Metabolomic Effects of the Dietary Inclusion of Hermetia illucens Larva Meal in Tilapia
Abstract
:1. Introduction
2. Results
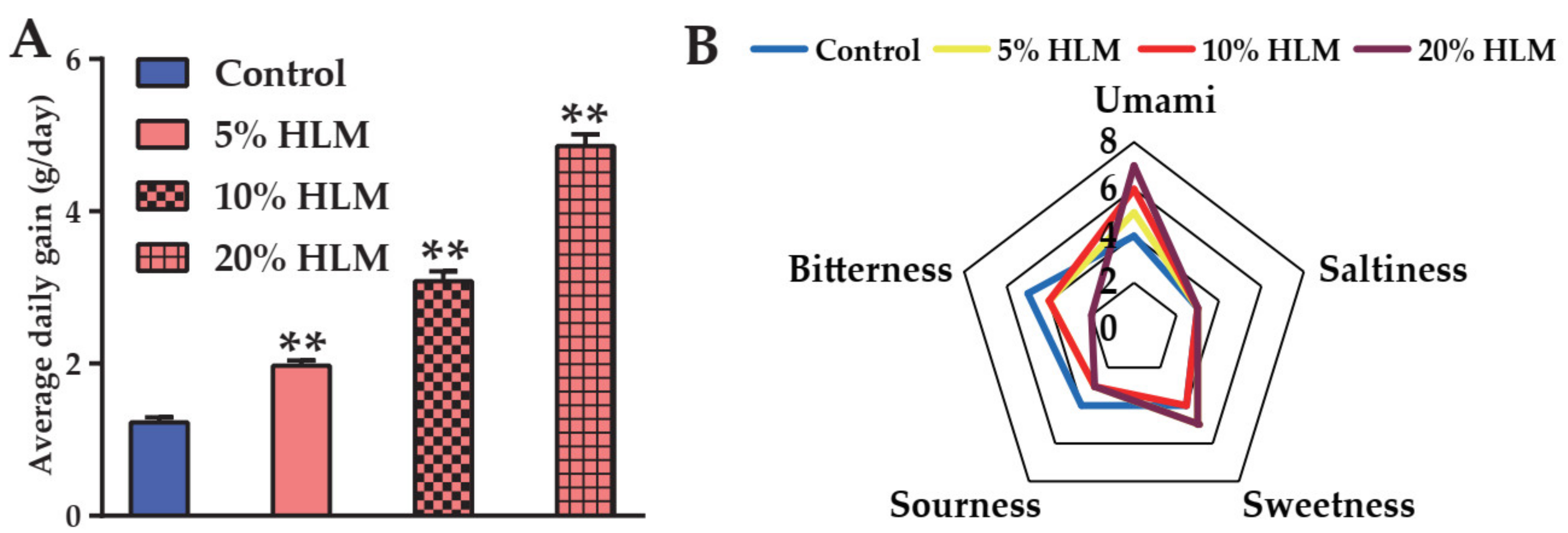
2.1. H. illucens Larva Meal Boosting the Production Performance and Sensory Taste of Tilapia
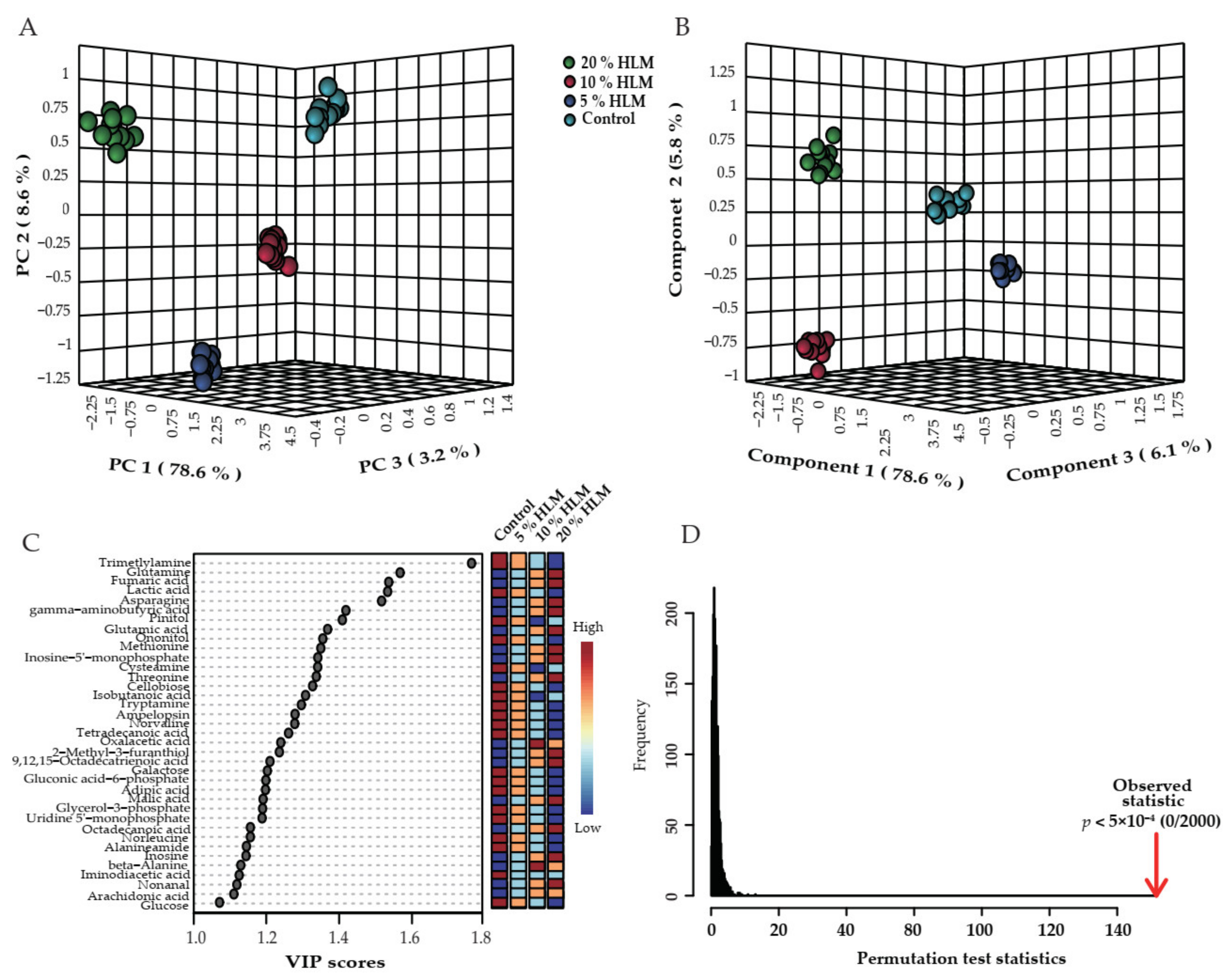
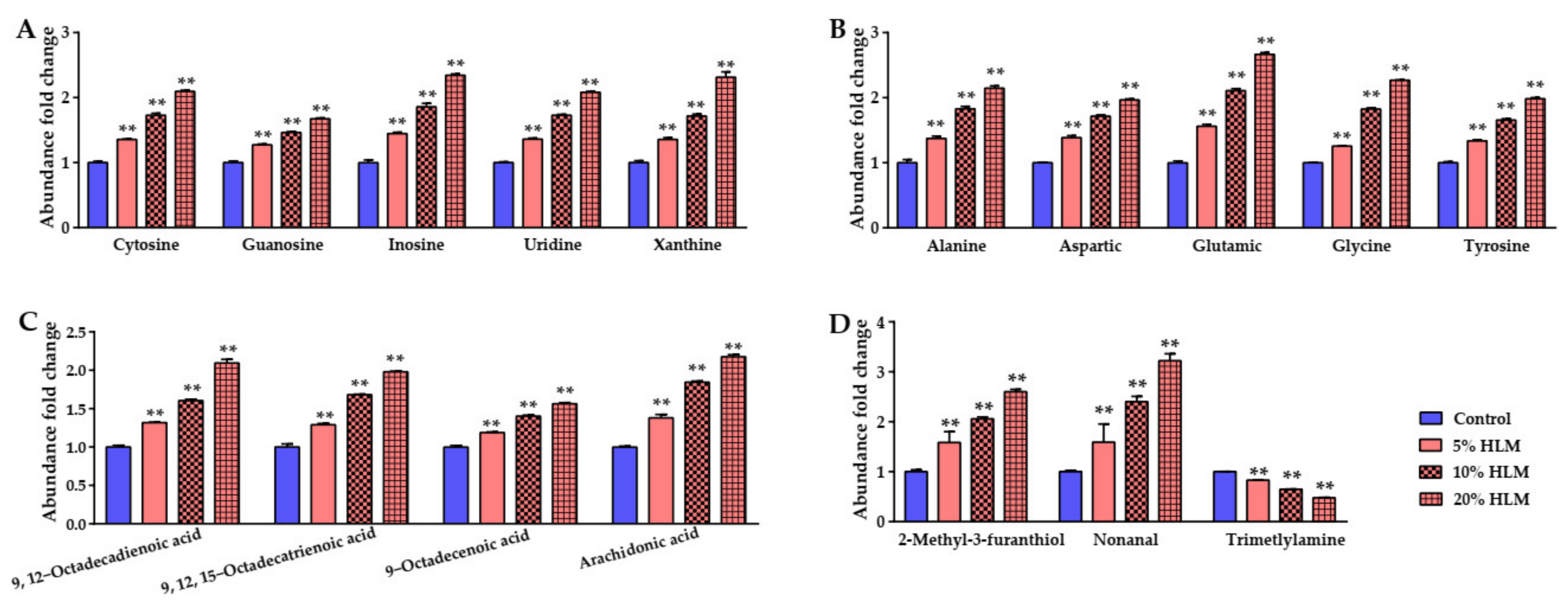
2.2. Metabolic Profile of Tilapias to H. illucens Larva Meal
2.3. Differential Metabolomes Responsible for H. illucens Larva Meal
2.4. Crucial Biomarkers Responsible for H. illucens Larva Meal
2.5. Differential Enriched Pathways Responsible for H. illucens Larva Meal
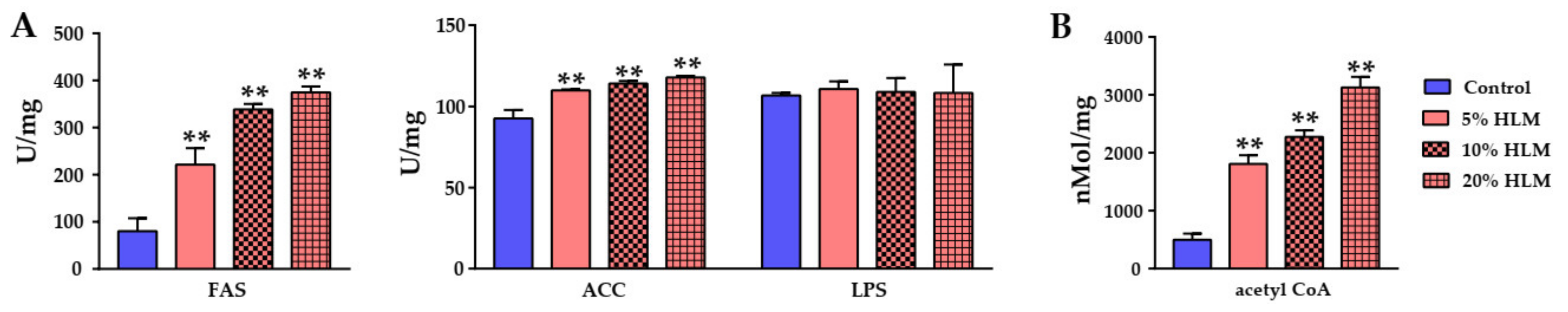
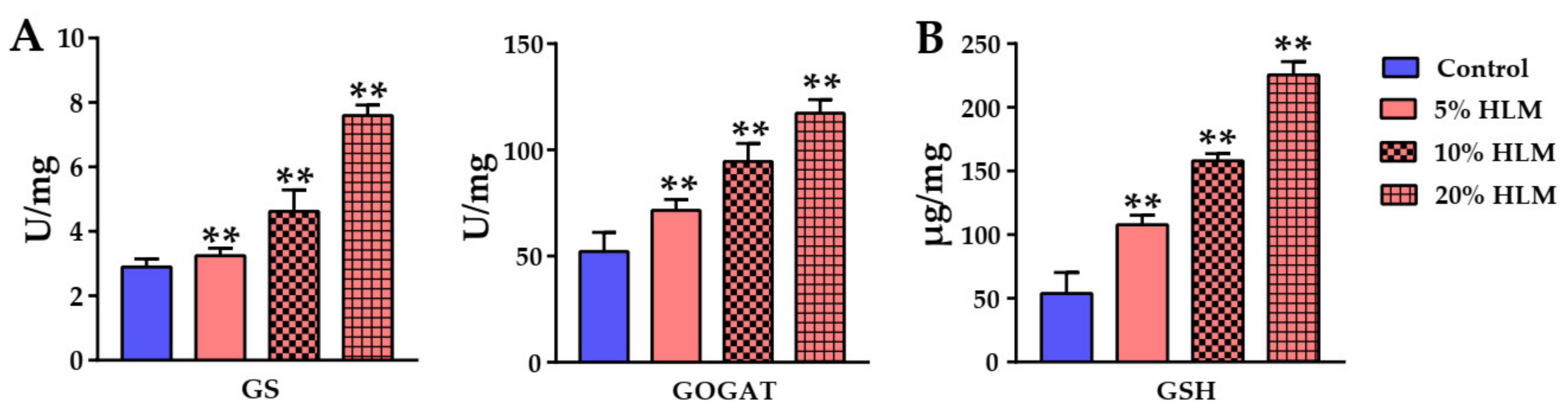
2.6. The Metabolomic Effects Responsible for H. illucens Larva Meal
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Chemical Reagents
4.3. Feed Preparation
4.4. Fish and Rearing Conditions
4.5. Sample Preparing for Gas Chromatography–Mass Spectrometry (GC-MS) Detection
4.6. Gas Chromatography–Mass Spectrometry Detection
4.7. Mass Spectra Processing for Gas Chromatography–Mass Spectrometry
4.8. Detection of the Enzyme Activity and Measurement of the Contents of Glutathione, Reduced Nicotinamide-Adenine Dinucleotid, and Adenosine Triphosphate
4.9. Bioinformatic Analyses
4.10. Sensory Evaluation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kevin, C.; Bromage, N.R. Reproductive physiology of female tilapia broodstock. Rev. Fish Biol. Fish. 2000, 10, 1–25. [Google Scholar]
