Metabolomic Study of Urine from Workers Exposed to Low Concentrations of Benzene by UHPLC-ESI-QToF-MS Reveals Potential Biomarkers Associated with Oxidative Stress and Genotoxicity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Population
2.2. Ethical Aspects of Research
2.3. Biochemical Biomarkers, Exposure, Oxidative Stress and Genotoxicity
2.4. Standards and Reagents
2.5. Sample Preparation
2.6. Analytical System—UHPLC-ESI-QTOF/MS
2.7. Detection and Identification of Non-Target Metabolites
2.8. Statistical Analysis
- -
- is the value of the oxidative stress biomarker of the ith individual;
- -
- is the kth metabolite of the ith individual;
- -
- is the coefficient of the kth metabolite;
- -
- is a random error, which follows normal distribution with mean 0 and standard deviation σ.
3. Results
3.1. Sociodemographic Characteristics and Results of Biological Monitoring
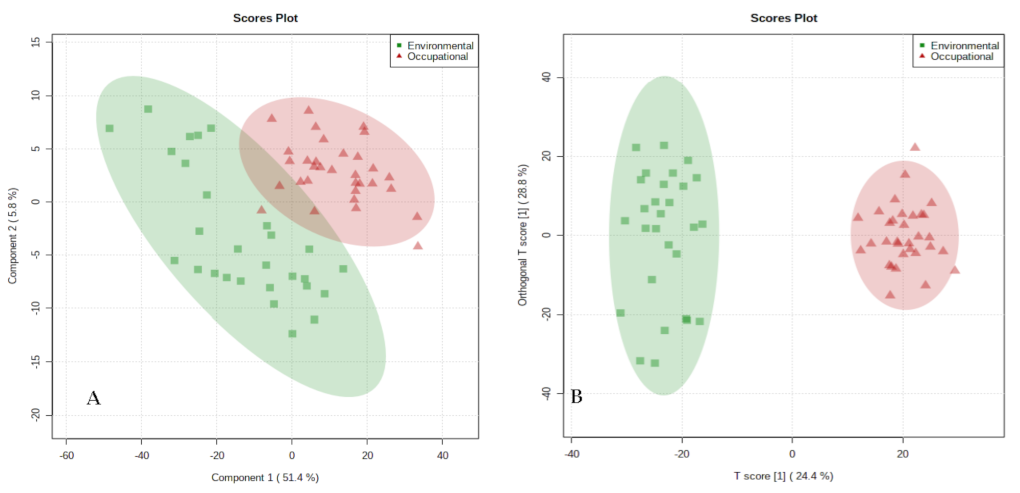
3.2. Identification of the Urinary Metabolic Profile of Individuals Occupationally and Environmentally Exposed to Benzene
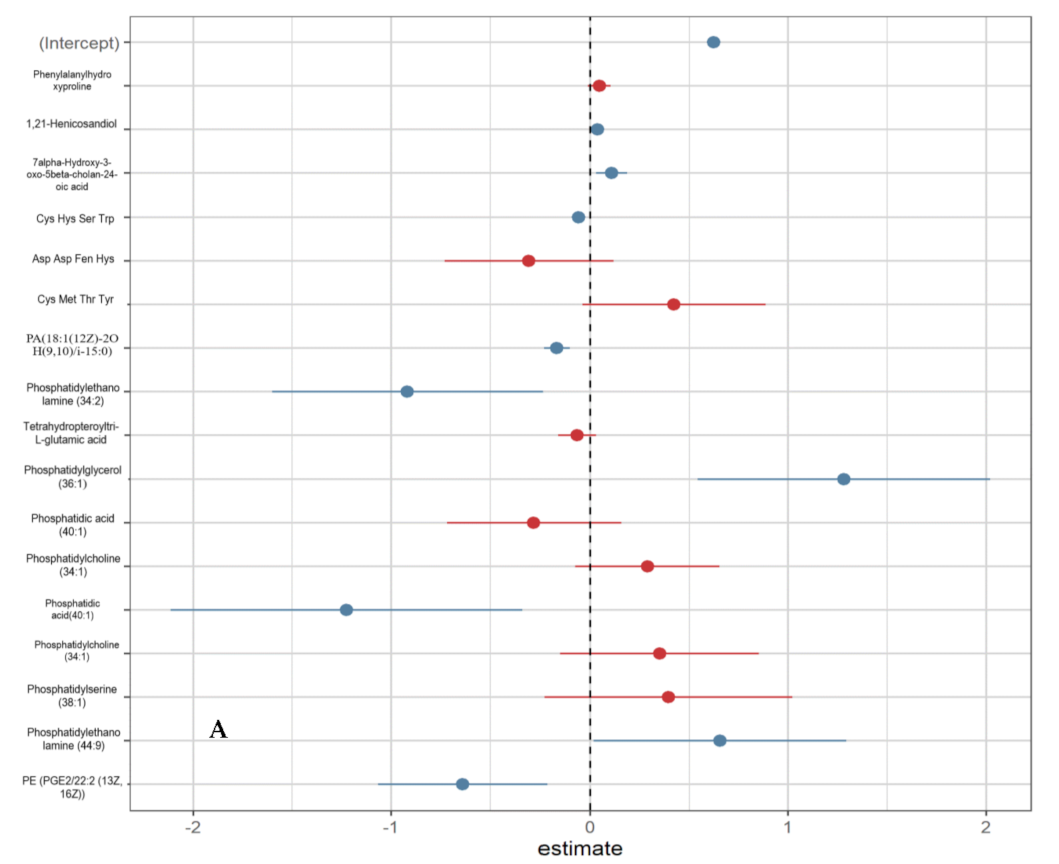
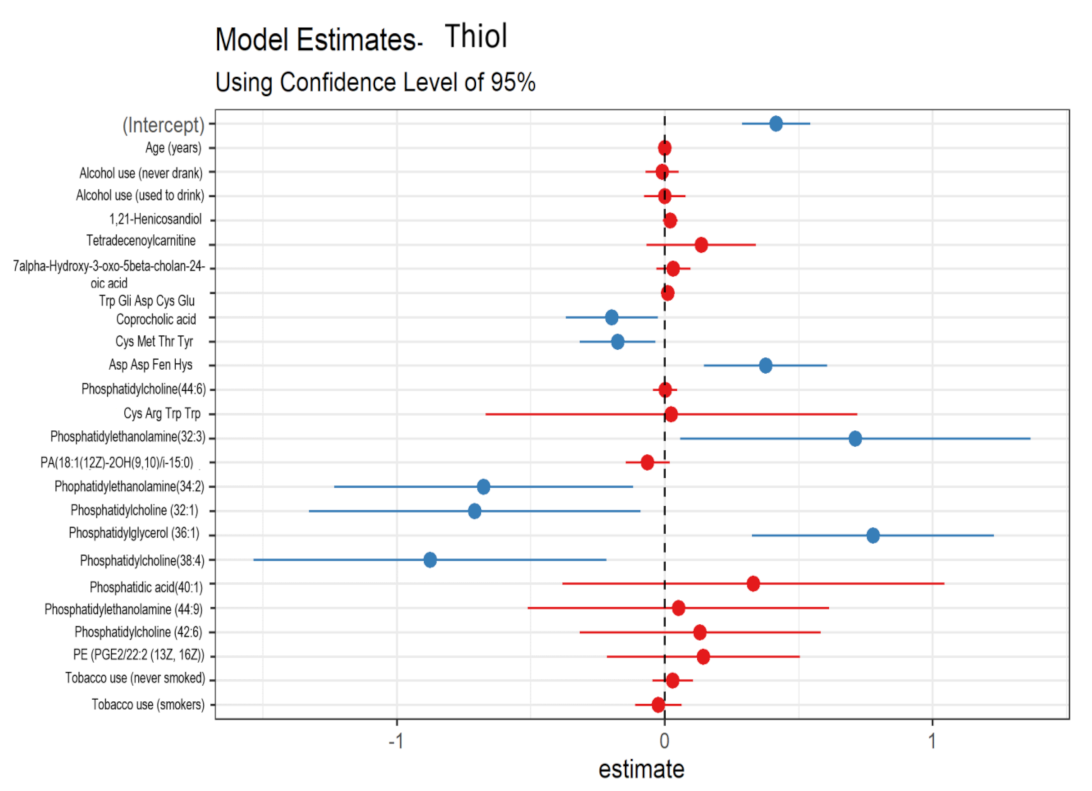
3.3. Urinary Metabolites Associated with Oxidative Stress
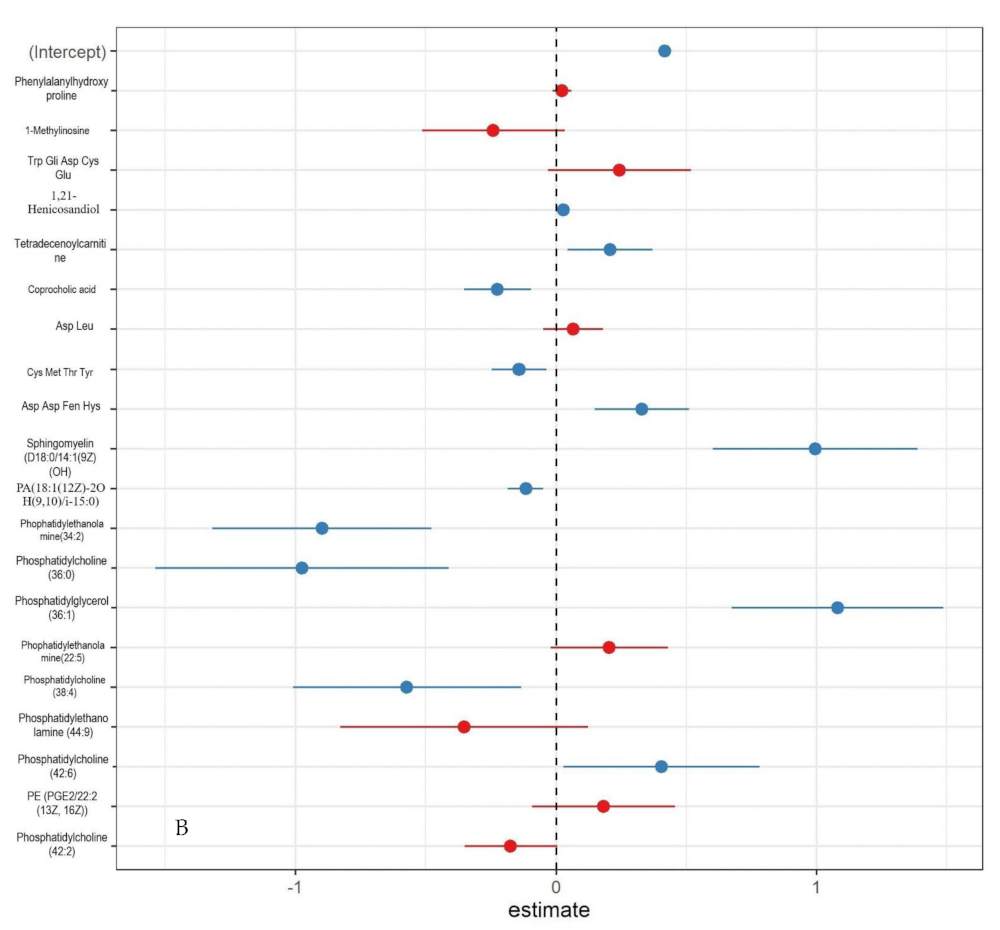
3.4. Urinary Metabolites Associated with Chromosomal Aberrations
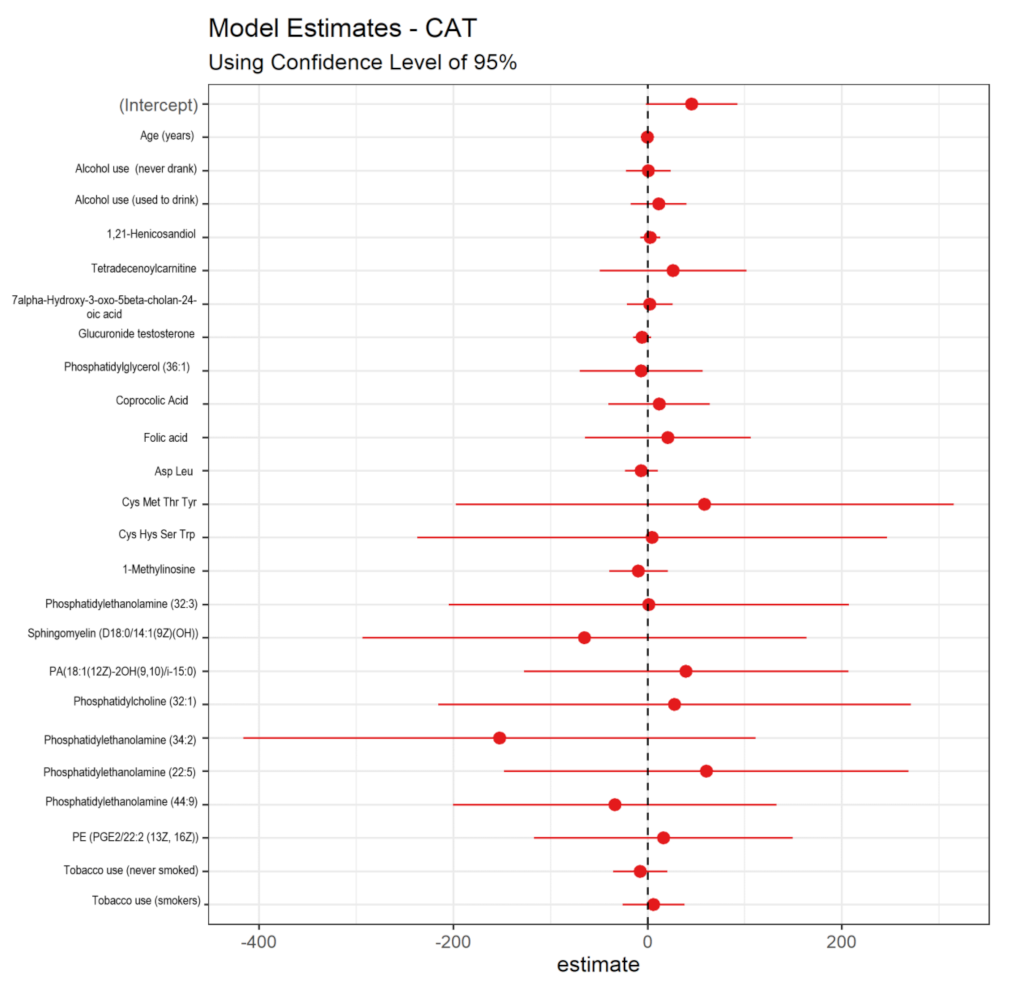
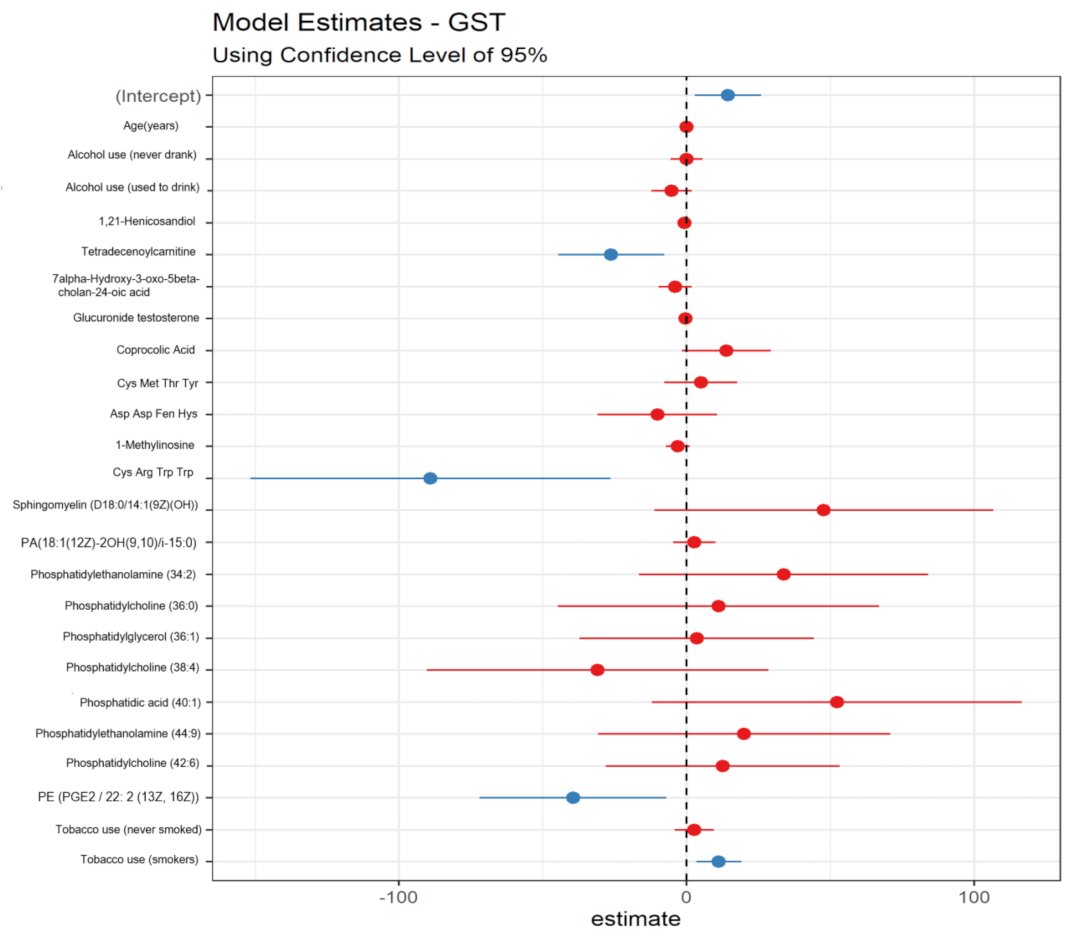
3.5. Potential Metabolomic Biomarkers Associated with Oxidative Damage and Benzene-Induced Chromosomal Aberrations
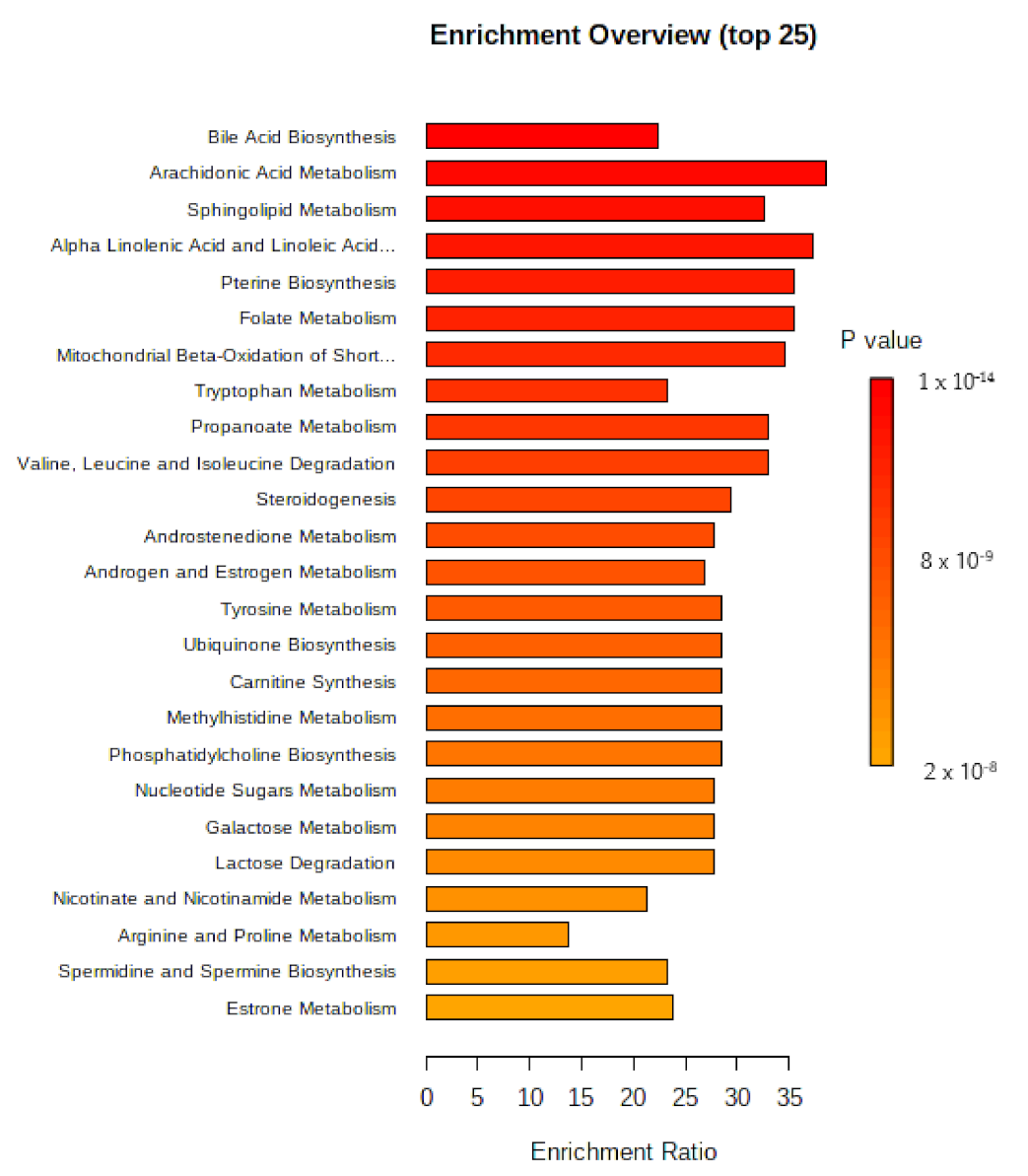
3.6. Metabolic Pathway Analysis
3.7. Discussion
3.8. Limitations of the Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wallace, L. Environmental Exposure to Benzene: An Update. Environ. Health Perspect. 1996, 104 (Suppl. 6), 1129–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- IARC—International Agency for Research on Cancer. Benzene/IARC Working Group on the Evaluation of Carcinogenic Risks to Humans; IARC: Lyon, France, 2018; Volume 120. [Google Scholar]
- Kuang, S.; Liang, W. Clinical Analysis of 43 Cases of Chronic Benzene Poisoning. Chem. Biol. Interact. 2005, 153, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Vermeulen, R.; Waidyanatha, S.; Johnson, B.A.; Lan, Q.; Smith, M.T.; Zhang, L.; Li, G.; Shen, M.; Yin, S.; et al. Modeling Human Metabolism of Benzene Following Occupational and Environmental Exposures. Cancer Epidemiol. Biomarkers Prev. 2006, 15, 2246–2252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rappaport, S.M.; Kim, S.; Thomas, R.; Johnson, B.A.; Bois, F.Y.; Kupper, L.L. Low-Dose Metabolism of Benzene in Humans: Science and Obfuscation. Carcinogenesis 2013, 34, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Arnold, S.M.; Angerer, J.; Boogaard, P.J.; Hughes, M.F.; O’Lone, R.B.; Robison, S.H.; Robert Schnatter, A. The Use of Biomoni toring Data in Exposure and Human Health Risk Assessment: Benzene Case Study. Crit. Rev. Toxicol. 2013, 43, 119–153. [Google Scholar] [CrossRef] [Green Version]
- Carrieri, M.; Spatari, G.; Tranfo, G.; Sapienza, D.; Scape, M.; Battista, G.; Manno, M. Biological Monitoring of Low Level Exposure to Benzene in an Oil Re Fi Nery: E Ff Ect of Modulating Factors. Toxicol. Lett. 2018, 298, 70–75. [Google Scholar] [CrossRef]
- Lovreglio, P.; Palma, G.D.; Barbieri, A.; Andreoli, R.; Drago, I.; Greco, L.; Gallo, E.; Diomede, L.; Ricossa, M.C.; Fostinelli, J.; et al. Biological Monitoring of Exposure to Low Concentrations of Benzene in Workers at a Metallurgical Coke Production Plant: New Insights into S-Phenylmercapturic Acid and Urinary Benzene. Biomarkers 2017, 23, 70–77. [Google Scholar] [CrossRef]
- Vineis, P.; Fecht, D. Environment, Cancer and Inequalities d the Urgent Need for Prevention. Eur. J. Cancer 2018, 103, 317–326. [Google Scholar] [CrossRef]
- Buesen, R.; Chorley, B.N.; Lima, S.; Daston, G.; Deferme, L.; Ebbels, T.; Gant, T.W.; Goetz, A.; Greally, J.; Hubesch, B.; et al. Applying ’ Omics Technologies in Chemicals Risk Assessment: Report of an ECETOC Workshop. Regul. Toxicol. Pharmacol. 2017, 91, 3–13. [Google Scholar] [CrossRef]
- Malinowska, J.M.; Viant, M.R. ScienceDirect Toxicology Confidence in Metabolite Identification Dictates the Applicability of Metabolomics to Regulatory Toxicology. Curr. Opin. Toxicol. 2020, 16, 32–38. [Google Scholar] [CrossRef]
