Antibiotic Isoflavonoids, Anthraquinones, and Pterocarpanoids from Pigeon Pea (Cajanus cajan L.) Seeds against Multidrug-Resistant Staphylococcus aureus
Abstract
:1. Introduction
2. Results
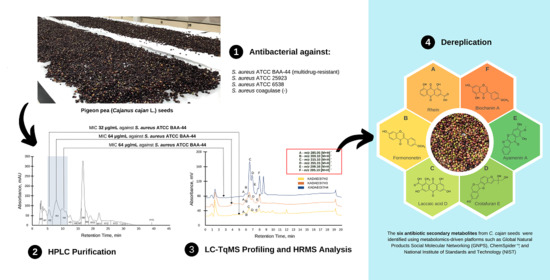
2.1. Preliminary Antibacterial Screening of C. cajan Extracts
2.2. Bioassay Guided Purification of Acetone Crude Extract
2.3. Purification of Ethyl Acetate Fraction (KADAME) by Flash Column Chromatography
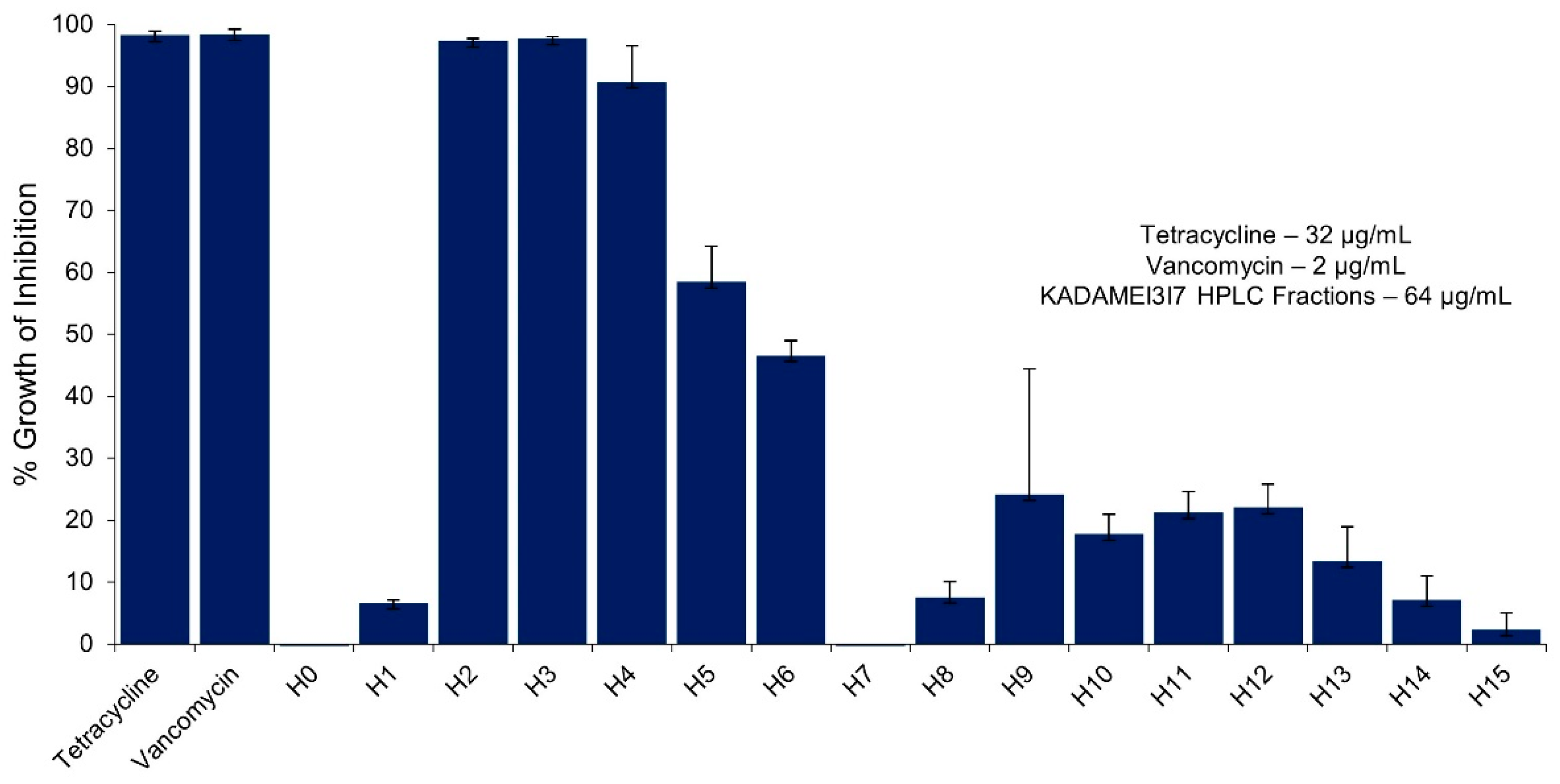
2.4. Antibiotic Kinetics Assay of KADAMEI3I7 Fraction
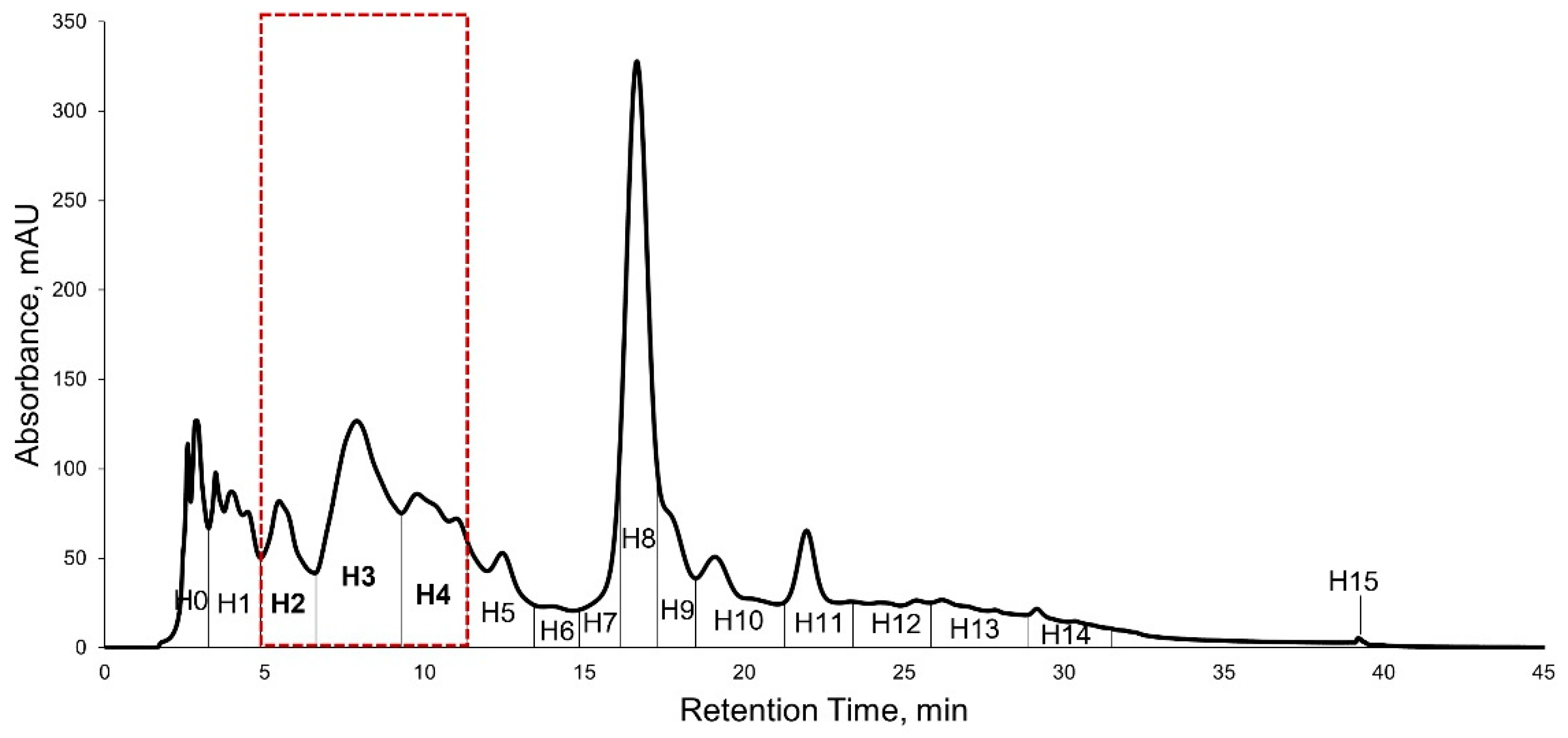
2.5. High Performance Liquid Chromatography (HPLC) Purification of KADAMEI3I7 and Antibacterial Assay against MDRSA
2.6. Liquid Chromatography-Triple Quadrupole Mass Spectrometry Profiling, MS/MS Analysis, and Dereplication by Global Natural Products Molecular Networking Social (GNPS)
2.7. High Resolution Mass Spectrometry (LCMS), MS/MS, and Dereplication Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Reagents and Standards
4.3. Test Pathogens
4.4. Phytochemical Extraction
4.5. Solvent Partitioning of C. cajan Seed Extracts
4.6. Solvent Partitioning of C. cajan Methanol Fraction
4.7. Purification of Ethyl Acetate Fraction by Accelerated Chromatographic Isolation (ACI™) Technology Biotage® Isolera Normal Phase Flash Column Chromatography
4.8. Purification of Fraction KADAMEI3 by Accelerated Chromatographic Isolation (ACI™) Technology Biotage® Isolera Reversed Phase Flash Column Chromatography
4.9. High-Performance Liquid Chromatography (HPLC) Purification of Bioactive Fraction
