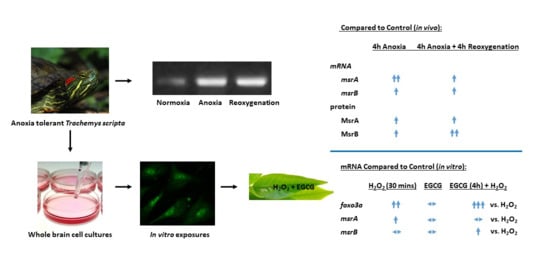
Differential Responses of Methionine Sulfoxide Reductases A and B to Anoxia and Oxidative Stress in the Freshwater Turtle Trachemys scripta
Abstract
:1. Introduction
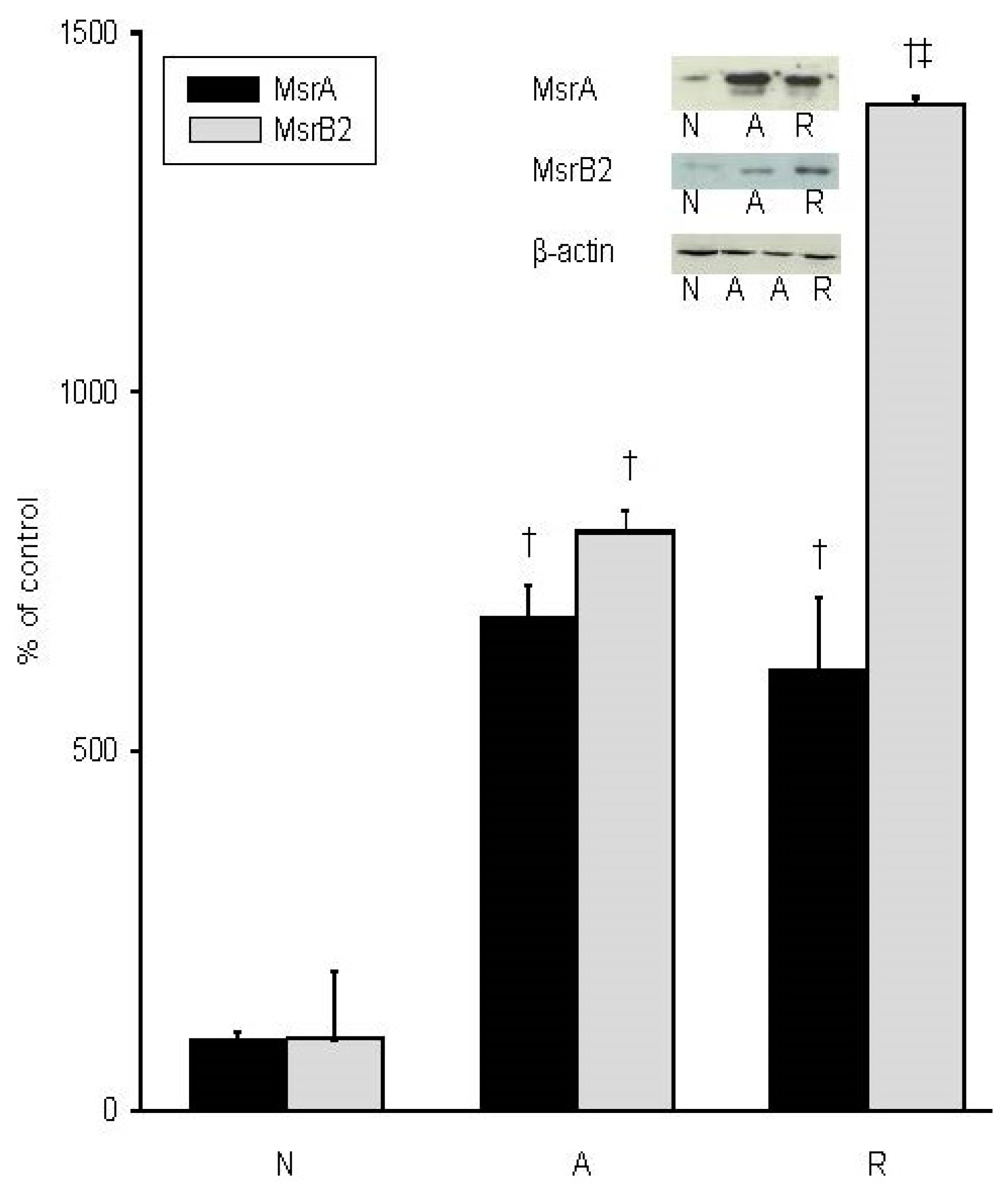
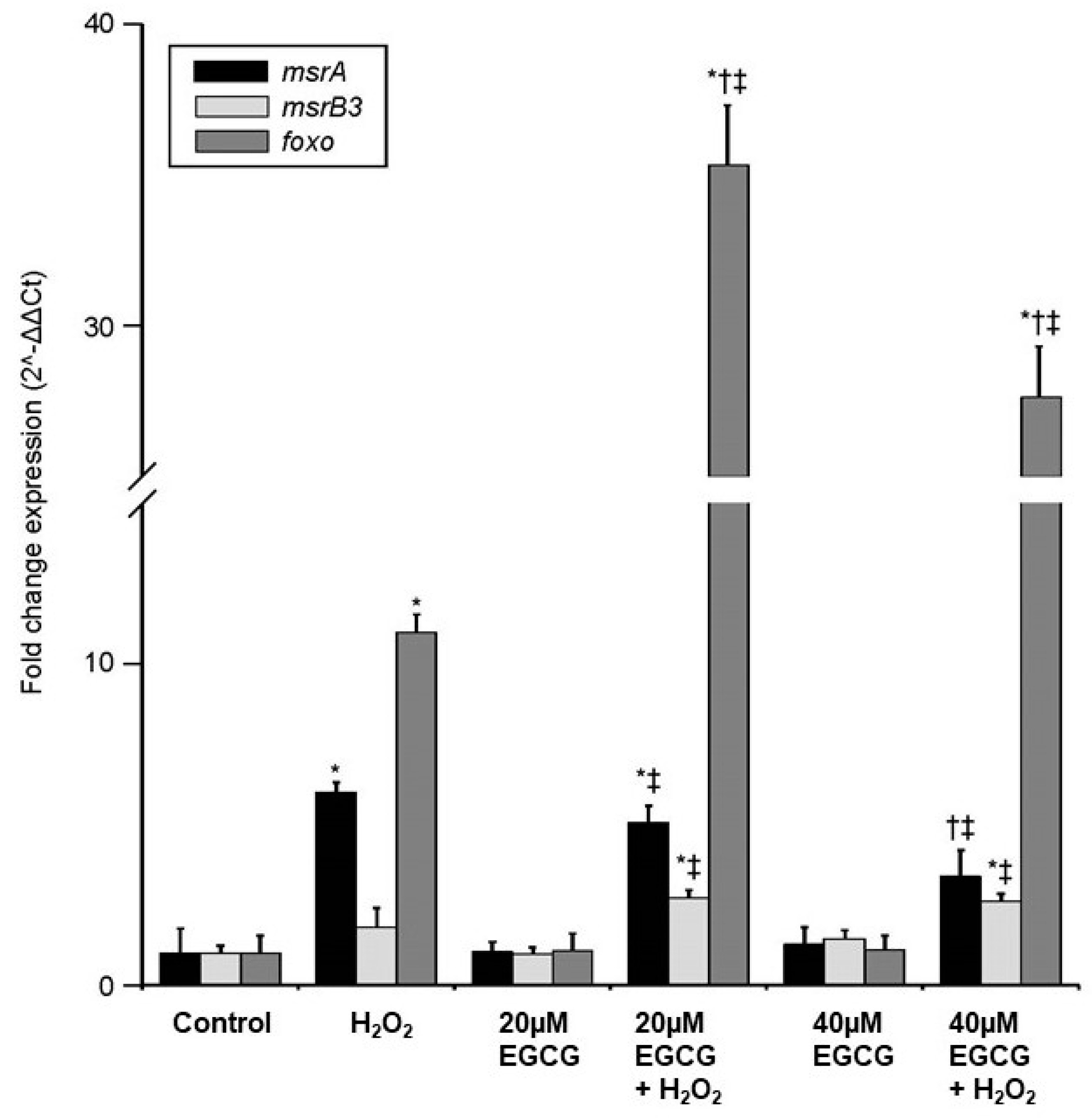
2. Results
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Protein Extraction from Whole Brain
4.3. Cell Culture Cultivation and Treatment
4.4. Protein Extraction from Culture
4.5. RT-PCR
4.6. Immunoblotting
4.7. Chemical Oxidative Stress
4.8. Pharmacological Stimulation of FOXO3a with EGCG
4.9. Reverse Transcription and QPCR
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lutz, P.L.; Nilsson, G.E.; Prentice, H.M. The Brain without Oxygen, 3rd ed.; Kluwer Academic Publishers: Boston, MA, USA, 2003; ISBN 1-4020-1165-2. [Google Scholar]
- Hochachka, P.W. Defense strategies against hypoxia and hypothermia. Science 1986, 231, 234–241. [Google Scholar] [CrossRef]
- Fernandes, J.A.; Lutz, P.L.; Tannenbaum, A.; Todorov, A.T.; Liebovitch, L.; Vertes, R. Electroencephalogram activity in the anoxic turtle brain. Am. J. Physiol. 1997, 273, R911–R919. [Google Scholar] [CrossRef] [PubMed]
- Warren, D.E.; Jackson, D.C. The metabolic consequences of repeated anoxic stress in the western painted turtle, Chrysemys picta bellii. Comp. Biochem. Physiol. Part. A Mol. Integr. Physiol. 2017, 203, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, M.E.; Lee, E.J.; Choy, Y. High levels of ascorbic acid, not glutathione, in the CNS of anoxia-tolerant reptiles contrasted with levels in anoxia-intolerant species. J. Neurochem. 1995, 64, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Milton, S.L.; Nayak, G.; Kesaraju, S.; Kara, L.; Prentice, H.M. Suppression of reactive oxygen species production enhances neuronal survival in vitro and in vivo in the anoxia-tolerant turtle Trachemys scripta. J. Neurochem. 2007, 101, 993–1001. [Google Scholar] [CrossRef] [PubMed]
- Willmore, W.G.; Storey, K.B. Antioxidant systems and anoxia tolerance in a freshwater turtle Trachemys scripta elegans. Mol. Cell. Biochem. 1997, 170, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Willmore, W.G.; Storey, K.B. Glutathione systems and anoxia tolerance in turtles. Am. J. Physiol. 1997, 273, R219–R225. [Google Scholar] [CrossRef] [PubMed]
- Milton, S.L.; Dirk, L.J.; Kara, L.F.; Prentice, H.M. Adenosine modulates ERK1/2, PI3K/Akt, and p38MAPK activation in the brain of the anoxia-tolerant turtle Trachemys scripta. J. Cereb. Blood Flow Metab. 2008, 28, 1469–1477. [Google Scholar] [CrossRef] [Green Version]
- Nayak, G.H.; Prentice, H.M.; Milton, S.L. Neuroprotective signaling pathways are modulated by adenosine in the anoxia tolerant turtle. J. Cereb. Blood Flow Metab. 2011, 31, 467–475. [Google Scholar] [CrossRef] [Green Version]
- Kesaraju, S.; Nayak, G.; Prentice, H.M.; Milton, S.L. Upregulation of Hsp72 mediates anoxia/reoxygenation neuroprotection in the freshwater turtle via modulation of ROS. Brain Res. 2014, 1582, 247–256. [Google Scholar] [CrossRef] [Green Version]