- de Castro, M.D.L.; Capote, F.P. The Analytical Process to Search for Metabolomics Biomarkers. J. Pharm. Biomed. Anal. 2017, 147, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yin, Y. Strategies for Large-Scale Targeted Metabolomics Quantification by Liquid Chromatography-Mass Spectrometry. Analyst 2016, 141, 6362–6373. [Google Scholar] [CrossRef] [PubMed]
- Koelmel, J.P.; Napolitano, M.P.; Ulmer, C.Z.; Vasiliou, V.; Garrett, T.J.; Yost, R.A.; Prasad, M.N.V.; Godri Pollitt, K.J.; Bowden, J.A. Environmental Lipidomics: Understanding the Response of Organisms and Ecosystems to a Changing World. Metabolomics 2020, 16, 56. [Google Scholar] [CrossRef] [PubMed]
- Niedzwiecki, M.M.; Walker, D.I.; Vermeulen, R.; Chadeau-hyam, M.; Jones, D.P.; Miller, G.W. The Exposome: Molecules to Populations. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 107–127. [Google Scholar] [CrossRef]
- Tzoulaki, I.; Ebbels, T.M.D.; Valdes, A.; Elliott, P.; Ioannidis, J.P.A. Practice of Epidemiology Design and Analysis of Metabolomics Studies in Epidemiologic Research: A Primer on -Omic Technologies. Am. J. Epidemiol. 2014, 180, 129–139. [Google Scholar] [CrossRef] [Green Version]
- Boyles, M.S.P.; Ranninger, C.; Reischl, R.; Rurik, M.; Tessadri, R.; Kohlbacher, O.; Duschl, A.; Huber, C.G. Copper Oxide Nanoparticle Toxicity Profiling Using Untargeted Metabolomics. Part. Fibre Toxicol. 2016, 13, 49. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhang, M.; Bo, L.; Li, S.; Hu, L.; Zhao, X. Metabolomic Analysis of the Toxic Effect of Chronic Exposure of Cadmium on Rat Urine. Environ. Sci. Pollut. Res. 2018, 25, 3765–3774. [Google Scholar] [CrossRef]
- Fabian, E.; Bordag, N.; Herold, M.; Kamp, H.; Krennrich, G.; Looser, R.; Ma-hock, L.; Mellert, W.; Montoya, G.; Peter, E.; et al. Metabolite pro Fi Les of Rats in Repeated Dose Toxicological Studies after Oral and Inhalative Exposure. Toxicol. Lett. 2016, 255, 11–23. [Google Scholar] [CrossRef]
- García-Sevillano, M.A.; García-Barrera, T.; Navarro, F.; Abril, N.; Pueyo, C. Combination of Direct Infusion Mass Spectrometry and Gas Chromatography Mass Spectrometry for Toxicometabolomic Study of Red Blood Cells and Serum of Mice Mus Musculus after Mercury Exposure. J. Chromatogr. B 2015, 985, 75–84. [Google Scholar] [CrossRef]
- Hou, Y.; Cao, C.; Bao, W.; Yang, S.; Shi, H.; Hao, D. The Plasma Metabolic Profiling of Chronic Acephate Exposure in Rats via an Ultra-Performance Liquid Chromatography-Mass Spectrometry Based. Mol. Biosyst. 2015, 11, 506–515. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, H.; Geng, N.; Ren, X.; Zhang, B.; Gong, Y.; Chen, J. A Metabolomics Strategy to Assess the Combined Toxicity of Polycyclic Aromatic Hydrocarbons ( PAHs ) and Short-Chain Chlorinated Paraf Fi Ns. Environ. Pollut. 2018, 234, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Blair, I.A.; Mesaros, C. Analytical Methods for Mass Spectrometry-Based Metabolomics Studies. Adv. Exp. Med. Biol. 2019, 1140, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larive, C.K.; Barding, G.A.; Dinges, M.M. NMR Spectroscopy for Metabolomics and Metabolic Profiling. Anal. Chem. 2015, 87, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhang, A.; Wang, X. Advances in Mass Spectrometry-Based Metabolomics for Investigation of Metabolites. RSC Adv. 2018, 8, 22335–22350. [Google Scholar] [CrossRef] [Green Version]
- Gika, H.; Virgiliou, C.; Theodoridis, G.; Plumb, R.S.; Wilson, I.D. Untargeted LC/MS-Based Metabolic Phenotyping (Metabonomics/Metabolomics): The State of the Art. J. Chromatogr. B 2019, 1117, 136–147. [Google Scholar] [CrossRef]
- Gorrochategui, E.; Jaumot, J.; Lacorte, S.; Tauler, R. Data Analysis Strategies for Targeted and Untargeted LC-MS Metabolomic Studies: Overview and Workflow. Trends Anal. Chem. 2016, 82, 425–442. [Google Scholar] [CrossRef]
- Costa-Amaral, I.C.; Carvalho, L.V.B.; Santos, M.V.C.; Teixeira, L.R.; Collins, A.R.; de Cássia, O.C.; Mattos, O.C.; Sarcinelli, P.N. Environmental Assessment and Evaluation of Oxidative Stress and Genotoxicity Biomarkers Related to Chronic Occupational Exposure to Benzene. Int. J. Environ. Res. Public Health 2019, 16, 2240. [Google Scholar] [CrossRef] [Green Version]
- Mahieu, N.G.; Genenbacher, J.L.; Patti, G.J. A Roadmap for the XCMS Family of Software Solutions in Metabolomics. Curr. Opin. Chem. Biol. 2016, 30, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A Web-Based Platform to Process Untargeted Metabolomic Data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; Anderson de Lima Morais, D.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venable, W.B.; Ripley, B. Modern Applied Statistics with S, 4th ed.; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Akaike, H. Information Theory and an Extension of the Maximum Likelihood Principle. In Proceedings of the 2nd International Symposium on Information Theory; Akaike, H., Ed.; Akademiai Kiado: Budapest, Hungary, 1973; pp. 267–281. [Google Scholar]
