, 2020; Volume 2156, pp. 203–239. [Google Scholar]
- Luedemann, A.; Strassburg, K.; Erban, A.; Kopka, J. TagFinder for the quantitative analysis of gas chromatography-Mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 2008, 24, 732–737. [Google Scholar] [CrossRef]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmüller, E.; Dörmann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. GMD@CSB.DB: The Golm metabolome database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef] [Green Version]
- Erban, A.; Schauer, N.; Fernie, A.R.; Kopka, J. Nonsupervised Construction and Application. In Metabolomics: Methods and Protocols, 1st ed.; Weckwerth, W., Ed.; Humana Press: Totowa, NJ, USA, 2007; Volume 358, pp. 19–38. [Google Scholar]
- Erban, A.; Fehrle, I.; Martinez-Seidel, F.; Brigante, F.; Lucini Más, A.; Baroni, V.; Wunderlin, D.; Kopka, J. Discovery of food identity markers by metabolomics and machine learning technology. Sci. Rep. 2019, 9, 9697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, J.; Xia, J. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed] [Green Version]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tedesco, S.; Erban, A.; Gupta, S.; Kopka, J.; Fevereiro, P.; Kragler, F.; Pina, A. The Impact of Metabolic Scion–Rootstock Interactions in Different Grapevine Tissues and Phloem Exudates. Metabolites 2021, 11, 349. https://doi.org/10.3390/metabo11060349
Tedesco S, Erban A, Gupta S, Kopka J, Fevereiro P, Kragler F, Pina A. The Impact of Metabolic Scion–Rootstock Interactions in Different Grapevine Tissues and Phloem Exudates. Metabolites. 2021; 11(6):349. https://doi.org/10.3390/metabo11060349
Chicago/Turabian StyleTedesco, Sara, Alexander Erban, Saurabh Gupta, Joachim Kopka, Pedro Fevereiro, Friedrich Kragler, and Ana Pina. 2021. "The Impact of Metabolic Scion–Rootstock Interactions in Different Grapevine Tissues and Phloem Exudates" Metabolites 11, no. 6: 349. https://doi.org/10.3390/metabo11060349
APA StyleTedesco, S., Erban, A., Gupta, S., Kopka, J., Fevereiro, P., Kragler, F., & Pina, A. (2021). The Impact of Metabolic Scion–Rootstock Interactions in Different Grapevine Tissues and Phloem Exudates. Metabolites, 11(6), 349. https://doi.org/10.3390/metabo11060349