- Lèveque, C. Out of Africa: The success story of tilapias. Environ. Biol. Fishes 2002, 64, 461–464. [Google Scholar]
- Tacon, A.G.; Metian, M. Global overview on the use of fish meal and fish oil in industrially compounded aquafeeds: Trends and future prospects. Aquaculture 2008, 285, 146–158. [Google Scholar]
- Hai, N.V. Research findings from the use of probiotics in tilapia aquaculture: A review. Fish Shellfish. Immunol. 2015, 45, 592–597. [Google Scholar] [PubMed]
- Proskina, L.; Cerina, S. Faba beans and peas in poultry feed: Economic assessment. J. Sci. Food Agric. 2017, 97, 4391–4398. [Google Scholar] [PubMed]
- Chen, Y.; Ma, J.; Huang, H.; Zhong, H. Effects of the replacement of fishmeal by soy protein concentrate on growth performance, apparent digestibility, and retention of protein and amino acid in juvenile pearl gentian grouper. PLoS ONE 2019, 14, e0222780. [Google Scholar]
- Elahi, U.; Xu, C.C.; Wang, J.; Lin, J.; Wu, S.G.; Zhang, H.J.; Qi, G.H. Insect meal as a feed ingredient for poultry. Anim. Biosci. 2022, 35, 332–346. [Google Scholar]
- Sheppard, D.C.; Tomberlin, J.K.; Joyce, J.A.; Kiser, B.C.; Sumner, S.M. Rearing methods for the black soldier fly (Diptera: Stratiomyidae). J. Med. Entomol. 2002, 39, 695–698. [Google Scholar]
- Čičková, H.; Newton, G.L.; Lacy, R.C.; Kozánek, M. The use of fly larvae for organic waste treatment. Waste Manag. 2015, 35, 68–80. [Google Scholar]