- Hoo, Z.H.; Candlish, J.; Teare, D. What Is an ROC Curve? Emerg. Med. J. 2017, 34, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Shen, H.; Xu, W.; Xia, Y.; Barr, D.B.; Mu, X.; Wang, X.; Liu, L.; Huang, Q.; Tian, M. Urinary Metabolomics Revealed Arsenic Internal Dose-Related Metabolic Alterations: A Proof-of-Concept Study in a Chinese Male Cohort. Environ. Sci. Technol. 2014, 48, 12265–12274. [Google Scholar] [CrossRef]
- de Moraes Moreau, R.L.; Bastos de Diqueira, M.E.P. Toxicologia Analítica, 2nd ed.; Guanabara Koogan: Rio de Janeiro, Brazil, 2016. [Google Scholar]
- Barr, D.B.; Wilder, L.C.; Caudill, S.P.; Gonzalez, A.J.; Needham, L.L.; Pirkle, J.L. Urinary Creatinine Concentrations in the U.S. Population: Implications for Urinary Biologic Monitoring Measurements. Environ. Health Perspect. 2005, 113, 192–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brazil Ministério do Trabalho. Norma Regulamentadora (NR) 15—Atividades e Operações Insalubres; Anexo No. 13-A; Ministério do Trabalho: Rio do Janeiro, Brazil, 1995. [Google Scholar]
- Mendes, M.; Mesquita, J.; Machado, H.; Durand, A.; Costa-Amaral, I.C.; Valente, D.; Sydneia, A.; Arcuri, A.; Trevisan, E.A.; Sarcinelli, P.D.N.; et al. Normas Ocupacionais Do Benzeno: Uma Abordagem Sobre o Risco e Exposição Nos Postos de Revenda de Combustíveis. Rev. Bras. Saúde Ocup. 2017, 6369, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Mchale, C.M.; Zhang, L.; Smith, M.T. Current Understanding of the Mechanism of Benzene-Induced Leukemia in Humans: Implications for Risk Assessment. Carcinogenesis 2012, 33, 240–252. [Google Scholar] [CrossRef] [Green Version]
- Bolton, J.L.; Trush, M.A.; Penning, T.M.; Dryhurst, G.; Monks, T.J. Role of Quinones in Toxicology. Chem. Res. Toxicol. 2000, 13, 135–160. [Google Scholar] [CrossRef]
- Lodovici, M.; Bigagli, E. Oxidative Stress and Air Pollution Exposure. J. Toxicol. 2011, 2011, 487074. [Google Scholar] [CrossRef]
- Moro, A.M.; Charão, M.F.; Brucker, N.; Durgante, J.; Baierle, M.; Bubols, G.; Goethel, G.; Fracasso, R.; Nascimento, S.; Bulcão, R.; et al. Genotoxicity and Oxidative Stress in Gasoline Station Attendants. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2013, 754, 63–70. [Google Scholar] [CrossRef]
- Uzma, N.; Kumar, S.S.; Hazari, M.A.H. Exposure to Benzene Induces Oxidative Stress, Alters the Immune Response and Expression of P53 in Gasoline Filling Workers. Am. J. Ind. Med. 2010, 53, 1264–1270. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Tan, B.; Yin, Y.; Blachier, F.; Tossou, M.C.B.; Rahu, N. Oxidative Stress and Inflammation: What Polyphenols Can Do for Us? Oxid. Med. Cell. Longev. 2016, 2016, 7432797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative Stress, Inflammation, and Cancer: How Are They Linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. “Metabonomics”: Understanding the Metabolic Responses of Living Systems to Pathophysiological Stimuli via Multivariate Statistical Analysis of Biological NMR Spectroscopic Data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef]
- Klassen, A.; Faccio, A.T.; Canuto, G.A.B.; Cruz, P.L.R.; Ribeiro, H.C.; Tavares, M.F.M.; Sussulini, A. Metabolomics: From Fundamentals to Clinical Applications. In Metabolomics: Definitions and Significance in Systems Biology; Springer: Berlin/Heidelberg, Germany, 2017; Volume 965, pp. 3–17. [Google Scholar] [CrossRef]
- Canuto, G.A.B.; Costa, J.L.; Cruz, P.L.R.; Rosa, S.A.; Policy, E.D.; Fluid, O.; Seeds, O.P. Metabolômica: Definições, Estado-da-Arte e Aplicações Representativas. Quim. Nova 2018, 41, 75–91. [Google Scholar] [CrossRef]
- Santana, F.B. Uso de Espectroscopia No Infravermelho Médio e Análise Discriminante Por Quadrados Mínimos Parciais Na Determinação de Adulterações Em Óleos de Andiroba, Prímula e Rosa Mosqueta. Ph.D. Thesis, Universidade Federal de Uberlândia, Uberlândia, Brazil, 2015. [Google Scholar]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinforma. 2019, 68, 1–128. [Google Scholar] [CrossRef]
