Metabolic Analysis of Nucleosides/Bases in the Urine and Serum of Patients with Alcohol-Associated Liver Disease
Abstract
:1. Introduction
2. Methods and Samples
2.1. Chemicals and Reagents
2.2. Patient Recruitment and Sample Collection
2.3. Metabolite Extraction
2.4. LC-MS/MS Analysis
2.5. Data Analysis
3. Results
3.1. Modified Nucleosides/Bases Are Increased in the Urine of Patients with ALD
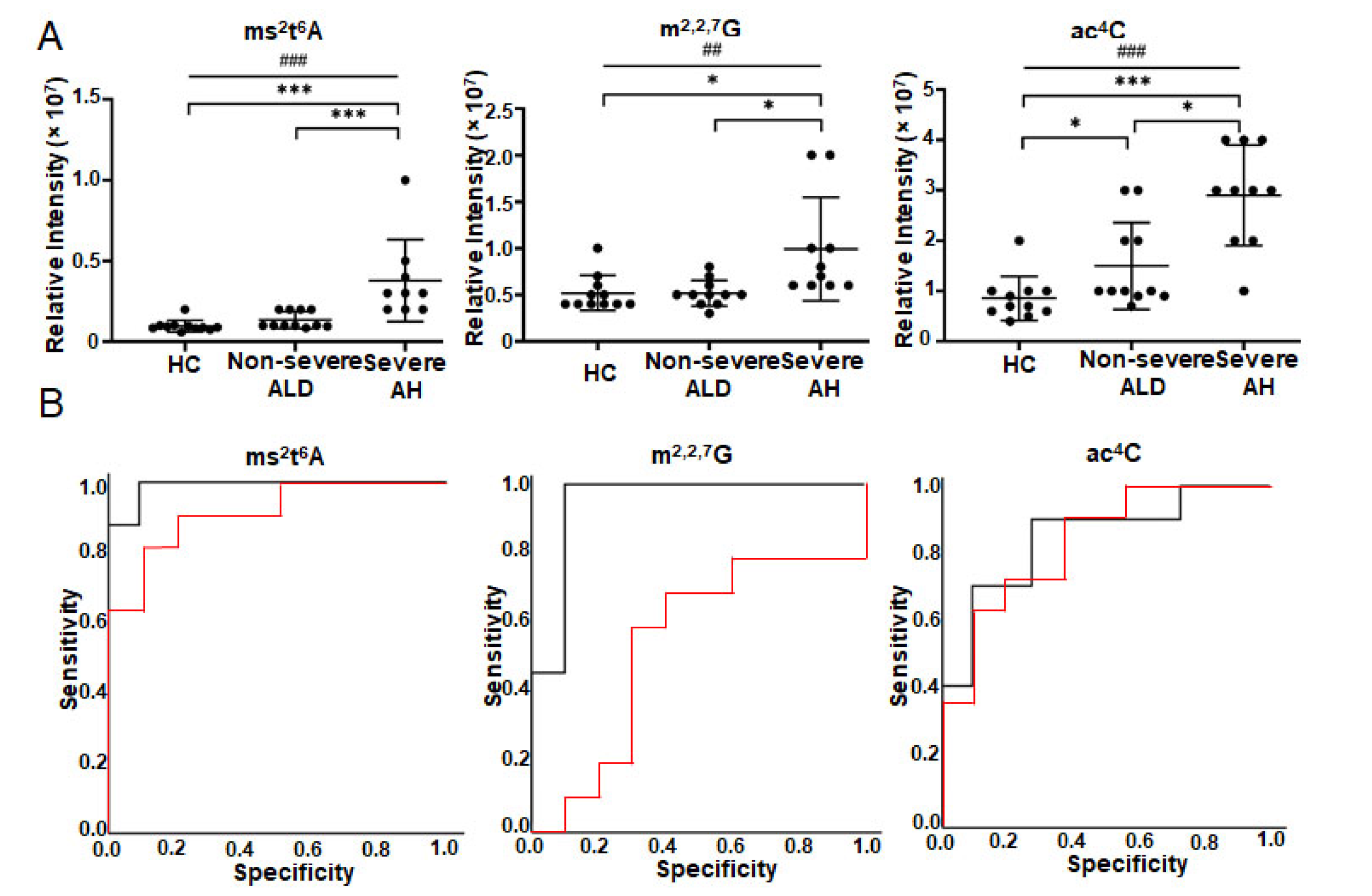
3.2. Some Free Modified Nucleosides/Bases in Patients’ Urine Are Strongly Associated with the Severity of ALD
3.3. Modified Nucleosides/Bases Are Changed in Serum of Patients with ALD
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crecy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef]
- Wei, J.; Liu, F.; Lu, Z.; Fei, Q.; Ai, Y.; He, P.C.; Shi, H.; Cui, X.; Su, R.; Klungland, A.; et al. Differential m(6)A, m(6)Am, and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 2018, 71, 973–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Chang, M.; Lv, H.; Zhang, Z.W.; Zhang, W.; He, X.; Wu, G.; Zhao, S.; Zhang, Y.; Wang, D.; et al. RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018, 19, 68. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Chen, Y.; Sun, B.; Wang, L.; Yang, Y.; Ma, D.; Lv, J.; Heng, J.; Ding, Y.; Xue, Y.; et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature 2017, 549, 273–276. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bogler, O.; et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef] [Green Version]
- Jonkhout, N.; Tran, J.; Smith, M.A.; Schonrock, N.; Mattick, J.S.; Novoa, E.M. The RNA modification landscape in human disease. RNA 2017, 23, 1754–1769. [Google Scholar] [CrossRef] [Green Version]
- Xu, K.; Sun, Y.; Sheng, B.; Zheng, Y.; Wu, X.; Xu, K. Role of identified RNA N6-methyladenosine methylation in liver. Anal. Biochem. 2019, 578, 45–50. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, L.; Chen, J.; Wang, Y. mRNA m(6)A methylation downregulates adipogenesis in porcine adipocytes. Biochem. Biophys. Res. Commun. 2015, 459, 201–207. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, Y.; Sun, B.F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.J.; Ping, X.L.; Chen, Y.S.; Wang, W.J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef]
- Imam, H.; Khan, M.; Gokhale, N.S.; McIntyre, A.B.R.; Kim, G.W.; Jang, J.Y.; Kim, S.J.; Mason, C.E.; Horner, S.M.; Siddiqui, A. N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci. USA 2018, 115, 8829–8834. [Google Scholar] [CrossRef] [Green Version]
