Metabolomics and Dual RNA-Sequencing on Root Nodules Revealed New Cellular Functions Controlled by Paraburkholderia phymatum NifA
Abstract
:1. Introduction
2. Results and Discussion
2.1. Metabolomics Analysis of Nodules Infected by a nifA Mutant Strain
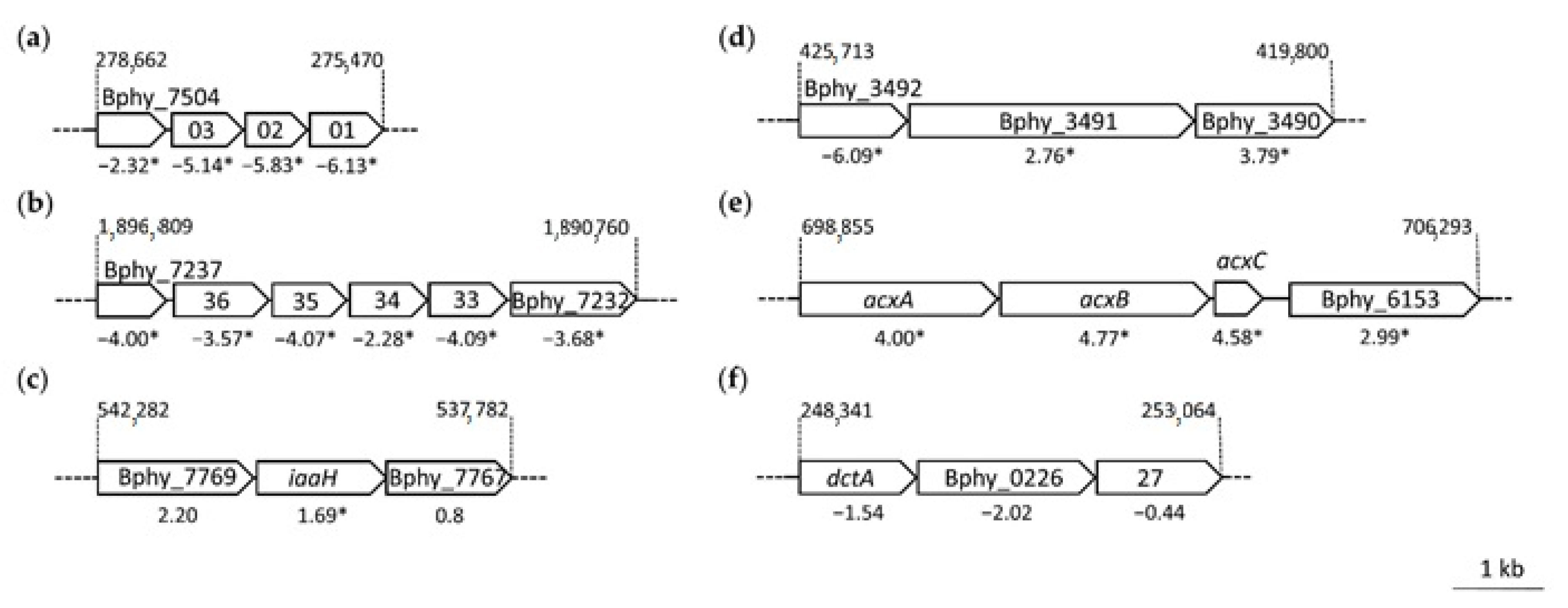
2.2. RNA-Sequencing and P. phymatum Differential Gene Expression
2.3. RNA-Sequencing and P. vulgaris Differential Gene Expression
2.4. The nifA Mutant Strain Produces a Higher Amount of Auxin Compounds
2.5. The nifA Mutant Strain Is Affected in Growth on Plates Containing Succinate, Malate, Glutamine and Glutamate as Only Carbon Sources
2.6. NifA Plays a Role in Osmotic Tolerance
3. Materials and Methods
3.1. Bacterial Strains, Media and Cultivation
3.2. Plant Growth Conditions
3.3. Metabolite Extraction and Data Analysis
3.4. RNA-Sequencing and Data Processing
3.5. Construction of P. phymatum STM815 Complemented Strains
3.6. Auxin Production
3.7. Carbon Source and Stress Survival Assay
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fowler, D.; Coyle, M.; Skiba, U.; Sutton, M.A.; Cape, J.N.; Reis, S.; Sheppard, L.J.; Jenkins, A.; Grizzetti, B.; Galloway, J.N.; et al. The global nitrogen cycle in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368. [Google Scholar] [CrossRef]
- Poole, P.; Allaway, D. Carbon and nitrogen metabolism in Rhizobium. Adv. Microb. Physiol. 2000, 43, 117–163. [Google Scholar] [CrossRef] [PubMed]
- Yurgel, S.N.; Kahn, M.L. Dicarboxylate transport by rhizobia. FEMS Microbiol. Rev. 2004, 28, 489–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udvardi, M.; Poole, P.S. Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 2013, 64, 781–805. [Google Scholar] [CrossRef] [Green Version]
- Masson-Boivin, C.; Giraud, E.; Perret, X.; Batut, J. Establishing nitrogen-fixing symbiosis with legumes: How many rhizobium recipes? Trends Microbiol. 2009, 17, 458–466. [Google Scholar] [CrossRef]
- Bontemps, C.; Elliott, G.N.; Simon, M.F.; Dos Reis Júnior, F.B.; Gross, E.; Lawton, R.C.; Neto, N.E.; De Fátima Loureiro, M.; De Faria, S.M.; Sprent, J.I.; et al. Burkholderia species are ancient symbionts of legumes. Mol. Ecol. 2010, 19, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Moulin, L.; Munive, A.; Dreyfus, B.; Boivin-Masson, C. Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature 2001, 411, 948. [Google Scholar] [CrossRef]
- Elliott, G.N.; Chen, W.M.; Chou, J.H.; Wang, H.C.; Sheu, S.Y.; Perin, L.; Reis, V.M.; Moulin, L.; Simon, M.F.; Bontemps, C.; et al. Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 2006, 173, 168–180. [Google Scholar] [CrossRef]
- Gyaneshwar, P.; Hirsch, A.M.; Moulin, L.; Chen, W.; Elliott, G.N.; Bontemps, C.; Santos, P.E.L.; Gross, E.; Bueno, F.; Sprent, J.I.; et al. Legume-nodulating Betaproteobacteria: Diversity, Host Range, and Future Prospects. Mol. Plant Microbe Interact. 2011, 24, 1276–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, R.P.N.; Tisseyre, P.; Melkonian, R.; Chaintreuil, C.; Miché, L.; Klonowska, A.; Gonzalez, S.; Bena, G.; Laguerre, G.; Moulin, L. Genetic diversity of Mimosa pudica rhizobial symbionts in soils of French Guiana: Investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol. Ecol. 2012, 79, 487–503. [Google Scholar] [CrossRef]
- Moulin, L.; Klonowska, A.; Caroline, B.; Booth, K.; Vriezen, J.A.C.; Melkonian, R.; James, E.K.; Young, J.P.W.; Bena, G.; Hauser, L.; et al. Complete Genome sequence of Burkholderia phymatum STM815T, a broad host range and efficient nitrogen-fixing symbiont of Mimosa species. Stand. Genomic Sci. 2014, 9, 763–774. [Google Scholar] [CrossRef] [Green Version]