4.10. Liquid Chromatography-Triple Quadrupole Mass Spectrometry (LC-TqMS) Profiling and Dereplication by Global Natural Products Social Molecular Networking (GNPS)
4.11. Liquid Chromatography-Quadrupole Time-of-Flight (LC-QToF) Mass Spectrometry and MS/MS
4.12. Antibacterial Testing
4.12.1. Agar Well Diffusion Assay
4.12.2. Thin Layer Chromatography (TLC) Bioautography
4.12.3. Minimum Inhibitory Concentration (MIC) Assay
4.12.4. Antibiotic Kinetics Assay
4.12.5. Microbroth Susceptibility Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barrett, T.C.; Mok, W.W.K.; Murawski, A.M. Enhanced antibiotic resistance development from fluoroquinolone persisters after a single exposure to antibiotic. Nat. Commun. 2019, 10, 1177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, J.; Raisen, C.L.; Ba, X.; Sadgrove, N.J.; Padilla-Gonzalez, G.F.; Simmonds, M.; Loncaric, I.; Kerschner, H.; Apfalter, P.; Hartl, R.; et al. Emergence of methicillin resistance predates the clinical use of antibiotics. Nature 2022, 602, 135–141. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Worldwide Country Situation Analysis: Response to Antimicrobial Resistance. 2015. Available online: https://apps.who.int/iris/handle/10665/163473 (accessed on 31 July 2021).
- Freiri, M.; Kumar, K.; Boutin, A. Antibiotic Resistance. J. Infect. Pub. Health 2017, 10, 369–378. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Antimicrobial Resistance Global Report on Surveillance. 2014. Available online: https://www.who.int/publications/i/item/9789241564748?msclkid=829eb582a8b911ec8eb8488f3ce19156 (accessed on 31 July 2021).