- Oien, D.; Moskovitz, J. Protein-carbonyl accumulation in the non-replicative senescence of the methionine sulfoxide reductase A (msrA) knockout yeast strain. Amino Acids 2007, 32, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-Y.; Gladyshev, V.N. Methionine sulfoxide reduction in mammals: Characterization of methionine-R-sulfoxide reductases. Mol. Biol. Cell 2004, 15, 1055–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weissbach, H.; Etienne, F.; Hoshi, T.; Heinemann, S.H.; Lowther, W.T.; Matthews, B.; St John, G.; Nathan, C.; Brot, N. Peptide methionine sulfoxide reductase: Structure, mechanism of action, and biological function. Arch. Biochem. Biophys. 2002, 397, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Delaye, L.; Becerra, A.; Orgel, L.; Lazcano, A. Molecular evolution of peptide methionine sulfoxide reductases (MsrA and MsrB): On the early development of a mechanism that protects against oxidative damage. J. Mol. Evol. 2007, 64, 15–32. [Google Scholar] [CrossRef]
- Minetti, G.; Balduini, C.; Brovelli, A. Reduction of DABS-L-methionine-DL-sulfoxide by protein methionine sulfoxide reductase from polymorphonuclear leukocytes: Stereospecificity towards the L-sulfoxide. Ital. J. Biochem. 1994, 43, 273–283. [Google Scholar]
- Achilli, C.; Ciana, A.; Minetti, G. The discovery of methionine sulfoxide reductase enzymes: An historical account and future perspectives. BioFactors 2015, 41, 135–152. [Google Scholar] [CrossRef]
- Lee, J.W.; Gordiyenko, N.V.; Marchetti, M.; Tserentsoodol, N.; Sagher, D.; Alam, S.; Weissbach, H.; Kantorow, M.; Rodriguez, I.R. Gene structure, localization and role in oxidative stress of methionine sulfoxide reductase A (MSRA) in the monkey retina. Exp. Eye Res. 2006, 82, 816–827. [Google Scholar] [CrossRef] [Green Version]
- Levine, R.L.; Mosoni, L.; Berlett, B.S.; Stadtman, E.R. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 15036–15040. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.; Hansel, A.; Kasperczyk, H.; Hoshi, T.; Heinemann, S.H. Activity, tissue distribution and site-directed mutagenesis of a human peptide methionine sulfoxide reductase of type B: hCBS1. FEBS Lett. 2002, 527, 91–94. [Google Scholar] [CrossRef] [Green Version]
- Lim, D.-H.; Han, J.Y.; Kim, J.-R.; Lee, Y.S.; Kim, H.-Y. Methionine sulfoxide reductase B in the endoplasmic reticulum is critical for stress resistance and aging in Drosophila. Biochem. Biophys. Res. Commun. 2012, 419, 20–26. [Google Scholar] [CrossRef]
- Dos Santos, S.L.; Petropoulos, I.; Friguet, B. The oxidized protein repair enzymes methionine sulfoxide reductases and their roles in protecting against oxidative stress, in ageing and in regulating protein function. Antioxidants 2018, 7, 191. [Google Scholar] [CrossRef]
- Moskovitz, J.; Bar-Noy, S.; Williams, W.M.; Requena, J.; Berlett, B.S.; Stadtman, E.R. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl. Acad. Sci. USA 2001, 98, 12920–12925. [Google Scholar] [CrossRef] [Green Version]
- Hansel, A.; Kuschel, L.; Hehl, S.; Lemke, C.; Agricola, H.J.; Hoshi, T.; Heinemann, S.H. Mitochondrial targeting of the human peptide methionine sulfoxide reductase (MSRA), an enzyme involved in the repair of oxidized proteins. FASEB J. 2002, 16, 911–913. [Google Scholar] [CrossRef] [PubMed]