- Lemaire, B.; Chimphango, S.B.M.; Stirton, C.; Rafudeen, S.; Honnay, O.; Smets, E.; Chen, W. Biogeographical patterns of legume-nodulating Burkholderia spp.: From African fynbos to continental scales. Appl. Environ. Microbiol. 2016, 82, 5099–5115. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.; Hirsch, A.M. Signals and responses: Choreographing the complex interaction between legumes and α- and β-rhizobia. Plant Signal. Behav. 2006, 1, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Ledermann, R.; Schulte, C.C.M.; Poole, P.S. How rhizobia adapt to the nodule environment. J. Bacteriol. 2021, 203. [Google Scholar] [CrossRef]
- Kuzma, M.M.; Hunt, S.; Layzell, D.B. Role of oxygen in the limitation and inhibition of nitrogenase activity and respiration rate in individual soybean nodules. Plant Physiol. 1993, 101, 161–169. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.; Kahn, D. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2004, 2, 621–631. [Google Scholar] [CrossRef]
- Fischer, H.M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 1994, 58, 352–386. [Google Scholar] [CrossRef]
- Lardi, M.; Liu, Y.; Purtschert, G.; de Campos, S.B.; Pessi, G. Transcriptome analysis of Paraburkholderia phymatum under nitrogen starvation and during symbiosis with Phaseolus vulgaris. Genes 2017, 8, 389. [Google Scholar] [CrossRef] [Green Version]
- Lardi, M.; Liu, Y.; Giudice, G.; Ahrens, C.H.; Zamboni, N.; Pessi, G. Metabolomics and transcriptomics identify multiple downstream targets of Paraburkholderia phymatum σ54 during symbiosis with Phaseolus vulgaris. Int. J. Mol. Sci. 2018, 19, 1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobik, C.; Meilhoc, E.; Batut, J. FixJ: A major regulator of the oxygen limitation response and late symbiotic functions of Sinorhizobium meliloti. J. Bacteriol. 2006, 188, 4890–4902. [Google Scholar] [CrossRef] [Green Version]
- Hauser, F.; Lindemann, A.; Vuilleumier, S.; Patrignani, A.; Schlapbach, R.; Fischer, H.M.; Hennecke, H. Design and validation of a partial-genome microarray for transcriptional profiling of the Bradyrhizobium japonicum symbiotic gene region. Mol. Genet. Genomics 2006, 275, 55–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauser, F.; Pessi, G.; Friberg, M.; Weber, C.; Rusca, N.; Lindemann, A.; Fischer, H.M.; Hennecke, H. Dissection of the Bradyrhizobium japonicum NifA+σ54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol. Genet. Genomics 2007, 278, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Salazar, E.; Javier Díaz-Mejía, J.; Moreno-Hagelsieb, G.; Martínez-Batallar, G.; Mora, Y.; Mora, J.; Encarnación, S. Characterization of the NifA-RpoN regulon in Rhizobium etli in free life and in symbiosis with Phaseolus vulgaris. Appl. Environ. Microbiol. 2010, 76, 4510–4520. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, A.; Reinhold-Hurek, B. Transcriptional profiling of nitrogen fixation and the role of NifA in the diazotrophic endophyte Azoarcus sp. strain BH72. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Lardi, M.; Murset, V.; Fischer, H.M.; Mesa, S.; Ahrens, C.H.; Zamboni, N.; Pessi, G. Metabolomic profiling of Bradyrhizobium diazoefficiens -induced root nodules reveals both host plant-specific and developmental signatures. Int. J. Mol. Sci. 2016, 17, 815. [Google Scholar] [CrossRef] [Green Version]