- Gokhale, N.S.; McIntyre, A.B.R.; McFadden, M.J.; Roder, A.E.; Kennedy, E.M.; Gandara, J.A.; Hopcraft, S.E.; Quicke, K.M.; Vazquez, C.; Willer, J.; et al. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host. Microbe. 2016, 20, 654–665. [Google Scholar] [CrossRef] [Green Version]
- Chan, T.H.; Lin, C.H.; Qi, L.; Fei, J.; Li, Y.; Yong, K.J.; Liu, M.; Song, Y.; Chow, R.K.; Ng, V.H.; et al. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut 2014, 63, 832–843. [Google Scholar] [CrossRef]
- Grover, Z.; Lewindon, P.; Clousten, A.; Shaag, A.; Elpeleg, O.; Coman, D. Hepatic Copper Accumulation: A Novel Feature in Transient Infantile Liver Failure Due to TRMU Mutations? JIMD Rep. 2015, 21, 109–113. [Google Scholar]
- Zeharia, A.; Shaag, A.; Pappo, O.; Mager-Heckel, A.M.; Saada, A.; Beinat, M.; Karicheva, O.; Mandel, H.; Ofek, N.; Segel, R.; et al. Acute infantile liver failure due to mutations in the TRMU gene. Am. J. Hum. Genet. 2009, 85, 401–407. [Google Scholar] [CrossRef] [Green Version]
- Bajaj, J.S.; Garcia-Tsao, G.; Reddy, K.R.; O′Leary, J.G.; Vargas, H.E.; Lai, J.C.; Kamath, P.S.; Tandon, P.; Subramanian, R.M.; Thuluvath, P.; et al. Admission Urinary and Serum Metabolites Predict Renal Outcomes in Hospitalized Patients With Cirrhosis. Hepatology 2021, 74, 2699–2713. [Google Scholar] [CrossRef]
- Shi, Q.M.; Xue, C.; Yuan, X.; He, Y.T.; Yu, Z.J. Gene signatures and prognostic values of m1A-related regulatory genes in hepatocellular carcinoma. Sci. Rep. 2020, 10, 15083. [Google Scholar] [CrossRef]
- Klinge, C.M.; Piell, K.M.; Petri, B.J.; He, L.Q.; Zhang, X.; Pan, J.M.; Rai, S.N.; Andreeva, K.; Rouchka, E.C.; Wahlang, B.; et al. Combined exposure to polychlorinated biphenyls and high-fat diet modifies the global epitranscriptomic landscape in mouse liver. Environ. Epigenetics 2021, 7, dvab008. [Google Scholar]
- He, L.; Wei, X.; Ma, X.; Yin, X.; Song, M.; Donninger, H.; Yaddanapudi, K.; McClain, C.J.; Zhang, X. Simultaneous Quantification of Nucleosides and Nucleotides from Biological Samples. J. Am. Soc. Mass Spectrom. 2019, 30, 987–1000. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Li, F.; Yin, X.; Bohman, P.; Kim, S.; McClain, C.J.; Feng, W.; Zhang, X. Profiling of Polar Metabolites in Mouse Feces Using Four Analytical Platforms to Study the Effects Of Cathelicidin-Related Antimicrobial Peptide in Alcoholic Liver Disease. J. Proteome Res. 2019, 18, 2875–2884. [Google Scholar] [CrossRef]