- Ramirez-Gomez, X.; Jimenez-Garcia, S.; Campos, V.B.; Campos, M.G. Plant metabolites in plant defense against pathogens. In Plant Diseases—Current Threats and Management Trends, 1st ed.; Topolovec-Pintaric, S., Ed.; IntechOpen: London, UK, 2019; pp. 2–10. [Google Scholar]
- Mazid, M.; Khan, T.; Mohammad, F. Role of secondary metabolites in defense mechanisms of plants. Biol. Med. 2011, 3, 232–249. [Google Scholar]
- Dixon, R.A. Natural products and plant disease resistance. Nature 2001, 411, 843–847. [Google Scholar] [CrossRef]
- Gonelimali, F.D.; Lin, J.; Miao, W.; Xuan, J.; Charles, F.; Chen, M.; Hatab, S.R. Antimicrobial properties and mechanism of action of some plant extracts against food pathogens and spoilage microorganisms. Front. Microbiol. 2018, 9, 10–12. [Google Scholar] [CrossRef]
- Boggia, R.; Zunin, P.; Turrini, F. Functional foods and food supplements. Appl. Sci. 2020, 10, 8538. [Google Scholar] [CrossRef]
- Fardet, A.; Chardigny, J.M. Plant-based foods as a source of lipotropes for human nutrition: A survey of in vivo studies. Crit. Rev. Food Sci. Nutr. 2013, 53, 535–590. [Google Scholar] [CrossRef]
- Kaur, R.; Tiwari, A.; Manish, M.; Mauurya, I.; Bhatnagar, R.; Singh, S. Common garlic (Allium sativum L.) has potent Anti-Bacillus anthracis activity. J. Ethnopharmacol. 2021, 264, 113230. [Google Scholar] [CrossRef]
- Peixoto Araujo, N.M.; Arruda, H.S.; Marques, D.R.P.; de Oliveira, W.Q.; Pereira, G.A.; Pastore, G.M. Functional and nutritional properties of selected Amazon fruits: A review. Food Res. Int. 2021, 147, 110520. [Google Scholar] [CrossRef]
- Souid, A.; Croce, C.M.D.; Frassinetti, S.; Gabriele, M.; Pozzo, L.; Ciardi, M.; Abdelly, C.; Hamed, K.B.; Magne, C.; Longo, V. Nutraceutical potential of leaf hydro-ethanolic extract of the edible halophyte. Molecules 2021, 26, 5380. [Google Scholar] [CrossRef] [PubMed]
- Mokhtar, M.; Bouamar, S.; Di Lorenzo, A.; Temporini, C.; Daglia, M.; Riazi, A. The Influence of ripeness on the phenolic content, antioxidant and antimicrobial activities of pumpkins (Cucurbita moschata Duchesne). Molecules 2021, 26, 3623. [Google Scholar] [CrossRef] [PubMed]
- Honkanen, P. Consumer Acceptance of (Marine) Functional Food. In The Nordic Network on Marine Functional Food, 1st ed.; Luten, J., Ed.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2009; Volume 1, pp. 141–154. [Google Scholar]
- Tecson-Mendoza, E.M. Development of functional foods in the Philippines. Food Sci. Technol. Res. 2007, 13, 179–186. [Google Scholar] [CrossRef] [Green Version]
- Suarez, A.F.L.; Tirador, A.D.G.; Villorente, Z.M.; Bagarinao, C.F.; Sollesta, J.V.N.; Dumancas, G.G.; Sun, Z.; Zhan, Z.Q.; Saludes, J.P.; Dalisay, D.S. The Isorhamnetin-containing fraction of Philippine honey produced by the stingless bee Tetragonula biroi is an antibiotic against multidrug-resistant Staphylococcus aureus. Molecules 2021, 26, 1688. [Google Scholar] [CrossRef]
- Sabido, E.M.; Tenebro, C.P.; Suarez, A.F.L.; Ong, S.D.C.; Trono, D.J.V.L.; Amago, D.S.; Evangelista, J.E., Jr.; Reynoso, A.M.Q.; Villalobos, I.G.M.; Alit, L.D.D.; et al. Marine sediment-derived Streptomyces strain produces angucycline antibiotics against multidrug-resistant Staphylococcus aureus harboring SCCmec type 1 gene. J. Mar. Sci. Eng. 2020, 8, 734. [Google Scholar] [CrossRef]
- Sabido, E.M.; Tenebro, C.P.; Trono, D.J.V.L.; Vicera, C.V.B.; Leonida, S.F.L.; Maybay, J.J.W.B.; Reyes-Salarda, R.; Amago, D.S.; Aguadera, A.M.V.; Octaviano, M.C.; et al. Insights into the variation in bioactivities of closely related Streptomyces Strains from marine sediments of the Visayan Sea against ESKAPE and ovarian cancer. Mar. Drugs 2021, 19, 441. [Google Scholar] [CrossRef]
- Tenebro, C.P.; Trono, D.; Vicera, C.; Sabido, E.M.; Ysulat, J.A., Jr.; Macaspac, A.; Tampus, K.A.; Fabrigar, T.; Saludes, J.P.; Dalisay, D.S. Multiple strain analysis of Streptomyces species from Philippine marine sediments reveals intraspecies heterogeneity in antibiotic activities. Sci. Rep. 2021, 11, 19046. [Google Scholar] [CrossRef]
- Orwa, C.; Mutua, A.; Kindt, R.; Jamnadass, R.; Anthony, S. Agroforestree Database: A Tree Reference and Selection Guide Version 4.0. 2009. Available online: http://apps.worldagroforestry.org/treedb2/speciesprofile.php?Spid=408 (accessed on 31 July 2021).