- Bellés-Sancho, P.; Lardi, M.; Liu, Y.; Hug, S.; Pinto-Carbó, M.A.; Zamboni, N.; Pessi, G. Paraburkholderia phymatum homocitrate synthase NifV plays a key role for nitrogenase activity during symbiosis with papilionoids and in free-living growth conditions. Cells 2021, 10, 952. [Google Scholar] [CrossRef]
- Barsomian, G.D.; Urzainqui, A.; Lohman, K.; Walker, G.C. Rhizobium meliloti mutants unable to synthesize anthranilate display a novel symbiotic phenotype. J. Bacteriol. 1992, 174, 4416–4426. [Google Scholar] [CrossRef] [Green Version]
- Prasad, C.K.; Vineetha, K.E.; Hassani, R.; Gupta, R.; Randhawa, G.S. Isolation and symbiotic characterization of aromatic amino acid auxotrophs of Sinorhizobium meliloti. Indian J. Exp. Biol. 2000, 38, 1041–1049. [Google Scholar]
- Flores-Tinoco, C.E.; Tschan, F.; Fuhrer, T.; Margot, C.; Sauer, U.; Christen, M.; Christen, B. Co-catabolism of arginine and succinate drives symbiotic nitrogen fixation. Mol. Syst. Biol. 2020, 16, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.; Delmotte, N.; Ahrens, C.H.; Omasits, U.; Schneider, K.; Danza, F.; Padhi, B.; Murset, V.; Braissant, O.; Vorholt, J.A.; et al. A link between arabinose utilization and oxalotrophy in Bradyrhizobium japonicum. Appl. Environ. Microbiol. 2014, 80, 2094–2101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, L.J.; Morris, P.; Hooker, J.E. Perception and modification of plant flavonoid signals by rhizosphere microorganisms. Environ. Microbiol. 2006, 8, 1867–1880. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Pandey, A.K. Chemistry and biological activities of flavonoids: An overview. Sci. World J. 2013, 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peer, W.A.; Murphy, A.S. Flavonoids and auxin transport: Modulators or regulators? Trends Plant Sci. 2007, 12, 556–563. [Google Scholar] [CrossRef]
- Fridman, Y.; Savaldi-Goldstein, S. Brassinosteroids in growth control: How, when and where. Plant Sci. 2013, 209, 24–31. [Google Scholar] [CrossRef] [PubMed]
- McGuiness, P.N.; Reid, J.B.; Foo, E. Brassinosteroids play multiple roles in nodulation of pea via interactions with ethylene and auxin. Planta 2020, 252, 1–8. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukasinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 2020, 32, 298–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadzieja, M.; Kelly, S.; Stougaard, J.; Reid, D. Epidermal auxin biosynthesis facilitates rhizobial infection in Lotus japonicus. Plant J. 2018, 95, 101–111. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, B.J.; Ross, J.J.; Reid, J.B. Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol. 2005, 138, 2396–2405. [Google Scholar] [CrossRef] [Green Version]
- Foo, E.; Ferguson, B.J.; Reid, J.B. The potential roles of strigolactones and brassinosteroids in the autoregulation of nodulation pathway. Ann. Bot. 2014, 113, 1037–1045. [Google Scholar] [CrossRef] [Green Version]
- Verbon, E.H.; Trapet, P.L.; Stringlis, I.A.; Kruijs, S.; Bakker, P.A.H.M.; Pieterse, C.M.J. Iron and Immunity. Annu. Rev. Phytopathol. 2017, 55, 355–375. [Google Scholar] [CrossRef]
- Yura, T.; Nakahigashi, K. Regulation of the heat-shock response. Curr. Opin. Microbiol. 1999, 2, 153–158. [Google Scholar] [CrossRef]
- Mitsui, H.; Sato, T.; Sato, Y.; Ito, N.; Minamisawa, K. Sinorhizobium meliloti RpoH1 is required for effective nitrogen-fixing symbiosis with alfalfa. Mol. Genet. Genomics 2004, 271, 416–425. [Google Scholar] [CrossRef]