- Sheppard, C. Housefly and lesser fly control utilizing the black soldier fly in manure management-systems for caged laying hens. Environ. Entomol. 1983, 12, 1439–1442. [Google Scholar]
- Green, T.R.; Popa, R. Enhanced ammonia content in compost leachate processed by black soldier fly larvae. Appl. Biochem. Biotechnol. 2012, 166, 1381–1387. [Google Scholar] [PubMed]
- Nguyen, T.T.; Tomberlin, J.K.; Vanlaerhoven, S. Ability of black soldier fly (Diptera: Stratiomyidae) larvae to recycle food waste. Environ. Entomol. 2015, 44, 406–410. [Google Scholar] [PubMed]
- Kim, W.; Bae, S.; Park, K.; Lee, S.; Choi, Y.; Han, S.; Koh, Y. Biochemical characterization of digestive enzymes in the black soldier fly, Hermetia illucens (Diptera: Stratiomyidae). J. Asia-Pac. Entomol. 2011, 14, 11–14. [Google Scholar]
- Liu, X.; Chen, X.; Wang, H.; Yang, Q.; Ur Rehman, K.; Li, W.; Cai, M.; Li, Q.; Mazza, L.; Zhang, J.; et al. Dynamic changes of nutrient composition throughout the entire life cycle of black soldier fly. PLoS ONE 2017, 12, e0182601. [Google Scholar]
- Wang, Y.S.; Shelomi, M. Review of black soldier fly (Hermetia illucens) as animal feed and human food. Foods 2017, 6, 91. [Google Scholar]
- Wachira, M.N.; Osuga, I.M.; Munguti, J.M.; Ambula, M.K.; Subramanian, S.; Tanga, C.M. Efficiency and improved profitability of insect-based aquafeeds for farming Nile Tilapia fish (Oreochromis niloticus L.). Animals 2021, 11, 2599. [Google Scholar]