- Saxena, K.B.; Mula, M.G.; Sugui, F.P.; Layaoen, H.L.; Domoguen, R.L.; Pascua, M.E.; Mula, R.P.; Dar, W.D.; Gowda, C.L.L.; Kumar, R.V.; et al. Pigeonpea—A Resilient Crop for the Philippine Drylands; International Crops Research Institute for the Semi-Arid Tropics Bulletin No. 85: Andhra Pradesh, India, 2010. [Google Scholar]
- Liu, X.L.; Zhang, X.J.; Fu, Y.J.; Zu, Y.G.; Wu, N.; Liang, L.; Efferth, T. Cajanol inhibits the growth of Escherichia coli and Staphylococcus aureus by acting on membrane and DNA damage. Planta Med. 2011, 77, 158–163. [Google Scholar] [CrossRef]
- Zu, Y.G.; Liu, X.L.; Fu, Y.J.; Wu, N.; Kong, Y.; Wink, M. Chemical composition of the SFE-CO extracts from Cajanus cajan (L.) Huth and their antimicrobial activity in vitro and in vivo. Phytomedicine 2010, 17, 1095–1101. [Google Scholar] [CrossRef]
- Panlasigui, L.N.; Panlilio, L.M.; Madrid, J.C. Glycaemic response in normal subjects to five different legumes commonly used in the Philippines. Int. J. Food Sci. Nutr. 1995, 46, 155–160. [Google Scholar] [CrossRef]
- Rama, R.L.C.M.; Fabi, J.C.Q.; Mateo, G.C.M.; Catubag, J.A.C.; Lozada, L.L.; Malimban, R.C.; Bulleceer, E.R. Protein efficiency ratio of pigeon pea (Cajanus cajan) and lima bean (Phaseolus lunatus): A sprague-dawley rat growth assay. Acta Med. Philipp. 2018, 52, 4–29. [Google Scholar] [CrossRef]
- Ayenan, M.A.T.; Ofori, K.; Ahoton, L.E.; Danquah, A. Pigeonpea [(Cajanus cajan (L.) Millsp.)] production system, farmers’ preferred traits and implications for variety development and introduction in Benin. Agric. Food Sec. 2017, 6, 48. [Google Scholar] [CrossRef]
- Contreras, S.M.; Urriza, G.I.P.; Nagumo, F.; Tejada, S.Q.; Gesite, A.; Creencia, R.P.; Dimaano, R.A.A. Evaluation of pigeon pea (Cajanus cajan) cover cropping and “no-till” as a soil and crop management practice in corn (Zea Mays) areas of Isabela, Philippines. Trop. Agric. Dev. 2014, 58, 66–74. [Google Scholar]
- Mendoza, E.M. Biochemical and nutritional qualities of several philippine indigenous food legumes. NAST 1991, 13, 841–857. [Google Scholar]
- Kong, Y.; Fu, Y.J.; Zu, Y.; Chang, F.; Chen, Y.; Liu, X.; Stelten, J.; Schiebel, H. Cajanuslactone, a new coumarin with anti-bacterial activity from pigeon pea [Cajanus cajan (L.) Millsp.] leaves. Food Chem. 2010, 121, 1150–1155. [Google Scholar] [CrossRef]
- Wu, J.; Li, B.; Xiao, W.; Hu, J.; Xie, J.; Yuan, J.; Wang, L. Longistylin A, a natural stilbene isolated from the leaves of Cajanus cajan, exhibits significant anti-MRSA activity. Int. J. Antimicrob. Agents 2020, 55, 105821. [Google Scholar] [CrossRef]
- Shamsi, T.N.; Parveen, R.; Afreen, S.; Azam, M.; Sen, P.; Sharma, Y.; Haque, Q.; Fatma, T.; Manzoor, N.; Fatima, S. Trypsin inhibitors from Cajanus cajan and Phaseolus limensis possess antioxidant, anti-inflammatory, and antibacterial activity. J. Diet. Suppl. 2018, 15, 939–950. [Google Scholar] [CrossRef]
- Sarkar, R.; Hazra, B.; Mandal, N. Anti-oxidative protection against iron overload-induced liver damage in mice by Cajanus cajan (L.) Millsp. leaf extract. Indian J. Exp. Biol. 2013, 51, 165–173. [Google Scholar]
- Hassan, E.M.; Matloub, A.A.; Aboutabl, M.E.; Ibrahim, N.A.; Mohamed, S.M. Assessment of anti-inflammatory, antinociceptive, immunomodulatory, and antioxidant activities of Cajanus cajan L. seeds cultivated in Egypt and its phytochemical composition. Pharm. Biol. 2015, 54, 1380–1391. [Google Scholar] [CrossRef] [Green Version]
- Pal, D.; Mishra, P.; Sachan, N.; Ghosh, A.K. Biological activities and medicinal properties of Cajanus cajan (L) Millsp. J. Adv. Pharm. Technol. Res. 2011, 2, 207–214. [Google Scholar] [CrossRef]
- Fu, Y.; Kadioglu, O.; Wiench, B.; Wei, Z.; Gao, C.; Luo, M.; Gu, C.; Zu, Y.; Efferth, T. Cell cycle arrest and induction of apoptosis by cajanin stilbene acid from Cajanus cajan in breast cancer cells. Phytomedicine 2015, 22, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Zu, Y.; Fu, Y.; Wang, W.; Wu, N.; Liu, W.; Kong, Y.; Schiebel, H.M.; Schwarz, G.; Schnitzler, P.; Reichling, J. Comparative study on the antiherpetic activity of aqueous and ethanolic extracts derived from Cajanus cajan (L.) Millsp. Forsch Komplementmed. 2010, 17, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.J.; Hsu, W.H.; Huang, J.J.; Wu, S.C. Effect of pigeon pea (Cajanus cajan L.) on high-fat diet-induced hypercholesterolemia in hamsters. Food Chem. Toxicol. 2013, 53, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Brito, S.A.; Rodrigues, F.F.; Campos, A.R.; da Costa, J.G. Evaluation of the antifungal activity and modulation between Cajanus cajan (L.) Millsp. leaves and roots ethanolic extracts and conventional antifungals. Pharmacogn. Mag. 2012, 8, 103–106. [Google Scholar] [PubMed]
- Ezike, A.C.; Akah, P.A.; Okoli, C.C.; Okpala, C.B. Experimental evidence for the antidiabetic activity of Cajanus cajan leaves in rats. J. Basic Clin. Pharm. 2010, 1, 81–84. [Google Scholar]
- Mahitha, B.; Archana, P.; Ebrahimzadeh, M.H.; Srikanth, K.; Rajinikanth, M.; Ramaswamy, N. In vitro antioxidant and pharmacognostic studies of leaf extracts of Cajanus cajan (L.) Millsp. Indian J. Pharm. Sci. 2015, 77, 170–177. [Google Scholar]
- Nwodo, U.U.; Ngene, A.A.; Iroegbu, C.U.; Onyedikachi, O.A.; Chigor, V.N.; Okoh, A.I. In vivo evaluation of the antiviral activity of Cajanus cajan on measles virus. Arch. Virol. 2011, 156, 1551–1557. [Google Scholar] [CrossRef] [Green Version]
- Tyavambiza, C.; Dube, P.; Goboza, M.; Meyer, S.; Madiehe, A.M.; Meyer, M. Wound healing activities and potential of selected african medicinal plants and their synthesized biogenic nanoparticles. Plants 2021, 10, 2635. [Google Scholar] [CrossRef]
- Maroyi, A. An ethnobotanical survey of medicinal plants used by the people in Nhema communal area, Zimbabwe. J. Ethnopharmacol. 2011, 136, 347–354. [Google Scholar] [CrossRef]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef]
- Ng, T.B.; Ye, X.J.; Wong, J.H.; Fang, E.F.; Chan, Y.S.; Pan, W.; Ye, X.Y.; Sze, S.C.; Zhang, K.Y.; Liu, F.; et al. Glyceollin, a soybean phytoalexin with medicinal properties. Appl. Microbiol. Biotechnol. 2011, 90, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Pedras, M.S.; Okanga, F.I.; Zaharia, I.L.; Khan, A.Q. Phytoalexins from crucifers: Synthesis, biosynthesis, and biotransformation. Phytochemistry 2000, 53, 161–176. [Google Scholar] [CrossRef]
- Dinore, J.M.; Farooqui, M. GC-MS and LC-MS: An integrated approach towards the phytochemical evaluation of methanolic extract of Pigeon Pea. Nat. Prod. Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Staphylococcus aureus subsp. aureus Rosenbach (ATCC® BAA-44™). 2021. Available online: https://www.atcc.org/products/baa-44 (accessed on 31 July 2021).