- Delory, M.; Hallez, R.; Letesson, J.J.; De Bolle, X. An RpoH-like heat shock sigma factor is involved in stress response and virulence in Brucella melitensis 16M. J. Bacteriol. 2006, 188, 7707–7710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Salazar, J.M.; Sandoval-Calderón, M.; Guo, X.; Castillo-Ramírez, S.; Reyes, A.; Loza, M.G.; Rivera, J.; Alvarado-Affantranger, X.; Sánchez, F.; González, V.; et al. The Rhizobium etli RpoH1 and RpoH2 sigma factors are involved in different stress responses. Microbiology 2009, 155, 386–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brahmachary, P.; Wang, G.; Benoit, S.L.; Weinberg, M.V.; Maier, R.J.; Hoover, T.R. The human gastric pathogen Helicobacter pylori has a potential acetone carboxylase that enhances its ability to colonize mice. BMC Microbiol. 2008, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Tellis, M.; Mathur, M.; Gurjar, G.; Kadoo, N.; Gupta, V. Identification and functionality prediction of pathogenesis-related protein 1 from legume family. Proteins Struct. Funct. Bioinforma. 2017, 85, 2066–2080. [Google Scholar] [CrossRef]
- Tronconi, M.A.; Fahnenstich, H.; Gerrard Weehler, M.C.; Andreo, C.S.; Flügge, U.I.; Drincovich, M.F.; Maurino, V.G. Arabidopsis NAD-malic enzyme functions as a homodimer and heterodimer and has a major impact on nocturnal metabolism. Plant Physiol. 2008, 146, 1540–1552. [Google Scholar] [CrossRef] [Green Version]
- Peng, M.; Bi, Y.M.; Zhu, T.; Rothstein, S.J. Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol. Biol. 2007, 65, 775–797. [Google Scholar] [CrossRef]
- Couzigou, J.M.; Mondy, S.; Sahl, L.; Gourion, B.; Ratet, P. To be or noot to be: Evolutionary tinkering for symbiotic organ identity. Plant Signal. Behav. 2013, 8, 3–6. [Google Scholar] [CrossRef] [Green Version]
- Shimada, S.; Komatsu, T.; Yamagami, A.; Nakazawa, M.; Matsui, M.; Kawaide, H.; Natsume, M.; Osada, H.; Asami, T.; Nakanoa, T. Formation and dissociation of the BSS1 protein complex regulates plant development via brassinosteroid signaling. Plant Cell 2015, 27, 375–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, D.E.; Ferguson, B.J.; Hayashi, S.; Lin, Y.H.; Gresshoff, P.M. Molecular mechanisms controlling legume autoregulation of nodulation. Ann. Bot. 2011, 108, 789–795. [Google Scholar] [CrossRef] [Green Version]
- Schofield, R.A.; Bi, Y.M.; Kant, S.; Rothstein, S.J. Over-expression of STP13, a hexose transporter, improves plant growth and nitrogen use in Arabidopsis thaliana seedlings. Plant Cell Environ. 2009, 32, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pii, Y.; Crimi, M.; Cremonese, G.; Spena, A.; Pandolfini, T. Auxin and nitric oxide control indeterminate nodule formation. BMC Plant Biol. 2007, 7, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosendahl, L.; Jochimsen, B.U. In vitro indole-3-acetic acid uptake in symbiosomes from soybean (Glycine max L.) root nodules. Symbiosis 1995, 19, 99–110. [Google Scholar]
- Pierre, O.; Engler, G.; Hopkins, J.; Brau, F.; Boncompagni, E.; Hérouart, D. Peribacteroid space acidification: A marker of mature bacteroid functioning in Medicago truncatula nodules. Plant Cell Environ. 2013, 36, 2059–2070. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.H. Experiments in Molecular Genetics. In The Quarterly Review of Biology; Cold Spring Harbor Lab Press: Cold Spring Harbor, NY, USA, 1972; pp. 352–355. ISBN 0879691069. [Google Scholar]
- Talbi, C.; Delgado, M.J.; Girard, L.; Ramírez-Trujillo, A.; Caballero-Mellado, J.; Bedmar, E.J. Burkholderia phymatum strains capable of nodulating Phaseolus vulgaris are present in moroccan soils. Appl. Environ. Microbiol. 2010, 76, 4587–4591. [Google Scholar] [CrossRef] [Green Version]