- Godzien, J.; Ciborowski, M.; Angulo, S.; Barbas, C. From Numbers to a Biological Sense: How the Strategy Chosen for Metabolomics Data Treatment May Affect Final Results. A Practical Example Based on Urine Fingerprints Obtained by LC-MS. Electrophoresis 2013, 34, 1–30. [Google Scholar] [CrossRef]
- Smith, M.T. Advances in Understanding Benzene Health Effects and Susceptibility. Annu. Rev. Public Health 2010, 31, 133–148. [Google Scholar] [CrossRef]
- Wishart, D.S. Emerging Applications of Metabolomics in Drug Discovery and Precision Medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef]
- Wild, C.P. Future Research Perspectives on Environment and Health: The Requirement for a More Expansive Concept of Translational Cancer Research. Environ. Health 2011, 10 (Suppl. 1), 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, R.; Zhang, J.; Xiong, M.; Chen, Y.; Yin, L.; Pu, Y. Metabonomics Biomarkers for Subacute Toxicity Screening for Benzene Exposure in Mice. J. Toxicol. Environ. Health 2012, 75, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Zhang, J.; Yin, L.; Pu, Y. Investigation into Variation of Endogenous Metabolites in Bone Marrow Cells and Plasma in C3H / He Mice Exposed to Benzene. Int. J. Mol. Sci. 2014, 15, 4994–5010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Sun, R.; Xu, K.; Pu, Y.; Huang, J.; Liu, M.; Chen, M.; Zhang, J.; Yin, L. Lipidomic Analysis Reveals Disturbances in Glycerophospholipid and Sphingolipid Metabolic Pathways in Benzene-Exposed Mice. Toxicol. Res. 2021, 10, 706–718. [Google Scholar] [CrossRef]
- Vaz, F.M.; Ferdinandusse, S. Bile Acid Analysis in Human Disorders of Bile Acid Biosynthesis. Mol. Aspects Med. 2017, 56, 10–24. [Google Scholar] [CrossRef]
- Jia, W.; Xie, G.; Jia, W. Bile Acid–Microbiota Cross-Talk in Gastrointestinal Inflammation and Carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef] [Green Version]
- Sun, R.; Xu, K.; Ji, S.; Pu, Y.; Man, Z.; Ji, J.; Chen, M.; Yin, L.; Zhang, J.; Pu, Y. Benzene Exposure Induces Gut Microbiota Dysbiosis and Metabolic Disorder in Mice. Sci. Total Environ. 2020, 705, 135879. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-Gut Microbiota Metabolic Interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [Green Version]
- Burtis, C.A.; Bruns, D.E.; Sawyer, B.G. Tietz Fundamentos de Química Clínica e Diagnóstico Molecular, 7th ed.; Elsevier: Rio de Janeiro, Brazil, 2016. [Google Scholar]
- Moro, A.M.; Brucker, N.; Charão, M.F.; Baierle, M.; Sauer, E.; Goethel, G.; Barth, A.; Nascimento, S.N.; Gauer, B.; Durgante, J.; et al. Biomonitoring of Gasoline Station Attendants Exposed to Benzene: Effect of Gender. Mutat. Res.—Genet. Toxicol. Environ. Mutagen. 2017, 813, 1–9. [Google Scholar] [CrossRef]
- Andrea, M.A.D.; Reddy, G.K. Hematological and Hepatic Alterations in Nonsmoking Residents Exposed to Benzene Following a Flaring Incident at the British Petroleum Plant in Texas City. Environ. Health 2014, 13, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.W.; Braidy, N.; Poljak, A.; Pickford, R.; Thambisetty, M.; Sachdev, P.S. Dysregulation of Lipids in Alzheimer’s Disease and Their Role as Potential Biomarkers. Alzheimer’s Dement. 2017, 13, 810–827. [Google Scholar] [CrossRef] [PubMed]
- Kurz, J.; Parnham, M.J.; Geisslinger, G.; Schiffmann, S. Ceramides as Novel Disease Biomarkers. Trends Mol. Med. 2019, 2019, 20–32. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid Metabolism in Cancer Signalling and Therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef] [Green Version]
- Mato, J.M.; Alonso, C.; Noureddin, M.; Lu, S.C. Biomarkers and Subtypes of Deranged Lipid Metabolism in Nonalcoholic Fatty Liver Disease. World J. Gastroenterol. 2019, 25, 3009–3020. [Google Scholar] [CrossRef]
- Pabst, T.; Kortz, L.; Fiedler, G.M.; Ceglarek, U.; Idle, J.R.; Beyoğlu, D. The Plasma Lipidome in Acute Myeloid Leukemia at Diagnosis in Relation to Clinical Disease Features. BBA Clin. 2017, 7, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Subrahmanyam, V.V.; Ross, D.; Eastmond, D.A.; Smith, M.T. Potential Role of Free Radicals in Benzene-Induced Myelotoxicity and Leukemia. Free Radic. Biol. Med. 1991, 11, 495–515. [Google Scholar] [CrossRef]
- Pirozzi, S.J.; Schlosser, M.J.; Kalf, G.F. Prevention of Benzene-Induced Myelotoxicity and Prostaglandin Synthesis in Bone Marrow of Mice by Inhibitors of Prostaglandin H Synthase. Immunopharmacology 1989, 18, 39–55. [Google Scholar] [CrossRef]