- Vougier, S.; Mary, J.; Friguet, B. Subcellular localization of methionine sulphoxide reductase A (MsrA): Evidence for mitochondrial and cytosolic isoforms in rat liver cells. Biochem. J. 2003, 373, 531–537. [Google Scholar] [CrossRef] [Green Version]
- Ruan, H.; Tang, X.D.; Chen, M.-L.; Joiner, M.-L.A.; Sun, G.; Brot, N.; Weissbach, H.; Heinemann, S.H.; Iverson, L.; Wu, C.-F.; et al. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc. Natl. Acad. Sci. USA 2002, 99, 2748–2753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moskovitz, J.; Berlett, B.S.; Poston, J.M.; Stadtman, E.R. The Yeast Peptide-Methionine Sulfoxide Reductase Functions as an Antioxidant in vivo. Proc. Natl. Acad. Sci. USA 1997, 94, 9585–9589. [Google Scholar] [CrossRef] [Green Version]
- Prentice, H.M.; Moench, I.A.; Rickaway, Z.T.; Dougherty, C.J.; Webster, K.A.; Weissbach, H. MsrA protects cardiac myocytes against hypoxia/reoxygenation induced cell death. Biochem. Biophys. Res. Commun. 2008, 366, 775–778. [Google Scholar] [CrossRef]
- Moskovitz, J.; Flescher, E.; Berlett, B.S.; Azare, J.; Poston, J.M.; Stadtman, E.R. Overexpression of Peptide-Methionine Sulfoxide Reductase in Saccharomyces cerevisiae and Human T Cells Provides them with High Resistance to Oxidative Stress. Proc. Natl. Acad. Sci. USA 1998, 95, 14071–14075. [Google Scholar] [CrossRef] [Green Version]
- Yermolaieva, O.; Xu, R.; Schinstock, C.; Brot, N.; Weissbach, H.; Heinemann, S.H.; Hoshi, T. Methionine sulfoxide reductase A protects neuronal cells against brief hypoxia/reoxygenation. Proc. Natl. Acad. Sci. USA 2004, 101, 1159–1164. [Google Scholar] [CrossRef] [Green Version]
- Kantorow, M.; Hawse, J.R.; Cowell, T.L.; Benhamed, S.; Pizarro, G.O.; Reddy, V.N.; Hejtmancik, J.F. Methionine sulfoxide reductase A is important for lens cell viability and resistance to oxidative stress. Proc. Natl. Acad. Sci. USA 2004, 101, 9654–9659. [Google Scholar] [CrossRef] [Green Version]
- Picot, C.R.; Petropoulos, I.; Perichon, M.; Moreau, M.; Nizard, C.; Friguet, B. Overexpression of MsrA protects WI-38 SV40 human fibroblasts against H2O2-mediated oxidative stress. Free Radic. Biol. Med. 2005, 39, 1332–1341. [Google Scholar] [CrossRef]
- Levine, R.L.; Berlett, B.S.; Moskovitz, J.; Mosoni, L.; Stadtman, E.R. Methionine residues may protect proteins from critical oxidative damage. Mech. Ageing Dev. 1999, 107, 323–332. [Google Scholar] [CrossRef]
- Nan, C.; Li, Y.; Jean-Charles, P.-Y.; Chen, G.; Kreymerman, A.; Prentice, H.; Weissbach, H.; Huang, X. Deficiency of methionine sulfoxide reductase A causes cellular dysfunction and mitochondrial damage in cardiac myocytes under physical and oxidative stresses. Biochem. Biophys. Res. Commun. 2010, 402, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Boschi-Muller, S. Molecular mechanisms of the methionine sulfoxide reductase system from Neisseria meningitidis. Antioxidants 2018, 7, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etienne, F.; Spector, D.; Brot, N.; Weissbach, H. A methionine sulfoxide reductase in Escherichia coli that reduces the R enantiomer of methionine sulfoxide. Biochem. Biophys. Res. Commun. 2003, 300, 378–382. [Google Scholar] [CrossRef]