- Vincent, J.M. A Manual for the Practical Study of the Root-Nodule Bacteria; IBP Handbk 15 Oxford and Edinburgh; Blackwell Scientific Publications: Hoboken, NJ, USA, 1970; ISBN 0632064102. [Google Scholar]
- Fuhrer, T.; Heer, D.; Begemann, B.; Zamboni, N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 2011, 83, 7074–7080. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, 388–396. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Pessi, G.; Ahrens, C.H.; Rehrauer, H.; Lindemann, A.; Hauser, F.; Fischer, H.M.; Hennecke, H. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol. Plant-Microbe Interact. 2007, 20, 1353–1363. [Google Scholar] [CrossRef] [Green Version]
- Lardi, M.; Aguilar, C.; Pedrioli, A.; Omasits, U.; Suppiger, A.; Cárcamo-Oyarce, G.; Schmid, N.; Ahrens, C.H.; Eberl, L.; Pessi, G. σ54-dependent response to nitrogen limitation and virulence in Burkholderia cenocepacia strain H111. Appl. Environ. Microbiol. 2015, 81, 4077–4089. [Google Scholar] [CrossRef] [Green Version]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression and sequence analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gravel, V.; Antoun, H.; Tweddell, R.J. Growth stimulation and fruit yield improvement of greenhouse tomato plants by inoculation with Pseudomonas putida or Trichoderma atroviride: Possible role of indole acetic acid (IAA). Soil Biol. Biochem. 2007, 39, 1968–1977. [Google Scholar] [CrossRef]
- Gordon, S.A.; Weber, R.P. Colorimetric estimation of indole-3-acetic acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Bellich, B.; Hug, S.; Eberl, L.; Cescutti, P.; Pessi, G. The exopolysaccharide cepacian plays a role in the establishment of the Paraburkholderia phymatum—Phaseolus vulgaris symbiosis. Front. Microbiol. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Elzer, P.H.; Roop, R.M.; Kovach, M.E.; Robertson, G.T.; Peterson, K.M.; Steven Hill, D.; Farris, M.A. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Phadnis, S.H.; Berg, D.E. Identification of base pairs in the outside end of insertion sequence IS50 that are needed for IS50 and Tn5 transposition. Proc. Natl. Acad. Sci. USA 1987, 84, 9118–9122. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Locus ID 1 | Description 1 | Old Locus ID 1 | Gene Name 1 | log2FC (nifA vs. wt) 2 |
|---|---|---|---|---|
| Genes up-regulated in nifA mutant bacteroids | ||||
| BPHY_RS41160 | hypothetical protein | Bphy_7816 | 4.47 | |
| BPHY_RS38115 | hypothetical protein | Bphy_7726 | 2.53 | |
| BPHY_RS38105 | ATP-grasp domain-containing protein | Bphy_7724 | 2.26 | |
| BPHY_RS36585 | DNA-binding protein | Bphy_7384 | 2.24 | |
| BPHY_RS38320 | indoleacetamide hydrolase | Bphy_7768 | iaaH | 1.69 |
| Genes down-regulated in nifA mutant bacteroids | ||||
| BPHY_RS37800 | branched-chain amino acid ABC transporter substrate-binding protein | Bphy_7652 | −2.10 | |
| BPHY_RS37125 | GntR family transcriptional regulator | Bphy_7505 | −2.18 | |
| BPHY_RS37205 | hypothetical protein | Bphy_7521 | −2.25 | |
| BPHY_RS37050 | biosynthetic-type acetolactate synthase large subunit | Bphy_7490 | ilvB | −2.27 |
| BPHY_RS37120 | ABC transporter substrate-binding protein | Bphy_7504 | −2.32 | |
| BPHY_RS38545 | iron-sulfur cluster assembly accessory protein | Bphy_7815 | −2.33 | |
| BPHY_RS38560 | porin | Bphy_7819 | −2.47 | |
| BPHY_RS37795 | aminotransferase class III-fold pyridoxal phosphate-dependent enzyme | Bphy_7651 | −2.51 | |
| BPHY_RS37925 | hypothetical protein | Bphy_7682 | −2.52 | |