- Vatsalya, V.; Cave, M.C.; Kong, M.; Gobejishvili, L.; Falkner, K.C.; Craycroft, J.; Mitchell, M.; Szabo, G.; McCullough, A.; Dasarathy, S.; et al. Keratin 18 Is a Diagnostic and Prognostic Factor for Acute Alcoholic Hepatitis. Clin. Gastroenterol. Hepatol. 2020, 18, 2046–2054. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate a Practical and Powerful Approach to Multiple Testing. J. R Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Mccloskey, J.A.; Nishimura, S. Modified Nucleosides in Transfer-Rna. Acc. Chem. Res. 1977, 10, 403–410. [Google Scholar] [CrossRef]
- Metzger, S.; Lippert, B. Self-complementarity of 7,9-dimethylguanine: A base pair containing three hydrogen bonds. Angew. Chem. Int. Ed. 1996, 35, 1228–1229. [Google Scholar] [CrossRef]
- Sigel, R.K.O.; Freisinger, E.; Abbate, M.; Lippert, B. Hydrogen bonding patterns of 7,9-dimethylguanine and its transplatinum(II) complexes. Inorg. Chim. Acta 2002, 339, 355–365. [Google Scholar] [CrossRef] [Green Version]
- Ramanathan, A.; Robb, G.B.; Chan, S.H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef] [Green Version]
- Yamagata, Y.; Fukumoto, S.; Hamada, K.; Fujiwara, T.; Tomita, K. A Novel Guanine-Guanine Base-Pairing Crystal-Structure of a Complex between 7-Methylguanosine and Its Iodide. Nucleic Acids Res. 1983, 11, 6475–6486. [Google Scholar] [CrossRef] [Green Version]
- Arragain, S.; Handelman, S.K.; Forouhar, F.; Wei, F.Y.; Tomizawa, K.; Hunt, J.F.; Douki, T.; Fontecave, M.; Mulliez, E.; Atta, M. Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA. J. Biol. Chem. 2010, 285, 28425–28433. [Google Scholar] [CrossRef] [Green Version]
- McCrate, N.E.; Varner, M.E.; Kim, K.I.; Nagan, M.C. Molecular dynamics simulations of human tRNA Lys,3 UUU: The role of modified bases in mRNA recognition. Nucleic Acids Res. 2006, 34, 5361–5368. [Google Scholar] [CrossRef]
- Wei, F.Y.; Suzuki, T.; Watanabe, S.; Kimura, S.; Kaitsuka, T.; Fujimura, A.; Matsui, H.; Atta, M.; Michiue, H.; Fontecave, M.; et al. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Investig. 2011, 121, 3598–3608. [Google Scholar] [CrossRef] [Green Version]
- Vangaveti, S.; Ranganathan, S.V.; Agris, P.F. Physical Chemistry of a Single tRNA-Modified Nucleoside Regulates Decoding of the Synonymous Lysine Wobble Codon and Affects Type 2 Diabetes. J. Phys. Chem. B 2022, 126, 1168–1177. [Google Scholar] [CrossRef]