- Al Obaid, I.A.; Udo, E.E.; Jacob, L.E.; Johny, M. Isolation and characterization of coagulase-negative methicillin-resistant Staphylococcus aureus from patients in an intensive care unit. Med. Princ. Pract. 1999, 8, 230–236. [Google Scholar] [CrossRef]
- Olver, W.J.; Carmichael, I.C.; Ziglam, H.M.; Morrison, D. Bacteraemia with tube-coagulase-negative methicillin-resistant Staphylococcus aureus. J. Hosp. Infect. 2005, 60, 87–88. [Google Scholar] [CrossRef]
- Bayston, R. Coagulase-negative methicillin-resistant Staphylococcus aureus. J. Hosp. Infect. 2006, 62, 127. [Google Scholar] [CrossRef]
- Bonvegna, M.; Grego, E.; Sona, B.; Stella, M.C.; Nebbia, P.; Mannelli, A.; Tomassone, L. Occurrence of methicillin-resistant coagulase-negative staphylococci (MRcons) and methicillin-resistant Staphylococcus aureus (MRSA) from pigs and farm environment in northwestern Italy. Antibiotics 2021, 10, 676. [Google Scholar] [CrossRef]
- Sinlapasorn, S.; Lulitanond, A.; Angkititrakul, S.; Chanawong, A.; Wilailuckana, C.; Tavichakorntrakool, R.; Chindawong, K.; Seelaget, C.; Krasaesom, M.; Chartchai, S.; et al. SCCmec IX in methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci from pigs and workers at pig farms in Khon Kaen, Thailand. J. Med. Microbiol. 2015, 64, 1087–1093. [Google Scholar] [CrossRef] [Green Version]
- Akineden, Ö.; Hassan, A.; Schneider, E.; Usleber, E. A coagulase-negative variant of Staphylococcus aureus from bovine mastitis milk. J. Dairy Res. 2011, 78, 38–42. [Google Scholar] [CrossRef]
- Wei, Z.; Zu, Y.; Fu, Y.; Wang, W.; Luo, M.; Zhao, C.; Pan, Y. Ionic liquids-based microwave-assisted extraction of active components from pigeon pea leaves for quantitative analysis. Sep. Purif. Technol. 2013, 102, 75–81. [Google Scholar] [CrossRef]
- Nix, A.; Paull, C.A.; Colgrave, M. The flavonoid profile of pigeonpea, Cajanus cajan: A review. SpringerPlus 2015, 4, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsamo, D.; Tamokou, J.D.; Kengne, I.C.; Ngnokam, C.; Djamalladine, M.D.; Voutquenne-Nazabadioko, L.; Ngnokam, D. Antimicrobial and antioxidant secondary metabolites from Trifolium baccarinii Chiov. (Fabaceae) and their mechanisms of antibacterial action. Biomed. Res. Int. 2021, 2021, 3099428. [Google Scholar] [PubMed]
- Rufatto, L.C.; Luchtenberg, P.; Garcia, C.; Thomassigny, C.; Bouttier, S.; Henriques, J.A.P.; Moura, S. Brazilian red propolis: Chemical composition and antibacterial activity determined using bioguided fractionation. Microbiol. Res. 2018, 214, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.H.; Lee, P.H.; Wein, Y.S. Four new compounds from the seeds of Cassia fistula. J. Nat. Prod. 2002, 65, 1165–1167. [Google Scholar] [CrossRef]
- Weng, J.R.; Tsao, L.T.; Yen, M.H.; Wang, J.P.; Lin, C.N. Anti-inflammatory constituents and new pterocarpanoid of Crotalaria pallida. J. Nat. Prod. 2003, 66, 404–407. [Google Scholar] [CrossRef]
- Gosh, J.; Lohot, V.; Singhal, V.; Ghosal, S.; Sharma, K.K. Pigeon pea-Lac insect interaction: Effect of lac culture on grain yield and biochemical parameters in pigeonpea. Indian J. Genet. 2014, 74, 644–650. [Google Scholar] [CrossRef]
- Regar, N.L.; Lekha, H.S.; Ahir, K.C. Biology of lac insect, Kerria lacca (Kerr) on pigeonpea, Cajanus cajan (L.). J. Entomol. Zool. Stud. 2021, 9, 5. [Google Scholar]
- Lechner, D.; Gibbons, S.; Bucar, F. Plant phenolic compounds as ethidium bromide efflux inhibitors in Mycobacterium smegmatis. J. Antimicrob. Chemother. 2008, 62, 345–348. [Google Scholar] [CrossRef] [Green Version]
- Sklenickova, O.; Flesar, J.; Kokoska, L.; Vlkova, E.; Halamova, K.; Malik, J. Selective growth inhibitory effect of biochanin A against intestinal tract colonizing bacteria. Molecules 2010, 15, 1270–1279. [Google Scholar] [CrossRef]
- Nikolic, I.L.; Savic, I.M.; Popsavin, M.M.; Rakic, S.J.; Mihajilov-Krstev, T.M.; Ristic, I.S.; Eric, S.P.; Savić-Gajic, I.M. Preparation, characterization and antimicrobial activity of inclusion complex of biochanin A with (2-hydroxypropyl)-β-cyclodextrin. J. Pharm. Pharmacol. 2018, 70, 1485–1493. [Google Scholar] [CrossRef]