| BPHY_RS37805 | IS630 family transposase | Bphy_7653 | −2.57 | |
| BPHY_RS37025 | DMT family transporter | Bphy_7485 | −2.58 | |
| BPHY_RS38285 | carbon starvation protein A | Bphy_7759 | −2.62 | |
| BPHY_RS37790 | M81 family peptidase | Bphy_7649 | −2.71 | |
| BPHY_RS37915 | hypothetical protein | Bphy_7680 | −2.84 | |
| BPHY_RS41075 | hypothetical protein | Bphy_7694 | −2.95 | |
| BPHY_RS37775 | ABC transporter permease | Bphy_7646 | −3.00 | |
| BPHY_RS36465 | hypothetical protein | Bphy_7358 | −3.01 | |
| BPHY_RS37930 | hypothetical protein | −3.13 | ||
| BPHY_RS38520 | nitrogenase molybdenum-iron protein alpha chain | Bphy_7809 | −3.16 | |
| BPHY_RS37765 | ABC transporter substrate-binding protein | Bphy_7644 | −3.16 | |
| BPHY_RS38135 | nitrogenase iron-molybdenum cofactor biosynthesis protein | Bphy_7730 | nifN | −3.16 |
| BPHY_RS37090 | pyridoxal-phosphate dependent enzyme | Bphy_7498 | −3.19 | |
| BPHY_RS41155 | hypothetical protein | Bphy_7814 | −3.27 | |
| BPHY_RS41070 | cytolethal distending toxin subunit B family protein | Bphy_7691 | −3.30 | |
| BPHY_RS37780 | ABC transporter ATP-binding protein | Bphy_7647 | −3.40 | |
| BPHY_RS40855 | IS110 family transposase | Bphy_7484 | −3.40 | |
| BPHY_RS37770 | ABC transporter permease | Bphy_7645 | −3.49 | |
| BPHY_RS37045 | ISL3 family transposase | Bphy_7489 | −3.60 | |
| BPHY_RS37920 | hypothetical protein | Bphy_7681 | −3.62 | |
| BPHY_RS38485 | electron transfer flavoprotein subunit alpha/FixB family protein | Bphy_7803 | fixB | −3.63 |
| BPHY_RS41535 | IS3 family transposase | −3.63 | ||
| BPHY_RS36015 | HyaD/HybD family hydrogenase maturation endopeptidase | Bphy_7262 | hybD | −3.70 |
| BPHY_RS36485 | hypothetical protein | −3.83 | ||
| BPHY_RS41135 | electron transfer flavoprotein alpha/beta-subunit | Bphy_7804 | −3.90 | |
| BPHY_RS36645 | alpha/beta fold hydrolase | Bphy_7397 | −3.91 | |
| BPHY_RS38310 | cytochrome P450 | Bphy_7766 | −4.00 | |
| BPHY_RS40820 | IS5/IS1182 family transposase | −4.06 | ||
| BPHY_RS38020 | radical SAM protein | Bphy_7706 | −4.07 | |
| BPHY_RS37785 | ABC transporter ATP-binding protein | Bphy_7648 | −4.10 | |
| BPHY_RS37970 | 7-carboxy-7-deazaguanine synthase QueE | Bphy_7690 | −4.11 | |
| BPHY_RS37005 | GntR family transcriptional regulator | Bphy_7481 | −4.44 | |
| BPHY_RS36735 | ArgP/LysG family DNA-binding transcriptional regulator | Bphy_7420 | −4.51 | |
| BPHY_RS37085 | Lrp/AsnC family transcriptional regulator | Bphy_7497 | −4.58 | |
| BPHY_RS38495 | FAD-binding oxidoreductase | Bphy_7805 | −4.62 | |
| BPHY_RS36730 | sodium:solute symporter | Bphy_7419 | −4.66 | |
| BPHY_RS35975 | carbamoyltransferase | Bphy_7254 | hypF | −4.74 |
| BPHY_RS38270 | hypothetical protein | −4.78 | ||
| BPHY_RS37910 | IS6 family transposase | Bphy_7679 | −4.80 | |
| BPHY_RS36725 | fatty acid desaturase | Bphy_7418 | −4.87 | |
| BPHY_RS38515 | nitrogenase iron protein | Bphy_7808 | nifH | −4.88 |
| BPHY_RS36010 | HypC/HybG/HupF family hydrogenase formation chaperone | Bphy_7261 | hypC | −4.95 |
| BPHY_RS38025 | glycosyltransferase | Bphy_7707 | −4.98 | |
| BPHY_RS38365 | putative nitrogen fixation protein | Bphy_7777 | nifT | −5.01 |
| BPHY_RS35985 | hydrogenase maturation nickel metallochaperone | Bphy_7256 | hypA | −5.14 |
| BPHY_RS37115 | histidine ABC transporter permease | Bphy_7503 | hisQ | −5.14 |
| BPHY_RS38425 | class I SAM-dependent methyltransferase | Bphy_7790 | −5.14 | |
| BPHY_RS36060 | hydrogenase expression/formation protein | Bphy_7271 | hypE | −5.17 |