- Sulciner, M.L.; Gartung, A.; Gilligan, M.M.; Serhan, C.N.; Panigrahy, D. Targeting Lipid Mediators in Cancer Biology. Cancer Metastasis Rev. 2018, 37, 557–572. [Google Scholar] [CrossRef]
- Fabbri, A.; Magalotti, D.; Marchesini, G.; Brizi, M.; Bianchi, G.; Zoli, M.; Interna, M.; Orsola, P.S. Effects of Systemic Prostaglandin E 1 on Splanchnic and Peripheral Haemodynamics in Control Subjects and in Patients with Cirrhosis. Prostaglandins Other Lipid Mediat. 1998, 55, 209–218. [Google Scholar] [CrossRef]
- Gao, P.; Hou, L.; Denslow, N.D.; Xiang, P.; Ma, L.Q. Human Exposure to Polycyclic Aromatic Hydrocarbons: Metabolomics Perspective. Environ. Int. 2018, 119, 466–477. [Google Scholar] [CrossRef]
- Liu, X.L.; Fan, D.M. Protective Effects of Prostaglandin E1 on Hepatocytes. World J. Gastroenterol. 2000, 6, 326–329. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, S.; Merrill, A.H. Sphingolipid Metabolism and Cell Growth Regulation. FASEB J. 1996, 10, 1388–1397. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.J.; Davies, S.; Darley, R.L.; Tonks, A. Reactive Oxygen Species Rewires Metabolic Activity in Acute Myeloid Leukemia. Front. Oncol. 2021, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Musharraf, S.G.; Siddiqui, A.J.; Shamsi, T.; Naz, A. SERUM Metabolomics of Acute Lymphoblastic Leukaemia and Acute Myeloid Leukaemia for Probing Biomarker Molecules. Hematol. Oncol. 2016, 35, 769–777. [Google Scholar] [CrossRef]
- Moon, S.M.; Lee, S.A.; Hong, J.H.; Kim, J.S.; Kim, D.K.; Kim, C.S. Oleamide Suppresses Inflammatory Responses in LPS-Induced RAW264.7 Murine Macrophages and Alleviates Paw Edema in a Carrageenan-Induced Inflammatory Rat Model. Int. Immunopharmacol. 2018, 56, 179–185. [Google Scholar] [CrossRef]
- Naumoska, K.; Jug, U.; Metličar, V.; Vovk, I. Oleamide, a Bioactive Compound, Unwittingly Introduced into the Human Body through Some Plastic Food/Beverages and Medicine Containers. Foods 2020, 9, 549. [Google Scholar] [CrossRef]
- Morgan, G.J.; Smith, M.T. Metabolic Enzyme Polymorphisms and Susceptibility to Acute Leukemia in Adults. Am. J. Pharmacogenomics 2002, 2, 79–92. [Google Scholar] [CrossRef]
- Bittinger, M.A.; Nguyen, L.P.; Bradfield, C.A. Aspartate Aminotransferase Generates Proagonists of the Aryl Hydrocarbon Receptor. Mol. Pharmacol. 2003, 64, 550–556. [Google Scholar] [CrossRef] [Green Version]
- Cummings, J.R.; Sakata, T. Physiological and Clinical Aspects of Short-Chain Fatty Acids; Cambridge University Press: Cambridge, UK, 1995. [Google Scholar]
- Spatari, G.; Saitta, S.; Cimino, F.; Sapienza, D.; Quattrocchi, P.; Carrieri, M.; Barbaro, M.; Saija, A.; Gangemi, S. Increased Serum Levels of Advanced Oxidation Protein Products and Glycation End Products in Subjects Exposed to Low-Dose Benzene. Int. J. Hyg. Environ. Health 2012, 215, 389–392. [Google Scholar] [CrossRef]
- Avery, S.V. Molecular Targets of Oxidative Stress. Biochem. J. 2011, 434, 201–210. [Google Scholar] [CrossRef] [Green Version]
- Rothman, N.; Vermeulen, R.; Zhang, L.; Hu, W.; Yin, S.; Rappaport, S.M.; Smith, M.T.; Jones, D.P.; Rahman, M.; Lan, Q.; et al. Metabolome-Wide Association Study of Occupational Exposure to Benzene. Carcinogenesis 2021, 42, 1326–1336. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhang, L.; Wang, J.; Zhang, W.; Ren, J.; Chen, Y.; Zhang, Y.; Gao, A. Plasma Metabolomics Study Reveals the Critical Metabolic Signatures for Benzene-Induced Hematotoxicity. JCI Insight 2022, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Cao, M.; Zhang, J.; Yang, W.; Wei, H.; Meng, X.; Yin, L. Benzene Exposure Alters Expression of Enzymes Involved in Fatty Acid β -Oxidation in Male C3H/He Mice. Int. J. Environ. Res. Public Health 2016, 13, 1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, R.; Man, Z.; Ji, J.; Ji, S.; Xu, K.; Pu, Y.; Yu, L.; Zhang, J.; Yin, L.; Pu, Y. l-Carnitine protects against 1,4-benzoquinone-induced apoptosisand DNA damage by suppressing oxidative stress and promoting fatty acid oxidation in K562 cells. Environ. Toxicol. 2020, 35, 1033–1042. [Google Scholar] [CrossRef]




| Time (min) | Rate (mL/min) | % A | % B |
|---|---|---|---|
| 0 | 0.200 | 96.0 | 4.0 |
| 2 | 0.200 | 96.0 | 4.0 |
| 7 | 0.200 | 81.7 | 18.3 |
| 12 | 0.223 | 50.0 | 50 |
| 14 | 0.400 | 0.1 | 99.9 |
| 16 | 0.480 | 0.1 | 99.9 |
| 19 | 0.480 | 96.0 | 4.0 |
| 19.10 | 0.200 | 96.0 | 4.0 |
| 20 | 0.200 | 96.0 | 4.0 |
| Metabolites | M | VIP a Score | p b Values | Chemical Category |
|---|---|---|---|---|
| Phenylalanylhydroxyproline | 279.13 | 2.42 | 0.0000 | Peptide |