- Baniasadi, H.; Gowda, G.A.N.; Gu, H.W.; Zeng, A.; Zhuang, S.; Skill, N.; Maluccio, M.; Raftery, D. Targeted metabolic profiling of hepatocellular carcinoma and hepatitis C using LC-MS/MS. Electrophoresis 2013, 34, 2910–2917. [Google Scholar] [CrossRef]


| Variables | HC (n = 19) | Alcohol-Associated Liver Disease (ALD) | p-Value a | |||
|---|---|---|---|---|---|---|
| Non-Severe ALD (n = 21) | Severe AH (n = 25) | Total Patients (n = 46) | ||||
| AUD (n = 8) | Moderate AH (n = 13) | |||||
| Age (years) | 36 (24–60) | 51 (39–67) | 50 (34–65) | 47 (27–66) | 49 (27–67) | - |
| Male (Female) | 13 (6) | 4 (4) | 7 (6) | 20 (5) | 31 (15) | - |
| BMI | N/A | 31.1 (27.8–33.4) | 25.9 (20.6–41.1) | 29.7 (22.4–50.5) | 29.7 (20.6–50.5) | - |
| T. Bilirubin (mg/dL) | 0.7 (0.4–1.3) | 1.5 (0.8–2.6) | 4.2 (1.2–18.2) | 12.9 (3.7–34.2) | 6.7 (0.8–34.2) | <0.001 |
| INR | N/A | 1.2 (1.1–1.7) | 1.5 (1.2–2.8) | 2.0 (1.0–3.2) | 1.7 (1.0–3.2) | - |
| AST (U/L) | 27 (19–66) | 59 (21–120) | 119 (53–347) | 88 (16–190) | 90 (16–347) | <0.001 |
| ALT (U/L) | 25 (16–109) | 36.8 (14.0–60.0) | 48 (18–194) | 35 (16–66) | 39 (14–194) | 0.015 |
| Alkaline phosphatase (IU/L) | 52 (37–62) | 124 (89–232) | 173 (80–518) | 144 (71–336) | 148 (71–518) | <0.001 |
| Albumin (g/dL) | 4.2 (3.8–4.3) | 3.9 (2.6–4.9) | 2.8 (1.9–4.5) | 2.4 (1.4–4.3) | 2.7 (1.4–4.9) | <0.001 |
| Creatinine (mg/dL) | 0.88 (0.69–1.07) | 0.69 (0.36–1.40) | 0.68 (0.32–1.30) | 0.89 (0.39–5.68) | 0.79 (0.32–5.68) | 0.082 |
| MELD score | N/A | 9.2 (6.0–11.0) | 16 (12–19) | 24 (20–39) | 18 (6–39) | - |
| Name | tR (min) | q-Value a | ROC Analysis | Non-Severe AH vs. HC | Severe AH vs. HC | Severe AH vs. Non-Severe ALD | -Value e | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-Value b | AUC c | 95% CI c | Fold-Change d | q-Value a | Fold-Change d | q-Value a | Fold-Change d | q-Value a | ||||
| 7,9-dimethylguanine | 6.35 | <0.001 | <0.001 | 0.934 | 0.863–1.000 | 3.20 | <0.001 | 18.63 | <0.001 | 5.82 | <0.001 | 0.71 |
| m2,2G | 5.52 | <0.001 | 0.001 | 0.791 | 0.662–0.920 | 1.31 | 0.071 | 2.32 | <0.001 | 1.77 | 0.008 | 0.48 |
| m1A | 6.68 | 0.001 | 0.002 | 0.757 | 0.610–0.903 | 2.47 | 0.774 | 7.99 | 0.001 | 3.24 | 0.004 | 0.47 |
| m1I | 5.42 | <0.001 | 0.002 | 0.763 | 0.624–0.903 | 1.42 | 0.043 | 2.62 | <0.001 | 1.84 | 0.002 | 0.57 |
| m3C/m4C/m5C | 6.86 | 0.007 | 0.003 | 0.235 | 0.100–0.370 | 1.08 | 0.547 | 0.24 | 0.094 | 0.22 | 0.003 | −0.48 |
| C | 9.98 | <0.001 | 0.003 | 0.752 | 0.609–0.896 | 0.97 | 0.208 | 2.63 | <0.001 | 2.72 | 0.048 | - |