- Li, S.L.; Ji, H.; Zhang, B.X.; Tian, J.J.; Zhou, J.S.; Yu, H.B. Influence of black soldier fly (Hermetia illucens) larvae oil on growth performance, body composition, tissue fatty acid composition and lipid deposition in juvenile jian carp (Cyprinus carpio var. Jian). Aquaculture 2016, 465, 43–52. [Google Scholar]
- Tippayadara, N.; Dawood, M.; Krutmuang, P.; Hoseinifar, S.H.; Doan, H.V.; Paolucci, M. Replacement of fish meal by black soldier fly (Hermetia illucens) larvae meal: Effects on growth, haematology, and skin mucus immunity of Nile Tilapia, Oreochromis niloticus. Animals 2021, 11, 193. [Google Scholar]
- Kroeckel, S.; Harjes, A.G.E.; Roth, I.; Katz, H.; Wuertz, S.; Susenbeth, A.; Schulz, C. When a turbot catches a fly: Evaluation of a prepupae meal of the black soldier fly (Hermetia illucens) as fish meal substitute—Growth performance and chitin degradation in juvenile turbot (Psetta maxima). Aquaculture 2012, 364, 345–352. [Google Scholar]
- Fawole, F.J.; Labh, S.N.; Hossain, M.S.; Overturf, K.; Small, B.C.; Welker, T.L.; Hardy, R.W.; Kumar, V. Insect (black soldier fly larvae) oil as a potential substitute for fish or soy oil in the fish meal-based diet of juvenile rainbow trout (Oncorhynchus mykiss). Anim. Nutr. 2021, 7, 1360–1370. [Google Scholar]
- Aniebo, A.O.; Erondu, E.S.; Owen, O.J. Replacement of fish meal with maggot meal in African catfish (Clarias gariepinus) diets. Rev. Cient. UDO Agric. 2009, 9, 653–656. [Google Scholar]