- Kwak, G.H.; Kim, J.R.; Kim, H.Y. Expression, subcellular localization, and antioxidant role of mammalian methionine sulfoxide reductases in Saccharomyces cerevisiae. BMB Rep. 2009, 42, 113–118. [Google Scholar] [CrossRef] [Green Version]
- Kryukov, G.V.; Abhilash Kumar, R.; Koc, A.; Sun, Z.; Gladyshev, V.N. Selenoprotein R is a Zinc-Containing Stereo-Specific Methionine Sulfoxide Reductase. Source 2002, 99, 4245–4250. [Google Scholar] [CrossRef] [Green Version]
- Abhilash Kumar, R.; Koc, A.; Cerny, R.L.; Gladyshev, V.N. Reaction mechanism, evolutionary analysis, and role of zinc in Drosophila methionine-R-sulfoxide reductase. J. Biol. Chem. 2002, 277, 37527–37535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.-Y.; Gladyshev, V.N. Characterization of mouse endoplasmic reticulum methionine-R-sulfoxide reductase. Biochem. Biophys. Res. Commun. 2004, 320, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Koc, A.; Gasch, A.P.; Rutherford, J.C.; Kim, H.-Y.; Gladyshev, V.N. Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and -independent components of aging. Proc. Natl. Acad. Sci. USA 2004, 101, 7999–8004. [Google Scholar] [CrossRef] [Green Version]
- Shchedrina, V.A.; Kabil, H.; Vorbruggen, G.; Lee, B.C.; Turanov, A.A.; Hirosawa-Takamori, M.; Kim, H.-Y.; Harshman, L.G.; Hatfield, D.L.; Gladyshev, V.N. Analyses of fruit flies that do not express selenoproteins or express the mouse selenoprotein, methionine sulfoxide reductase B1, reveal a role of selenoproteins in stress resistance. J. Biol. Chem. 2011, 286, 29449–29461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shchedrina, V.A.; Vorbrüggen, G.; Lee, B.C.; Kim, H.Y.; Kabil, H.; Harshman, L.G.; Gladyshev, V.N. Overexpression of methionine-R-sulfoxide reductases has no influence on fruit fly aging. Mech. Ageing Dev. 2009, 130, 429–443. [Google Scholar] [CrossRef] [Green Version]
- Cabreiro, F.; Picot, C.R.; Perichon, M.; Friguet, B.; Petropoulos, I. Overexpression of methionine sulfoxide reductases A and B2 protects MOLT-4 cells against zinc-induced oxidative stress. Antioxid. Redox Signal. 2009, 11, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Brunet, A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 2005, 24, 7410–7425. [Google Scholar] [CrossRef] [Green Version]
- Accili, D.; Arden, K.C. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell 2004, 117, 421–426. [Google Scholar] [CrossRef] [Green Version]
- Arden, K.C. FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene 2008, 27, 2345–2350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Yu, T.; Huang, P. Post-translational modifications of FOXO family proteins (Review). Mol. Med. Rep. 2016, 14, 4931–4941. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, G.; Khan, A.K.; Rashid, R.; Muneer, S.; Hasan, S.M.F.; Chen, J. FOXO Transcriptional Factors and Long-Term Living. Oxid. Med. Cell. Longev. 2017, 2017. [Google Scholar] [CrossRef]
- Li, M.; Chiu, J.F.; Mossman, B.T.; Fukagawa, N.K. Down-regulation of manganese-superoxide dismutase through phosphorylation of FOXO3a by Akt in explanted vascular smooth muscle cells from old rats. J. Biol. Chem. 2006, 281, 40429–40439. [Google Scholar] [CrossRef] [Green Version]