| DHU | 6.64 | 0.001 | 0.003 | 0.748 | 0.610–0.886 | 1.11 | 0.767 | 3.15 | 0.001 | 2.85 | 0.005 | 0.45 |
| 1-Methylguanine | 9.62 | 0.033 | - | - | - | 1.12 | 0.189 | 2.27 | 0.033 | 2.04 | 0.192 | - |
| A | 5.72 | 0.039 | - | - | - | 0.93 | 0.664 | 0.66 | 0.039 | 0.71 | 0.201 | - |
| Uracil | 6.58 | 0.034 | - | - | - | 2.08 | 0.086 | 3.35 | 0.034 | 1.61 | 0.921 | - |
| Name | tR (min) | q-Value a | ROC Analysis | Non-Severe ALD vs. HC | Severe ALD vs. HC | Severe AH vs. Non-Severe ALD | -Value e | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-Value b | AUC c | 95% CI c | Fold-Change d | q-Value a | Fold-Change d | q-Value a | Fold-Change d | q-Value a | ||||
| 7,9-Dimethylguanine | 4.95 | <0.001 | <0.001 | 0.969 | 0.909–1.000 | 5.35 | <0.001 | 19.20 | <0.001 | 3.59 | <0.001 | 0.66 |
| ms2t6A | 18.98 | <0.001 | 0.001 | 0.796 | 0.665–0.927 | 1.28 | 0.314 | 1.86 | <0.001 | 1.45 | 0.119 | 0.50 |
| m2,2G | 10.85 | <0.001 | <0.001 | 0.856 | 0.740–0.971 | 1.02 | 0.974 | 1.56 | <0.001 | 1.53 | 0.013 | 0.50 |
| m2,2,7G | 9.60 | 0.013 | 0.001 | 0.791 | 0.645–0.937 | 1.09 | 0.691 | 2.02 | 0.013 | 1.86 | 0.161 | - |
| 5′-Deoxyuridine | 4.26 | 0.031 | 0.006 | 0.738 | 0.591–0.885 | 1.09 | 0.931 | 1.65 | 0.031 | 1.50 | 0.130 | 0.47 |
| DHU | 4.27 | 0.009 | 0.006 | 0.737 | 0.589–0.885 | 1.10 | 0.691 | 1.67 | 0.009 | 1.51 | 0.130 | 0.44 |
| MTA | 12.84 | 0.009 | 0.011 | 0.737 | 0.582–0.891 | 1.43 | 0.818 | 2.54 | 0.009 | 1.77 | 0.111 | - |
| mcm5s2U | 12.92 | 0.027 | 0.002 | 0.779 | 0.636–0.922 | 0.78 | 0.999 | 1.43 | 0.092 | 1.84 | 0.027 | - |
| m1I | 8.91 | <0.001 | 0.017 | 0.713 | 0.555–0.871 | 1.16 | 0.136 | 1.47 | <0.001 | 1.27 | 0.284 | 0.48 |
| m6A | 9.15 | 0.073 | 0.005 | 0.233 | 0.062–0.404 | 1.59 | 1.000 | 0.72 | 0.130 | 0.45 | 0.073 | - |
| m3U | 9.01 | 0.136 | 0.016 | 0.719 | 0.559–0.880 | 0.89 | 0.776 | 1.14 | 0.663 | 1.28 | 0.136 | - |
| I | 7.12 | 0.018 | - | - | - | 1.72 | 0.018 | 1.93 | 0.119 | 1.12 | 0.875 | - |
| Ψ | 4.36 | 0.013 | - | - | - | 1.04 | 0.139 | 1.52 | 0.013 | 1.46 | 0.728 | - |
| G | 7.10 | 0.009 | - | - | - | 1.62 | 0.300 | 2.08 | 0.009 | 1.28 | 0.330 | - |
| Name | tR (min) | q-Value a | ROC Analysis | Non-Severe ALD vs. HC | Severe ALD vs. HC | Severe AH vs. Non-Severe ALD | -Value e | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-Value b | AUC c | 95% CI c | Fold-Change d | q-Value a | Fold-Change d | q-Value a | Fold-Change d | q-Value a | ||||