| Tetrahydropteroyltri-L glutamic acid | 703.25 | 2.29 | 0.0001 | Tetrahydrofolic acid |
| Trp Gln Asp Cys Glu | 679.22 | 2.22 | 0.0000 | Peptide |
| Testosterone glucuronide | 465.24 | 2.19 | 0.0000 | Steroid glucuronide |
| 7alpha-Hydroxy-3-oxo-5beta-cholan-24-oic acid | 390.27 | 2.18 | 0.0000 | Bile acid |
| Phosphatidylcholine(44:6) | 889.65 | 2.15 | 0.0000 | Phosphatidylcholine |
| Asp Asp Phe Hys | 532.19 | 2.14 | 0.0000 | Peptide |
| Phosphatidylcholine(42:6) | 861.62 | 2.11 | 00000 | Glycerophospholipid |
| Phosphatidylcholine(42:2) | 869.68 | 2.10 | 0.0000 | Glycerophospholipid |
| Phosphatidylcholine(22:2) | 825.66 | 2.10 | 0.0000 | Glycerophospholipid |
| Folic acid | 441.13 | 2.10 | 0.0000 | Pterins |
| Phosphatidylethanolamine PGE2/22:2(13Z,16Z) | 867.56 | 2.10 | 0.0000 | Glycerophospholipids |
| Phosphatidylcholine(40:3) | 891.64 | 2.10 | 0.0000 | Phosphatidylcholine |
| Cys Hys Ser Trp | 531.19 | 2.10 | 0.0000 | Peptide |
| 1,21-Henicosanediol | 328.33 | 2.09 | 0.0000 | Long chain fatty alcohol |
| Heptadecanoic carnitine | 413.35 | 2.08 | 0.0000 | Acyl carnitine |
| Phosphatidylglycerol (36:1) | 776.55 | 2.06 | 0.0000 | Phosphatidylglycerol |
| Phophatidylethanolamine(44:9) | 841.56 | 2.06 | 0.0000 | Phosphatidylethanolamine |
| 1-(9Z-heptadecenoyl)-2-(7Z, 10Z, 13Z, 16Z-docosatetraenoyl) -glycero-3-phosphoserine | 823.53 | 2.06 | 0.0000 | Glycerophospholipids |
| 1-Methylinosine | 283.28 | 2.05 | 0.0001 | Purine nucleoside |
| Cys Met Thr Tyr | 517.85 | 2.05 | 0.0000 | Peptide |
| Tetradecenoylcarnitine | 369.28 | 2.05 | 0.0000 | Acyl carnitine |
| Phosphatidylethanolamine(22:5) | 778.09 | 2.05 | 0.0000 | Glycerophospholipids |
| Phosphatidylcholine(36:0) | 775.64 | 2.05 | 0.0000 | Glycerophospholipids |
| Coprocholic acid | 450.33 | 2.04 | 0.0000 | Bile acid |
| Asp Leu | 494.24 | 2.04 | 0.0000 | Peptide |
| Phosphatidylserine(38:1) | 817.58 | 2.04 | 0.0000 | Phosphatidylserine |
| Phosphatidic acid(40:1) | 758.58 | 2.03 | 0.0000 | 1,2-diacylglycerol-3-phosphate |
| Phosphatidic acid PA(18:1(12Z)-2OH(9,10)/i-15:0) | 692.46 | 2.03 | 0.0000 | Glycerophospholipids |
| Phosphatidylcholine(32:1) | 731.54 | 2.02 | 0.0000 | Glycerophospholipids |
| Phosphatidylcholine(34:1) | 759.57 | 2.02 | 0.0000 | Glycerophospholipids |
| Phosphatidylethanolamine(34:2) | 715.51 | 2.02 | 0.0000 | Phosphatidylethanolamine |
| Cys Arg Trp Trp | 649.27 | 2.01 | 0.0000 | Peptide |
| Sphingomyelin (D18: 0/14: 1 (9Z) (OH)) | 688.51 | 2.01 | 0.0000 | Sphingolipid |
| Sphingomyelin (d18:1/16:0) | 702.56 | 2.01 | 0.0000 | Sphingolipid |
| Phosphatidylcholine(38:4) | 795.61 | 2.01 | 0.0000 | Glycerophosphocholine |
| Phosphatidylethanolamine(35:0) | 733.56 | 2.01 | 0.0000 | Glycerophospholipids |
| Phosphatidylethanolamine(32:3) | 671.48 | 2.00 | 0.0000 | Glycerophosphoethanolamine |
| CAT | GST | THIOL | MDA | SOD | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P¹ | p Value | P¹ | p Value | P¹ | p Value | P¹ | p Value | P¹ | p Value | |
| Phenylalanylhydroxyproline | −0.068 | 0.605 | 0.205 | 0.116 | 0.083 | 0.531 | −0.342 | 0.007 | −0.012 | 0.928 |
| Sphingomyelin (d18:1/16:0) | −0.019 | 0.887 | 0.162 | 0.218 | 0.149 | 0.256 | −0.356 | 0.005 | −0.002 | 0.987 |
| 1,21-Henicosanediol | 0.008 | 0.949 | −0.017 | 0.897 | 0.047 | 0.721 | 0.007 | 0.957 | 0.007 | 0.960 |
| Tetradecenoylcarnitine | 0.114 | 0.386 | 0.264 | 0.041 | 0.314 | 0.014 | −0.272 | 0.036 | −0.215 | 0.099 |
| 7alpha-Hydroxy-3-oxo-5beta-cholan-24-oic acid | 0.091 | 0.490 | 0.275 | 0.033 | 0.333 | 0.009 | −0.216 | 0.098 | −0.183 | 0.161 |
| Testosterone glucuronide | 0.034 | 0.796 | 0.268 | 0.039 | 0.273 | 0.035 | −0.306 | 0.017 | −0.267 | 0.039 |
| Phosphatidylglycerol (36:1) | −0.146 | 0.267 | −0.079 | 0.548 | 0.153 | 0.242 | −0.083 | 0.528 | 0.274 | 0.034 |
| Coprocholic acid | 0.135 | 0.303 | 0.302 | 0.019 | 0.238 | 0.067 | −0.305 | 0.018 | −0.280 | 0.030 |
| Folic acid | 0.064 | 0.627 | 0.281 | 0.030 | 0.331 | 0.010 | −0.276 | 0.033 | −0.251 | 0.053 |
| Asp Leu | 0.097 | 0.459 | 0.246 | 0.058 | 0.236 | 0.069 | −0.310 | 0.016 | −0.223 | 0.087 |
| Cys Met Thr Tyr | 0.141 | 0.282 | 0.266 | 0.040 | 0.214 | 0.100 | −0.303 | 0.019 | −0.236 | 0.070 |
| 1-(9Z-heptadecenoyl)-2-(7Z,10Z, 13Z, 16Z-docosatetraenoyl)-glycero-3-phosphoserine | 0.086 | 0.514 | 0.295 | 0.022 | 0.281 | 0.030 | −0.275 | 0.034 | −0.260 | 0.045 |
| Cys Hys Ser Trp | 0.030 | 0.817 | 0.275 | 0.033 | 0.254 | 0.051 | −0.248 | 0.056 | −0.261 | 0.044 |