- Chung, Y.W.; Kim, H.K.; Kim, I.Y.; Yim, M.B.; Chock, P.B. Dual function of protein kinase C (PKC) in 12-O-tetradecanoylphorbol-13- acetate (TPA)-induced manganese superoxide dismutase (MnSOD) expression: Activation of creb and foxo3a by PKC-α phosphorylation and by PKC-mediated inactivation of akt, respectively. J. Biol. Chem. 2011, 286, 29681–29690. [Google Scholar] [CrossRef] [Green Version]
- Awad, H.; Nolette, N.; Hinton, M.; Dakshinamurti, S. AMPK and FoxO1 regulate catalase expression in hypoxic pulmonary arterial smooth muscle. Pediatr. Pulmonol. 2014, 49, 885–897. [Google Scholar] [CrossRef]
- Yun, H.; Park, S.; Kim, M.J.; Yang, W.K.; Im, D.U.; Yang, K.R.; Hong, J.; Choe, W.; Kang, I.; Kim, S.S.; et al. AMP-activated protein kinase mediates the antioxidant effects of resveratrol through regulation of the transcription factor FoxO1. FEBS J. 2014, 281, 4421–4438. [Google Scholar] [CrossRef]
- Venkatesan, B.; Mahimainathan, L.; Das, F.; Ghosh-Choudhury, N.; Choudhury, G.G. Downregulation of catalase by reactive oxygen species via PI 3 kinase/Akt signaling in mesangial cells. J. Cell. Physiol. 2007, 211, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Krivoruchko, A.; Storey, K.B. Anoxia-responsive regulation of the FoxO transcription factors in freshwater turtles, Trachemys scripta elegans. Biochim. Biophys. Acta 2013, 1830, 4990–4998. [Google Scholar] [CrossRef] [PubMed]
- Minniti, A.N.; Cataldo, R.; Trigo, C.; Vasquez, L.; Mujica, P.; Leighton, F.; Inestrosa, N.C.; Aldunate, R. Methionine sulfoxide reductase A expression is regulated by the DAF-16/FOXO pathway in Caenorhabditis elegans. Aging Cell 2009, 8, 690–705. [Google Scholar] [CrossRef]
- Chung, H.; Kim, A.K.; Jung, S.A.; Kim, S.W.; Yu, K.; Lee, J.H. The Drosophila homolog of methionine sulfoxide reductase A extends lifespan and increases nuclear localization of FOXO. FEBS Lett. 2010, 584, 3609–3614. [Google Scholar] [CrossRef] [Green Version]
- Reiterer, M.; Milton, S.L. Induction of foxo3a protects turtle neurons against oxidative stress. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2020, 243, 110671. [Google Scholar] [CrossRef]
- Bartholome, A.; Kampkötter, A.; Tanner, S.; Sies, H.; Klotz, L.O. Epigallocatechin gallate-induced modulation of FoxO signaling in mammalian cells and C. elegans: FoxO stimulation is masked via PI3K/Akt activation by hydrogen peroxide formed in cell culture. Arch. Biochem. Biophys. 2010, 501, 58–64. [Google Scholar] [CrossRef]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef] [Green Version]
- Jahn, A.; Scherer, B.; Fritz, G.; Honnen, S. Statins Induce a DAF-16/Foxo-dependent longevity phenotype via JNK-1 through Mevalonate depletion in C. elegans. Aging Dis. 2020, 11, 60–72. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.C.; Dikiy, A.; Kim, H.-Y.; Gladyshev, V.N. Functions and evolution of selenoprotein methionine sulfoxide reductases. Biochim. Biophys. Acta 2009, 1790, 1471–1477. [Google Scholar] [CrossRef] [Green Version]
- Martins, R.; Lithgow, G.J.; Link, W. Long live FOXO: Unraveling the role of FOXO proteins in aging and longevity. Aging Cell 2016, 15, 196–207. [Google Scholar] [CrossRef]