| ms2t6A | 19.01 | <0.001 | <0.001 | 0.989 | 0.953–1.000 | 1.62 | 0.173 | 2.97 | <0.001 | 1.83 | <0.001 | 0.74 |
| ac4C | 9.63 | <0.001 | 0.007 | 0.845 | 0.672–1.000 | 1.86 | 0.036 | 3.99 | <0.001 | 2.15 | 0.028 | 0.59 |
| m2,2,7G | 9.55 | 0.022 | 0.001 | 0.944 | 0.832–1.000 | 1.23 | 1.000 | 1.75 | 0.022 | 1.43 | 0.022 | 0.50 |
| m2,2G | 10.84 | 0.033 | 0.102 | 0.722 | 0.469–0.975 | 1.21 | 0.265 | 1.54 | 0.033 | 1.28 | 0.494 | - |
| m1I | 8.89 | <0.001 | 0.014 | 0.828 | 0.647–1.000 | 1.36 | 0.265 | 1.93 | <0.001 | 1.42 | 0.040 | 0.47 |
| m3U | 8.98 | 0.018 | 0.014 | 0.828 | 0.644–1.000 | 1.03 | 1.000 | 1.70 | 0.018 | 1.66 | 0.022 | 0.44 |
| m5U | 7.88 | 0.179 | 0.029 | 0.218 | 0.007–0.429 | 0.97 | 1.000 | 0.79 | 0.362 | 0.82 | 0.179 | - |
| ncm5U | 5.56 | 0.010 | - | 0.727 | 0.502–0.953 | 1.08 | 0.531 | 1.51 | 0.010 | 1.39 | 0.129 | 0.44 |
| m4C | 5.18 | 0.173 | 0.017 | 0.170 | 0.000–0.364 | 1.65 | 0.316 | 0.77 | 1.000 | 0.47 | 0.173 | −0.40 |
| C | 4.25 | <0.001 | 0.007 | 0.860 | 0.689–1.000 | 1.95 | 0.087 | 4.06 | <0.001 | 2.08 | 0.086 | 0.67 |
| I | 7.09 | <0.001 | - | - | - | 0.14 | <0.001 | 0.13 | <0.001 | 0.91 | 1.000 | - |
| m5Um | 10.96 | 0.010 | - | - | - | 1.53 | 0.071 | 1.72 | 0.010 | 1.13 | 0.577 | - |
| G | 7.08 | <0.001 | - | - | - | 0.09 | <0.001 | 0.10 | <0.001 | 1.10 | 1.000 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Vatsalya, V.; Ma, X.; Klinge, C.M.; Cave, M.C.; Feng, W.; McClain, C.J.; Zhang, X. Metabolic Analysis of Nucleosides/Bases in the Urine and Serum of Patients with Alcohol-Associated Liver Disease. Metabolites 2022, 12, 1187. https://doi.org/10.3390/metabo12121187
He L, Vatsalya V, Ma X, Klinge CM, Cave MC, Feng W, McClain CJ, Zhang X. Metabolic Analysis of Nucleosides/Bases in the Urine and Serum of Patients with Alcohol-Associated Liver Disease. Metabolites. 2022; 12(12):1187. https://doi.org/10.3390/metabo12121187
Chicago/Turabian StyleHe, Liqing, Vatsalya Vatsalya, Xipeng Ma, Carolyn M. Klinge, Matthew C. Cave, Wenke Feng, Craig J. McClain, and Xiang Zhang. 2022. "Metabolic Analysis of Nucleosides/Bases in the Urine and Serum of Patients with Alcohol-Associated Liver Disease" Metabolites 12, no. 12: 1187. https://doi.org/10.3390/metabo12121187
APA StyleHe, L., Vatsalya, V., Ma, X., Klinge, C. M., Cave, M. C., Feng, W., McClain, C. J., & Zhang, X. (2022). Metabolic Analysis of Nucleosides/Bases in the Urine and Serum of Patients with Alcohol-Associated Liver Disease. Metabolites, 12(12), 1187. https://doi.org/10.3390/metabo12121187