| Asp Asp Fen Hys | 0.054 | 0.680 | 0.287 | 0.026 | 0.283 | 0.028 | −0.246 | 0.059 | −0.275 | 0.034 |
| Cys Arg Trp Trp | 0.113 | 0.391 | 0.297 | 0.021 | 0.291 | 0.024 | −0.265 | 0.041 | −0.262 | 0.043 |
| Phosphatidylethanolamine(32:3) | 0.103 | 0.433 | 0.308 | 0.017 | 0.303 | 0.019 | −0.265 | 0.041 | −0.264 | 0.042 |
| Sphingomyelin (D18: 0/14: 1 (9Z) (OH)) | 0.087 | 0.511 | 0.312 | 0.015 | 0.284 | 0.028 | −0.256 | 0.049 | −0.270 | 0.037 |
| Trp Gln Asp Cys Glu | 0.039 | 0.770 | 0.294 | 0.022 | 0.321 | 0.012 | −0.330 | 0.010 | −0.213 | 0.103 |
| Tetrahydropteroyltri-L-glutamic acid | −0.005 | 0.969 | 0.300 | 0.020 | 0.240 | 0.065 | −0.348 | 0.006 | −0.278 | 0.031 |
| Phosphatidic acid PA(18:1(12Z)-2OH(9,10)/i-15:0) | 0.095 | 0.472 | 0.304 | 0.018 | 0.268 | 0.038 | −0.253 | 0.051 | −0.256 | 0.048 |
| Phosphatidylcholine(32:1) | 0.096 | 0.464 | 0.306 | 0.017 | 0.286 | 0.027 | −0.258 | 0.047 | −0.259 | 0.045 |
| Phosphatidylethanolamine(34:2) | 0.107 | 0.414 | 0.295 | 0.022 | 0.277 | 0.032 | −0.256 | 0.049 | −0.261 | 0.044 |
| 1-Methylinosine | 0.044 | 0.739 | 0.292 | 0.024 | 0.247 | 0.057 | −0.196 | 0.132 | −0.401 | 0.002 |
| Phosphatidylethanolamine(35:0) | 0.121 | 0.358 | 0.291 | 0.024 | 0.274 | 0.034 | −0.253 | 0.051 | −0.251 | 0.053 |
| Heptadecanoic carnitine | 0.064 | 0.625 | 0.301 | 0.019 | 0.290 | 0.025 | −0.266 | 0.040 | −0.252 | 0.052 |
| Phosphatidylcholine(36:0) | 0.081 | 0.539 | 0.293 | 0.023 | 0.291 | 0.024 | −0.269 | 0.038 | −0.262 | 0.043 |
| Phophatidylethanolamine(22:5) | 0.096 | 0.468 | 0.297 | 0.021 | 0.268 | 0.038 | −0.255 | 0.049 | −0.279 | 0.031 |
| Phosphatidylcholine(38:4) | 0.091 | 0.489 | 0.308 | 0.017 | 0.266 | 0.040 | −0.256 | 0.048 | −0.251 | 0.053 |
| Phosphatidic acid(40:1) | 0.090 | 0.493 | 0.299 | 0.021 | 0.287 | 0.026 | −0.273 | 0.035 | −0.253 | 0.051 |
| Phosphatidylcholine(34:1) | 0.093 | 0.480 | 0.295 | 0.022 | 0.275 | 0.034 | −0.261 | 0.044 | −0.262 | 0.043 |
| Phosphatidylserine(38:1) | 0.088 | 0.503 | 0.296 | 0.022 | 0.277 | 0.032 | −0.259 | 0.045 | −0.252 | 0.052 |
| Phophatidylethanolamine(44:9) | 0.089 | 0.501 | 0.304 | 0.018 | 0.294 | 0.023 | −0.262 | 0.043 | −0.262 | 0.043 |
| Phosphatidylcholine(22:2) | 0.100 | 0.448 | 0.299 | 0.020 | 0.289 | 0.025 | −0.256 | 0.048 | −0.242 | 0.063 |
| Phosphatidylcholine(42:6) | 0.082 | 0.536 | 0.288 | 0.026 | 0.288 | 0.026 | −0.261 | 0.044 | −0.257 | 0.048 |
| Phosphatidylcholine(40:3) | 0.113 | 0.391 | 0.297 | 0.021 | 0.291 | 0.024 | −0.265 | 0.041 | −0.262 | 0.043 |
| Phosphatidylethanolamine PGE2/22:2(13Z, 16Z) | 0.095 | 0.473 | 0.289 | 0.025 | 0.263 | 0.043 | −0.281 | 0.029 | −0.238 | 0.067 |
| Phosphatidylcholine(42:2) | 0.079 | 0.547 | 0.311 | 0.015 | 0.285 | 0.027 | −0.274 | 0.034 | −0.279 | 0.031 |
| Phosphatidylcholine(44:6) | 0.075 | 0.569 | 0.310 | 0.016 | 0.293 | 0.023 | −0.270 | 0.037 | −0.252 | 0.052 |
| Break Rate | Fragments Rate | Metaphase Rate with Premature Separation | ||||
|---|---|---|---|---|---|---|
| P¹ | p Value | P¹ | p Value | P¹ | p Value | |
| Phenylalanylhydroxyproline | −0.315 | 0.014 | −0.316 | 0.014 | 0.196 | 0.136 |
| Sphingomyelin (d18:1/16:0) | −0.271 | 0.036 | −0.263 | 0.042 | 0.150 | 0.256 |
| 1,21-Henicosanediol | −0.334 | 0.009 | −0.331 | 0.010 | 0.154 | 0.243 |
| Tetradecenoylcarnitine | −0.291 | 0.024 | −0.290 | 0.024 | 0.320 | 0.013 |
| 7alpha-Hydroxy-3-oxo-5beta-cholan-24-oic acid | −0.244 | 0.060 | −0.249 | 0.055 | 0.353 | 0.006 |
| Testosterone glucuronide | −0.272 | 0.035 | −0.272 | 0.035 | 0.300 | 0.021 |
| Phosphatidylglycerol (36:1) | 0083 | 0.528 | 0.089 | 0.498 | 0.130 | 0.327 |
| Coprocholic acid | −0.287 | 0.026 | −0.298 | 0.021 | 0.275 | 0.035 |
| Folic acid | −0.241 | 0.063 | −0.243 | 0.061 | 0.349 | 0.007 |
| Asp Leu | −0.303 | 0.019 | −0.308 | 0.017 | 0.319 | 0.014 |
| Cys Met Thr Tyr | −0.315 | 0.014 | −0.313 | 0.015 | 0.269 | 0.039 |
| 1-(9Z-heptadecenoyl)-2-(7Z,10Z, 13Z, 16Z-docosatetraenoyl)-glycero-3-phosphoserine | −0.282 | 0.029 | −0.290 | 0.025 | 0.304 | 0.019 |
| Cys Hys Ser Trp | −0.258 | 0.047 | −0.250 | 0.054 | 0.281 | 0.031 |
| Asp Asp Fen Hys | −0.279 | 0.031 | −0.277 | 0.032 | 0.299 | 0.021 |
| Cys Arg Trp Trp | −0.276 | 0.033 | −0.298 | 0.021 | 0.360 | 0.005 |
| Phosphatidylethanolamine(32:3) | −0.250 | 0.054 | −0.249 | 0.055 | 0.273 | 0.037 |
| Sphingomyelin (D18: 0/14: 1 (9Z)(OH)) | −0.247 | 0.057 | −0.248 | 0.056 | 0.280 | 0.032 |
| Trp Gln Asp Cys Glu | −0.256 | 0.049 | −0.256 | 0.048 | 0.286 | 0.028 |