- Nayak, G.; Prentice, H.M.; Milton, S.L. Lessons from nature: Signalling cascades associated with vertebrate brain anoxic survival. Exp. Physiol. 2016, 101, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Prentice, H.M.; Milton, S.L.; Scheurle, D.; Lutz, P.L. The upregulation of cognate and inducible heat shock proteins in the anoxic turtle brain. J. Cereb. Blood Flow Metab. 2004, 24, 826–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Breukelen, F.; Maier, R.; Hand, S.C. Depression of nuclear transcription and extension of mRNA half-life under anoxia in Artemia franciscana embryos. J. Exp. Biol. 2000, 203, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Fraser, K.P.; Houlihan, D.F.; Lutz, P.L.; Leone-Kabler, S.; Manuel, L.; Brechin, J.G. Complete suppression of protein synthesis during anoxia with no post-anoxia protein synthesis debt in the red-eared slider turtle Trachemys scripta elegans. J. Exp. Biol. 2001, 204, 4353–4360. [Google Scholar] [CrossRef]
- van Breukelen, F.; Martin, S.L. Reversible depression of transcription during hibernation. J. Comp. Physiol. B 2002, 172, 355–361. [Google Scholar] [CrossRef]
- Osborne, P.G.; Gao, B.; Hashimoto, M. Determination in vivo of newly synthesized gene expression in hamsters during phases of the hibernation cycle. Jpn. J. Physiol. 2004, 54, 295–305. [Google Scholar] [CrossRef] [Green Version]
- Kesaraju, S.; Schmidt-Kastner, R.; Prentice, H.M.; Milton, S.L. Modulation of stress proteins and apoptotic regulators in the anoxia tolerant turtle brain. J. Neurochem. 2009, 109, 1413–1426. [Google Scholar] [CrossRef] [Green Version]
- Storey, K.B. Anoxia tolerance in turtles: Metabolic regulation and gene expression. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2007, 147, 263–276. [Google Scholar] [CrossRef]
- Pamenter, M.E.; Richards, M.D.; Buck, L.T. Anoxia-induced changes in reactive oxygen species and cyclic nucleotides in the painted turtle. J. Comp. Physiol. B 2007, 177, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Lutz, P.L.; Prentice, H.M.; Milton, S.L. Is turtle longevity linked to enhanced mechanisms for surviving brain anoxia and reoxygenation? Exp. Gerontol. 2003, 38, 797–800. [Google Scholar] [CrossRef]
- Giraud-Billoud, M.; Rivera-Ingraham, G.A.; Moreira, D.C.; Burmester, T.; Castro-Vazquez, A.; Carvajalino-Fernández, J.M.; Dafre, A.; Niu, C.; Tremblay, N.; Paital, B.; et al. Twenty years of the ‘Preparation for Oxidative Stress’ (POS) theory: Ecophysiological advantages and molecular strategies. Comp. Biochem. Physiol. Part. A Mol. Integr. Physiol. 2019, 234, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.A.; Pizarro, G.O.; Sagher, D.; Deamicis, C.; Brot, N.; Hejtmancik, J.F.; Weissbach, H.; Kantorow, M. Methionine sulfoxide reductases B1, B2, and B3 are present in the human lens and confer oxidative stress resistance to lens cells. Investig. Ophthalmol. Vis. Sci. 2005, 46, 2107–2112. [Google Scholar] [CrossRef] [PubMed]
- Bruce, L.; Singkornrat, D.; Wilson, K.; Hausman, W.; Robbins, K.; Huang, L.; Foss, K.; Binninger, D. In vivo effects of methionine sulfoxide reductase deficiency in Drosophila melanogaster. Antioxidants 2018, 7, 155. [Google Scholar] [CrossRef] [Green Version]