- Dastidar, S.G.; Manna, A.; Kumar, K.A.; Mazumdar, K.; Dutta, N.K.; Chakrabarty, A.N.; Motohashi, N.; Shirataki, Y. Studies on the antibacterial potentiality of isoflavones. Int. J. Antimicrob. Agents 2004, 23, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Kuete, V.; Nono, E.C.; Mkounga, P.; Marat, K.; Hultin, P.G.; Nkengfack, A.E. Antimicrobial activities of the CH2Cl2-CH3OH (1:1) extracts and compounds from the roots and fruits of Pycnanthus angolensis (Myristicaceae). Nat. Prod. Res. 2011, 25, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Liang, J.C.; Wang, X.L.; Li, Z.H.; Wang, W.; Guo, N.; Wu, X.P.; Shen, F.G.; Xing, M.X.; Liu, L.H.; et al. In vitro synergy of biochanin a and ciprofloxacin against clinical isolates of Staphylococcus aureus. Molecules 2011, 16, 6656–6666. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Qi, C.; Zou, Y.; Kong, Y.; Ruan, Z.; Ding, H.; Xie, X.; Zhang, J. Biochanin A partially restores the activity of ofloxacin and ciprofloxacin against topoisomerase IV mutation-associated fluoroquinolone-resistant Ureaplasma species. J. Med. Microbiol. 2017, 66, 1545–1553. [Google Scholar] [CrossRef]
- Feng, J.; Sun, D.; Wang, L.; Li, X.; Guan, J.; Wei, L.; Yue, D.; Wang, X.; Zhao, Y.; Yang, H.; et al. Biochanin A as an α-hemolysin inhibitor for combating methicillin-resistant Staphylococcus aureus infection. World J. Microbiol. Biotechnol. 2021, 38, 6. [Google Scholar] [CrossRef]
- Zhao, X.; Tang, X.; Guo, N.; An, Y.; Chen, X.; Shi, C.; Wang, C.; Li, Y.; Li, S.; Xu, H.; et al. Biochanin A enhances the defense against Salmonella enterica Infection Through AMPK/ULK1/mTOR-Mediated autophagy and extracellular traps and reversing spi-1-dependent macrophage (MΦ) M2 polarization. Front. Cell Infect. Microbiol. 2018, 8, 318. [Google Scholar] [CrossRef] [Green Version]
- Hu, K.X.; Shi, X.C.; Xu, D.; Laborda, P.; Wu, G.C.; Liu, F.Q.; Laborda, P.; Wang, S.Y. Antibacterial mechanism of Biochanin A and its efficacy for the control of Xanthomonas axonopodis pv. glycines in soybean. Pest. Manag. Sci. 2021, 77, 1668–1673. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, J.; Zheng, X.; Chen, Y.; Wu, C.; Li, B.; Fu, J.; Cao, H.; Lu, Y.; Li, J.; et al. Targeting CpG DNA to screen and isolate anti-sepsis fraction and monomers from traditional Chinese herbs using affinity biosensor technology. Int. Immunopharmacol. 2009, 9, 1021–1031. [Google Scholar] [CrossRef]
- Zhou, Y.X.; Xia, W.; Yue, W.; Peng, C.; Rahman, K.; Zhang, H. Rhein: A Review of Pharmacological Activities. Evid. Based Complement. Alternat. Med. 2015, 20, 578107. [Google Scholar] [CrossRef] [Green Version]
- Chung, J.G.; Tsou, M.F.; Wang, H.H.; Lo, H.H.; Hsieh, S.E.; Yen, Y.S.; Wu, L.T.; Chang, S.H.; Ho, C.C.; Hung, C.F. Rhein affects arylamine N-acetyltransferase activity in Helicobacter pylori from peptic ulcer patients. J. Appl. Toxicol. 1998, 18, 117–123. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, S.; Yue, J.; Sun, S.; Lv, Q.; Jian, S.; Xie, Y.; Han, L.; Zhang, F.; Dai, Y.; et al. Antimicrobial activity of diacerein on 76 isolates of gram-positive cocci from bacterial keratitis patients and and in vivo study of diacerein eye drops on Staphylococcus aureus keratitis in mice. Antimicrob. Agents Chemother. 2019, 63, 1874–1918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, R.; Lee, D.Y.; Lee, J.W.; Choi, D.J.; Kim, G.S.; Lee, S.H.; Lee, Y.S. Potentiating activity of rhein in targeting of resistance genes in methicillin-resistant Staphylococcus aureus. Asian Pac. J. Trop. Med. 2018, 2, 14. [Google Scholar]
- Yu, L.; Xiang, H.; Fan, J.; Wang, D.; Yang, F.; Guo, N.; Deng, X. Global transcriptional response of Staphylococcus aureus to rhein, a natural plant product. J. Biotechnol. 2008, 135, 304–308. [Google Scholar] [CrossRef] [PubMed]
- Joung, D.K.; Joung, H.; Yang, D.W.; Kwon, D.Y.; Choi, J.G.; Woo, S.; Shin, D.W. Synergistic effect of rhein in combination with ampicillin or oxacillin against methicillin-resistant Staphylococcus aureus. Exp. Ther. Med. 2012, 3, 608–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jimtaisong, A.; Janthadee, R.; Nakrit, T. In vitro antioxidant activities of laccaic acids and its aluminum lake. Food Sci. Biotechnol. 2013, 22, 1055–1061. [Google Scholar] [CrossRef]
- Srivastava, S.; Chowdhury, A.R.; Maurya, S. Antimicrobial efficacy of methylated lac dye, an anthraquinone derivative. Indian J. Microbiol. 2017, 57, 470–476. [Google Scholar] [CrossRef]
- Moein, M.R.; Khan, S.I.; Ali, Z.; Ayatollahi, S.A.; Kobarfard, F.; Nasim, S.; Khan, I.A. Flavonoids from Iris songarica and their antioxidant and estrogenic activity. Planta Med. 2008, 74, 1492–1495. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.W.; Tsao, L.T.; Huang, L.J.; Kuo, S.C.; Weng, J.R.; Ko, H.H.; Wang, J.P. Inhibition of lipopolysaccharide-stimulated NO production by crotafuran B in RAW 264.7 macrophages involves the blockade of NF-kappaB activation through the increase in IkappaBalpha synthesis. Toxicol. Appl. Pharmacol. 2006, 210, 108–115. [Google Scholar] [CrossRef]
- ProteoWizard. Available online: http://proteowizard.sourceforge.net (accessed on 29 September 2021).