| Tetrahydropteroyltri-L-glutamic acid | −0.264 | 0.042 | −0.258 | 0.046 | 0.291 | 0.025 |
| Phosphatidic acid PA(18:1(12Z)-2OH(9,10)/i-15:0) | −0.277 | 0.032 | −0.283 | 0.028 | 0.357 | 0.006 |
| Phosphatidylcholine(32:1) | −0.266 | 0040 | −0.274 | 0.034 | 0.282 | 0.031 |
| Phosphatidylethanolamine(34:2) | −0.259 | 0.045 | −0.261 | 0.044 | 0.284 | 0.029 |
| 1-Methylinosine | −0.259 | 0.046 | −0.260 | 0.045 | 0.285 | 0.029 |
| Phosphatidylethanolamine(35:0) | −0.259 | 0.046 | −0.260 | 0.045 | 0.278 | 0.033 |
| Heptadecanoic carnitine | −0.257 | 0.047 | −0.260 | 0.045 | 0.294 | 0.024 |
| Phosphatidylcholine(36:0) | −0.264 | 0.042 | −0.268 | 0.039 | 0.303 | 0.020 |
| Phophatidylethanolamine(22:5) | −0.292 | 0.024 | −0.295 | 0.022 | 0.296 | 0.023 |
| Phosphatidylcholine(38:4) | −0.270 | 0.037 | −0.276 | 0.033 | 0.301 | 0.021 |
| Phosphatidic acid(40:1) | −0.259 | 0.046 | −0.262 | 0.043 | 0.299 | 0.021 |
| Phosphatidylcholine(34:1) | −0.253 | 0.051 | −0.258 | 0.047 | 0.312 | 0.016 |
| Phosphatidylserine(38:1) | −0.259 | 0.045 | −0.265 | 0.041 | 0.301 | 0.021 |
| Phophatidylethanolamine(44:9) | −0.272 | 0.035 | −0.279 | 0.031 | 0.282 | 0.030 |
| Phosphatidylcholine(22:2) | −0.258 | 0.047 | −0.259 | 0.046 | 0.280 | 0.032 |
| Phosphatidylcholine(42:6) | −0.250 | 0.054 | −0.255 | 0.049 | 0.298 | 0.022 |
| Phosphatidylcholine(40:3) | −0.255 | 0.049 | −0.261 | 0.044 | 0.306 | 0.018 |
| Phosphatidylethanolamine PGE2/22:2(13Z,16Z) | −0.258 | 0.046 | −0.263 | 0.042 | 0.276 | 0.034 |
| Phosphatidylcholine(42:2) | −0.276 | 0.033 | −0.283 | 0.029 | 0.287 | 0.027 |
| Phosphatidylcholine(44:6) | −0.257 | 0.047 | −0.267 | 0.039 | 0.321 | 0.013 |
| Metabolite | p Value b | Fold Change c | Pathway |
|---|---|---|---|
| 1,21-Henicosanediol | <0.0001 | 2.30 | Lipid transport and lipid metabolism Fatty acid metabolism Lipid peroxidation and cell signaling. |
| Tetradecenoylcarnitine | <0.0001 | 5.05 | Lipid transport and lipid metabolism Fatty acid metabolism Lipid peroxidation and cell signaling. |
| Coprocholic acid | <0.0001 | 5.33 | Lipid transport and metabolism Fatty acid metabolism |
| Cys Met Thr Tyr | <0.0001 | 4.76 | Product of incomplete decomposition of proteins or protein catabolism |
| Asp Leu | <0.0001 | 4.72 | Product of incomplete decomposition of proteins or protein catabolism |
| Phenylalanylhydroxyproline | <0.0001 | 2.27 | Product of incomplete decomposition of proteins or protein catabolism |
| Cys Hys Ser Trp | <0.0001 | 2.31 | Product of incomplete decomposition of proteins or protein catabolism. |
| Sphingomyelin (D18: 0/14: 1 (9Z) (OH)) | <0.0001 | 5.05 | Lipid metabolism and signaling cell. |
| PE (PGE2/22:2 (13Z, 16Z)) | <0.0001 | 4.73 | Cell signaling |
| Phophatidylethanolamine(44:9) | <0.0001 | 5.20 | Components of the lipid bilayer of cells Lipid metabolism Cell signaling |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendes, M.P.R.; Paiva, M.J.N.; Costa-Amaral, I.C.; Carvalho, L.V.B.; Figueiredo, V.O.; Gonçalves, E.S.; Larentis, A.L.; André, L.C. Metabolomic Study of Urine from Workers Exposed to Low Concentrations of Benzene by UHPLC-ESI-QToF-MS Reveals Potential Biomarkers Associated with Oxidative Stress and Genotoxicity. Metabolites 2022, 12, 978. https://doi.org/10.3390/metabo12100978
Mendes MPR, Paiva MJN, Costa-Amaral IC, Carvalho LVB, Figueiredo VO, Gonçalves ES, Larentis AL, André LC. Metabolomic Study of Urine from Workers Exposed to Low Concentrations of Benzene by UHPLC-ESI-QToF-MS Reveals Potential Biomarkers Associated with Oxidative Stress and Genotoxicity. Metabolites. 2022; 12(10):978. https://doi.org/10.3390/metabo12100978
Chicago/Turabian StyleMendes, Michele P. R., Maria José N. Paiva, Isabele C. Costa-Amaral, Leandro V. B. Carvalho, Victor O. Figueiredo, Eline S. Gonçalves, Ariane L. Larentis, and Leiliane C. André. 2022. "Metabolomic Study of Urine from Workers Exposed to Low Concentrations of Benzene by UHPLC-ESI-QToF-MS Reveals Potential Biomarkers Associated with Oxidative Stress and Genotoxicity" Metabolites 12, no. 10: 978. https://doi.org/10.3390/metabo12100978
APA StyleMendes, M. P. R., Paiva, M. J. N., Costa-Amaral, I. C., Carvalho, L. V. B., Figueiredo, V. O., Gonçalves, E. S., Larentis, A. L., & André, L. C. (2022). Metabolomic Study of Urine from Workers Exposed to Low Concentrations of Benzene by UHPLC-ESI-QToF-MS Reveals Potential Biomarkers Associated with Oxidative Stress and Genotoxicity. Metabolites, 12(10), 978. https://doi.org/10.3390/metabo12100978