- Salmon, A.B.; Pérez, V.I.; Bokov, A.; Jernigan, A.; Kim, G.; Zhao, H.; Levine, R.L.; Richardson, A. Lack of methionine sulfoxide reductase A in mice increases sensitivity to oxidative stress but does not diminish life span. FASEB J. 2009, 23, 3601–3608. [Google Scholar] [CrossRef] [Green Version]
- Moskovitz, J.; Malik, A.; Hernandez, A.; Band, M.; Avivi, A. Methionine sulfoxide reductases and methionine sulfoxide in the subterranean mole rat (Spalax): Characterization of expression under various oxygen conditions. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2012, 161, 406–414. [Google Scholar] [CrossRef]
- Shams, I.; Avivi, A.; Nevo, E. Hypoxic stress tolerance of the blind subterranean mole rat: Expression of erythropoietin and hypoxia-inducible factor 1α. Proc. Natl. Acad. Sci. USA 2004, 101, 9698–9703. [Google Scholar] [CrossRef] [Green Version]
- Shams, I.; Avivi, A.; Nevo, E. Oxygen and carbon dioxide fluctuations in burrows of subterranean blind mole rats indicate tolerance to hypoxic-hypercapnic stresses. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2005, 142, 376–382. [Google Scholar] [CrossRef]
- Bergeron, M.; Yu, A.Y.; Solway, K.E.; Semenza, G.L.; Sharp, F.R. Induction of hypoxia-inducible factor-1 (HIF-1) and its target genes following focal ischaemia in rat brain. Eur. J. Neurosci. 1999, 11, 4159–4170. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci. STKE 2007, 2007, cm8. [Google Scholar] [CrossRef]
- Prentice, H.M.; Milton, S.L.; Scheurle, D.; Lutz, P.L. Gene transcription of brain voltage-gated potassium channels is reversibly regulated by oxygen supply. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003, 285, R1317–R1321. [Google Scholar] [CrossRef] [Green Version]
- Kesaraju, S. Molecular Mechanisms of Neuroprotection in the Anoxia Tolerant Freshwater Turtle; Florida Atlantic University: Boca Raton, FL, USA, 2008. [Google Scholar]
- Rissanen, E.; Tranberg, H.K.; Sollid, J.; Nilsson, G.E.; Nikinmaa, M. Temperature regulates hypoxia-inducible factor-1 (HIF-1) in a poikilothermic vertebrate, crucian carp (Carassius carassius). J. Exp. Biol. 2006, 209, 994–1003. [Google Scholar] [CrossRef] [Green Version]
- Ji, C.; Zhang, N.; Jiang, H.; Meng, X.; Ge, H.; Yang, X.; Xu, X.; Qian, K.; Park, Y.; Zheng, Y.; et al. 20-hydroxyecdysone regulates expression of methioninesulfoxide reductases through transcription factor FOXO in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 2021, 131. [Google Scholar] [CrossRef] [PubMed]
- Kops, G.J.P.L.; Dansen, T.B.; Polderman, P.E.; Saarloos, I.; Wirtz, K.W.A.; Coffer, P.J.; Huang, T.T.; Bos, J.L.; Medema, R.H.; Burgering, B.M.T. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 2002, 419, 316–321. [Google Scholar] [CrossRef]
- Honda, Y.; Honda, S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 1999, 13, 1385–1393. [Google Scholar] [CrossRef] [Green Version]