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- GNPS: Global Natural Products Social Molecular Networking. Available online: http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=51db892e72684fab80aa723b37829fbd (accessed on 28 October 2021).
- GNPS: Global Natural Products Social Molecular Networking. Available online: http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=97aaa037ef734d0fae9e3a4bddaa1928 (accessed on 30 October 2021).
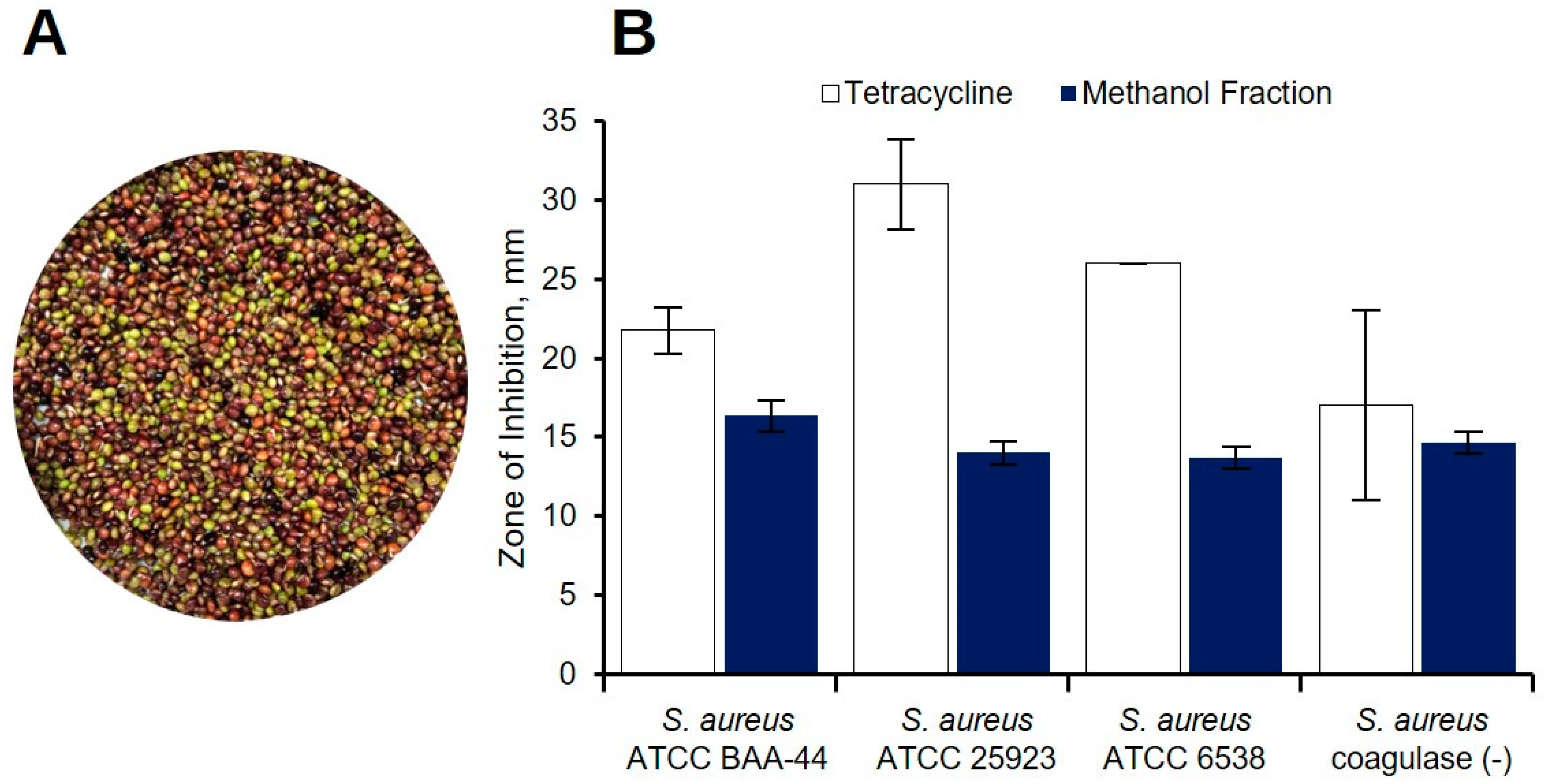



| Solvent/ Sample | Zone of Inhibition, mm | |||
|---|---|---|---|---|
| S. aureus ATCC BAA-44 | S. aureus ATCC 25923 | S. aureus ATCC 6538 | S. aureus Coagulase (−) | |
| Acetone a | 13.2 ± 1.7 | 11.8 ± 1.1 | 11.7 ± 0.5 | – |
| Methanol a | 13.3 ± 2.1 | 10.0 | – | – |
| 95% Ethanol b | 12.8 ± 0.3 | Not Tested | Not Tested | Not Tested |
| Tetracycline | 23.5 ± 2.7 c | 29.0 ± 2.5 c | 26.8 ± 1.0 c | 14.0 d |
| DMSO | – | – | – | – |
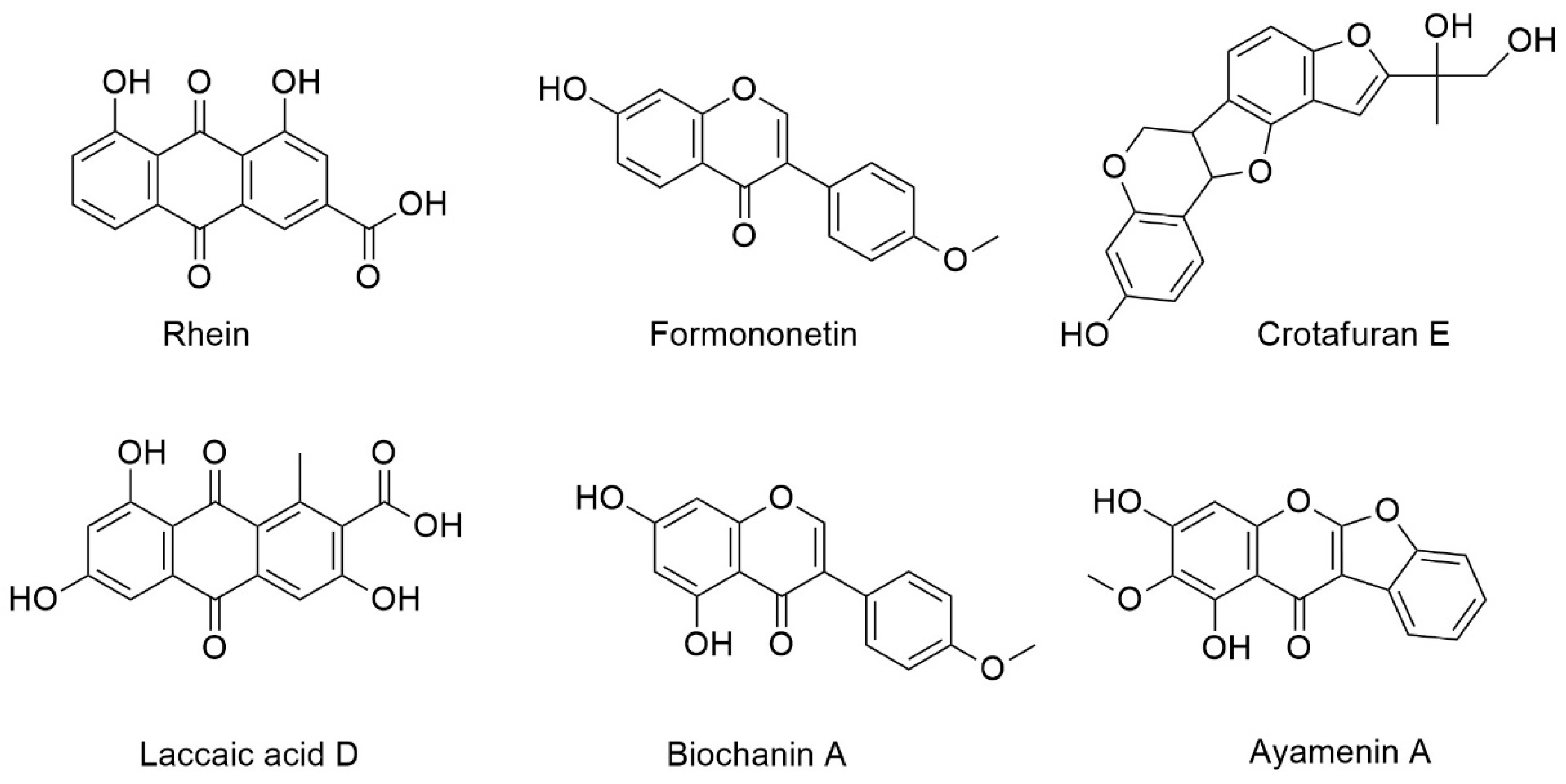
| Compound | Retention Time (min) | Measured Mass m/z | Ion | Formula Predictor | Δppm | DBE * | Chemical ID by MS/MS |
|---|---|---|---|---|---|---|---|
| A | 5.78 | 285.0392 | [M + H]+ | C15H8O6 | −0.47 | 12 | Rhein a |
| B | 5.86 | 269.0809 | [M + H]+ | C16H12O4 | 0.16 | 11 | Formononetin b,c |
| [M + H]+ | 0.16 | 11 | Isoformononetin a | ||||
| C | 6.11 | 315.0499 | [M + H]+ | C16H10O7 | 0.06 | 12 | Laccaic acid D a |
| D | 6.4 | 355.1178 | [M + H]+ | C20H18O6 | 0.49 | 12 | Crotafuran E a |
| E | 6.8 | 299.0549 | [M + H]+ | C16H10O6 | -0.23 | 12 | Ayamenin A a |