| BPHY_RS37065 | porin | Bphy_7493 | −5.33 | |
| BPHY_RS38140 | nitrogen fixation protein | Bphy_7731 | nifX | −5.35 |
| BPHY_RS36005 | hydrogenase | Bphy_7260 | hyaE | −5.43 |
| BPHY_RS36050 | hypothetical protein | −5.51 | ||
| BPHY_RS36020 | Ni/Fe-hydrogenase, b-type cytochrome subunit | Bphy_7263 | cybH | −5.63 |
| BPHY_RS36740 | SDR family oxidoreductase | Bphy_7421 | −5.68 | |
| BPHY_RS40825 | glutamine amidotransferase class-II | Bphy_7471 | −5.75 | |
| BPHY_RS38160 | hypothetical protein | Bphy_7735 | −5.82 | |
| BPHY_RS37075 | hypothetical protein | Bphy_7495 | −5.83 | |
| BPHY_RS37110 | histidine ABC transporter permease | Bphy_7502 | hisM | −5.83 |
| BPHY_RS41065 | cupin | Bphy_7689 | −5.83 | |
| BPHY_RS38225 | hypothetical protein | Bphy_7748 | −5.88 | |
| BPHY_RS38130 | nitrogenase iron-molybdenum cofactor biosynthesis protein | Bphy_7729 | nifE | −5.89 |
| BPHY_RS37735 | NAD-dependent epimerase/dehydratase family protein | Bphy_7636 | −5.92 | |
| BPHY_RS41080 | AraC family transcriptional regulator | Bphy_7704 | −5.92 | |
| BPHY_RS37080 | MmgE/PrpD family protein | Bphy_7496 | −5.97 | |
| BPHY_RS37105 | ATP-binding cassette domain-containing protein | Bphy_7501 | −6.13 | |
| BPHY_RS36745 | histidinol dehydrogenase | Bphy_7422 | hisD | −6.13 |
| BPHY_RS36000 | hydrogenase expression/formation protein | Bphy_7259 | −6.16 | |
| BPHY_RS35990 | HupK protein | Bphy_7257 | hupK | −6.18 |
| BPHY_RS41530 | transposase | Bphy_7402 | −6.22 | |
| BPHY_RS37965 | class I SAM-dependent methyltransferase | Bphy_7688 | −6.25 | |
| BPHY_RS36055 | cysteine hydrolase | Bphy_7270 | −6.29 | |
| BPHY_RS37070 | ABC transporter substrate-binding protein | Bphy_7494 | −6.33 | |
| BPHY_RS40990 | IS5/IS1182 family transposase | Bphy_7635 | −6.34 | |
| BPHY_RS37100 | aminopeptidase P family protein | Bphy_7500 | −6.59 | |
| BPHY_RS38155 | nitrogen fixation protein | Bphy_7734 | nifQ | −6.59 |
| BPHY_RS35995 | [NiFe]-hydrogenase assembly, chaperone | Bphy_7258 | hybE | −6.61 |
| BPHY_RS41545 | FAD-dependent oxidoreductase | −6.68 | ||
| BPHY_RS41140 | hypothetical protein | Bphy_7806 | −6.82 | |
| BPHY_RS41095 | hypothetical protein | Bphy_7746 | −6.85 | |
| BPHY_RS39040 | pyridoxal-phosphate dependent enzyme | Bphy_7479 | −6.85 | |
| BPHY_RS38150 | ferredoxin III, nif-specific | Bphy_7733 | fdxB | −6.86 |
| BPHY_RS36650 | IS5/IS1182 family transposase | Bphy_7398 | −6.90 | |
| BPHY_RS36025 | nickel-dependent hydrogenase large subunit | Bphy_7264 | −6.96 | |
| BPHY_RS38145 | hypothetical protein | Bphy_7732 | −7.01 | |
| BPHY_RS38245 | ankyrin repeat domain-containing protein | Bphy_7751 | −7.11 | |
| BPHY_RS38170 | protein FixC | Bphy_7737 | fixC | −7.11 |
| BPHY_RS37030 | ISL3 family transposase | Bphy_7486 | −7.17 | |
| BPHY_RS37740 | SDR family NAD(P)-dependent oxidoreductase | Bphy_7637 | −7.21 | |
| BPHY_RS38230 | IS630 family transposase | Bphy_7749 | −7.34 | |
| BPHY_RS38255 | nitrogenase iron protein | Bphy_7753 | nifH | −7.40 |
| BPHY_RS36670 | aldehyde dehydrogenase family protein | Bphy_7406 | −7.41 | |
| BPHY_RS38525 | LysR family transcriptional regulator | Bphy_7810 | −7.42 | |
| BPHY_RS36065 | carbamoyltransferase | Bphy_7272 | hypF | −7.47 |
| BPHY_RS36035 | hypothetical protein | −7.51 | ||
| BPHY_RS36675 | maltose alpha-D-glucosyltransferase | Bphy_7407 | treS | −7.53 |
| BPHY_RS38220 | putative nitrogen fixation protein | Bphy_7747 | nifT | −7.55 |
| BPHY_RS36040 | diaminobutyrate-2-oxoglutarate transaminase | Bphy_7266 | ectB | −7.58 |