- Brunet, A.; Sweeney, L.B.; Sturgill, J.F.; Chua, K.F.; Greer, P.L.; Lin, Y.; Tran, H.; Ross, S.E.; Mostoslavsky, R.; Cohen, H.Y.; et al. Stress-Dependent Regulation of FOXO Transcription Factors by the SIRT1 Deacetylase. Science 2004, 303, 2011–2015. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, Y.; Furukawa-Hibi, Y.; Chen, C.; Horio, Y.; Isobe, K.; Ikeda, K.; Motoyama, N. SIRT1 is critical regulator of FOXO-mediated transcription in response to oxidative stress. Int. J. Mol. Med. 2005, 16, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, S.; Nakamura, N.; Vazquez, F.; Batt, D.B.; Perera, S.; Roberts, T.M.; Sellers, W.R. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/akt pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 2110–2115. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Liu, Y.; Liu, X.; Zhou, L.; Ma, X.; Liu, J.; Wang, L.; Guo, H. Activation of forkhead box O3a by mono(2-ethylhexyl)phthalate and its role in protection against mono(2-ethylhexyl)phthalate-induced oxidative stress and apoptosis in human cardiomyocytes. J. Appl. Toxicol. 2021, 41, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, S.; Finkel, T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science 2002, 295, 2450–2452. [Google Scholar] [CrossRef] [PubMed]
- Cudic, P.; Joshi, N.; Sagher, D.; Williams, B.T.; Stawikowski, M.J.; Weissbach, H. Identification of activators of methionine sulfoxide reductases A and B. Biochem. Biophys. Res. Commun. 2016, 469, 863–867. [Google Scholar] [CrossRef] [Green Version]
- Roesijadi, G.; Rezvankhah, S.; Binninger, D.M.; Weissbach, H. Ecdysone induction of MsrA protects against oxidative stress in Drosophila. Biochem. Biophys. Res. Commun. 2007, 354, 511–516. [Google Scholar] [CrossRef]
- Picot, C.R.; Perichon, M.; Lundberg, K.C.; Friguet, B.; Szweda, L.I.; Petropoulos, I. Alterations in mitochondrial and cytosolic methionine sulfoxide reductase activity during cardiac ischemia and reperfusion. Exp. Gerontol. 2006, 41, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Moskovitz, J.; Stadtman, E.R. Selenium-deficient diet enhances protein oxidation and affects methionine sulfoxide reductase (MsrB) protein level in certain mouse tissues. Proc. Natl. Acad. Sci. USA 2003, 100, 7486–7490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiterer, M.; Schmidt-Kastner, R.; Milton, S.L. Methionine sulfoxide reductase (Msr) dysfunction in human brain disease. Free Radic. Res. 2019, 53. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reiterer, M.; Bruce, L.; Milton, S. Differential Responses of Methionine Sulfoxide Reductases A and B to Anoxia and Oxidative Stress in the Freshwater Turtle Trachemys scripta. Metabolites 2021, 11, 458. https://doi.org/10.3390/metabo11070458
Reiterer M, Bruce L, Milton S. Differential Responses of Methionine Sulfoxide Reductases A and B to Anoxia and Oxidative Stress in the Freshwater Turtle Trachemys scripta. Metabolites. 2021; 11(7):458. https://doi.org/10.3390/metabo11070458
Chicago/Turabian StyleReiterer, Melissa, Lynsey Bruce, and Sarah Milton. 2021. "Differential Responses of Methionine Sulfoxide Reductases A and B to Anoxia and Oxidative Stress in the Freshwater Turtle Trachemys scripta" Metabolites 11, no. 7: 458. https://doi.org/10.3390/metabo11070458
APA StyleReiterer, M., Bruce, L., & Milton, S. (2021). Differential Responses of Methionine Sulfoxide Reductases A and B to Anoxia and Oxidative Stress in the Freshwater Turtle Trachemys scripta. Metabolites, 11(7), 458. https://doi.org/10.3390/metabo11070458