| F | 7 | 285.0757 | [M + H]+ | C16H12O5 | -0.16 | 11 | Biochanin A a,b,c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balida, L.A.P.; Regalado, J.T.A.; Teodosio, J.J.R.; Dizon, K.A.H.; Sun, Z.; Zhan, Z.Q.; Blancaflor, J.M.D.; Sollesta, J.V.N.; Villorente, Z.M.; Saludes, J.P.; et al. Antibiotic Isoflavonoids, Anthraquinones, and Pterocarpanoids from Pigeon Pea (Cajanus cajan L.) Seeds against Multidrug-Resistant Staphylococcus aureus. Metabolites 2022, 12, 279. https://doi.org/10.3390/metabo12040279
Balida LAP, Regalado JTA, Teodosio JJR, Dizon KAH, Sun Z, Zhan ZQ, Blancaflor JMD, Sollesta JVN, Villorente ZM, Saludes JP, et al. Antibiotic Isoflavonoids, Anthraquinones, and Pterocarpanoids from Pigeon Pea (Cajanus cajan L.) Seeds against Multidrug-Resistant Staphylococcus aureus. Metabolites. 2022; 12(4):279. https://doi.org/10.3390/metabo12040279
Chicago/Turabian StyleBalida, Lex Aliko P., Julia Theresa A. Regalado, Jade Joshua R. Teodosio, Kathryn Ann H. Dizon, Zhe Sun, Zhao Qi Zhan, Jenny Marie D. Blancaflor, Jan Vincent N. Sollesta, Zenith M. Villorente, Jonel P. Saludes, and et al. 2022. "Antibiotic Isoflavonoids, Anthraquinones, and Pterocarpanoids from Pigeon Pea (Cajanus cajan L.) Seeds against Multidrug-Resistant Staphylococcus aureus" Metabolites 12, no. 4: 279. https://doi.org/10.3390/metabo12040279
APA StyleBalida, L. A. P., Regalado, J. T. A., Teodosio, J. J. R., Dizon, K. A. H., Sun, Z., Zhan, Z. Q., Blancaflor, J. M. D., Sollesta, J. V. N., Villorente, Z. M., Saludes, J. P., & Dalisay, D. S. (2022). Antibiotic Isoflavonoids, Anthraquinones, and Pterocarpanoids from Pigeon Pea (Cajanus cajan L.) Seeds against Multidrug-Resistant Staphylococcus aureus. Metabolites, 12(4), 279. https://doi.org/10.3390/metabo12040279