| BPHY_RS38265 | nitrogenase molybdenum-iron protein subunit beta | Bphy_7755 | nifK | −7.67 |
| BPHY_RS38240 | zinc ribbon domain-containing protein | Bphy_7750 | −7.72 | |
| BPHY_RS38185 | nitrogenase stabilizing/protective protein | Bphy_7740 | nifW | −7.78 |
| BPHY_RS36655 | group II intron reverse transcriptase/maturase | Bphy_7399 | ltrA | −7.78 |
| BPHY_RS36045 | GNAT family N-acetyltransferase | Bphy_7268 | −8.06 | |
| BPHY_RS40995 | IS5/IS1182 family transposase | Bphy_7638 | −8.07 | |
| BPHY_RS37730 | multi anti extrusion protein | Bphy_7634 | matE | −8.21 |
| BPHY_RS37000 | ISL3 family transposase | Bphy_7480 | −8.22 | |
| BPHY_RS36990 | GHMP kinase | Bphy_7478 | −8.24 | |
| BPHY_RS38190 | homocitrate synthase | Bphy_7741 | nifV | −8.28 |
| BPHY_RS37725 | D-alanine-D-alanine ligase | Bphy_7633 | −8.45 | |
| BPHY_RS38165 | ferredoxin | Bphy_7736 | fixX | −8.93 |
| BPHY_RS38175 | electron transfer flavoprotein subunit alpha/FixB family protein | Bphy_7738 | fixB | −9.16 |
| BPHY_RS36680 | ABC transporter ATP-binding protein | Bphy_7408 | −9.33 | |
| BPHY_RS38195 | nitrogenase cofactor biosynthesis protein | Bphy_7742 | nifB | −9.83 |
| BPHY_RS38260 | nitrogenase molybdenum-iron protein alpha chain | Bphy_7754 | nifD | −10.05 |
| BPHY_RS36030 | twin-arginine translocation signal domain-containing protein | Bphy_7265 | hydA | −10.25 |
| BPHY_RS38180 | electron transfer flavoprotein beta subunit/FixA family protein | Bphy_7739 | fixA | −10.52 |
| BPHY_RS38505 | hypothetical protein | Bphy_7807 | −11.11 | |
| BPHY_RS38250 | hypothetical protein | Bphy_7752 | −11.54 | |
| BPHY_RS40755 | IS66 family transposase | Bphy_7404 | NA | |
| BPHY_RS37010 | IS3 family transposase | Bphy_7482 | NA | |
| BPHY_RS36905 | glycolate oxidase subunit G | Bphy_7454 | glcD | NA |
| BPHY_RS41325 | diaminobutyrate-2-oxoglutarate transaminase | Bphy_7273 | NA | |
| BPHY_RS38360 | nitrogen fixation protein | Bphy_7776 | nifZ | NA |
| BPHY_RS37060 | hypothetical protein | NA | ||
| BPHY_RS38235 | hypothetical protein | NA | ||
| BPHY_RS35970 | HypC/HybG/HupF family hydrogenase formation chaperone | Bphy_7253 | hypC | NA |
| BPHY_RS38210 | nitrogen fixation protein | Bphy_7745 | nifZ | NA |
| BPHY_RS40615 | hypothetical protein | Bphy_7267 | NA | |
| BPHY_RS40750 | hypothetical protein | NA | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellés-Sancho, P.; Lardi, M.; Liu, Y.; Eberl, L.; Zamboni, N.; Bailly, A.; Pessi, G. Metabolomics and Dual RNA-Sequencing on Root Nodules Revealed New Cellular Functions Controlled by Paraburkholderia phymatum NifA. Metabolites 2021, 11, 455. https://doi.org/10.3390/metabo11070455
Bellés-Sancho P, Lardi M, Liu Y, Eberl L, Zamboni N, Bailly A, Pessi G. Metabolomics and Dual RNA-Sequencing on Root Nodules Revealed New Cellular Functions Controlled by Paraburkholderia phymatum NifA. Metabolites. 2021; 11(7):455. https://doi.org/10.3390/metabo11070455
Chicago/Turabian StyleBellés-Sancho, Paula, Martina Lardi, Yilei Liu, Leo Eberl, Nicola Zamboni, Aurélien Bailly, and Gabriella Pessi. 2021. "Metabolomics and Dual RNA-Sequencing on Root Nodules Revealed New Cellular Functions Controlled by Paraburkholderia phymatum NifA" Metabolites 11, no. 7: 455. https://doi.org/10.3390/metabo11070455
APA StyleBellés-Sancho, P., Lardi, M., Liu, Y., Eberl, L., Zamboni, N., Bailly, A., & Pessi, G. (2021). Metabolomics and Dual RNA-Sequencing on Root Nodules Revealed New Cellular Functions Controlled by Paraburkholderia phymatum NifA. Metabolites, 11(7), 455. https://doi.org/10.3390/metabo11070455