- Bondari, K.; Sheppard, D.C. Soldier fly larvae as feed in commercial fish production. Aquaculture 1981, 24, 103–109. [Google Scholar]
- Diener, S.; Zurbrügg, C.; Tockner, K. Conversion of organic material by black soldier fly larvae: Establishing optimal feeding rates. Waste Manag. Res. 2009, 27, 603–610. [Google Scholar] [PubMed]
- Magalhães, R.; Sánchez-López, A.; Leal, R.S.; Martínez-Llorens, S.; Oliva-Teles, A.; Peres, H. Black soldier fly (Hermetia illucens) pre-pupae meal as a fish meal replacement in diets for European seabass (Dicentrarchus labrax). Aquaculture 2017, 476, 79–85. [Google Scholar]
- Yang, M.J.; Cheng, Z.X.; Jiang, M.; Zeng, Z.H.; Peng, B.; Peng, X.X.; Li, H. Boosted TCA cycle enhances survival of zebrafish to Vibrio alginolyticus infection. Virulence 2018, 9, 634–644. [Google Scholar]
- Yang, M.J.; Xu, D.; Yang, D.X.; Li, L.; Peng, X.X.; Chen, Z.G.; Li, H. Malate enhances survival of zebrafish against Vibrio alginolyticus infection in the same manner as taurine. Virulence 2020, 11, 349–364. [Google Scholar]
- Rawski, M.; Mazurkiewicz, J.; Kierończyk, B.; Józefiak, D. Black soldier fly full-fat larvae meal as an alternative to fish meal and fish oil in Siberian sturgeon nutrition: The effects on physical properties of the feed, animal growth performance, and feed acceptance and utilization. Animals 2020, 10, 2119. [Google Scholar]
- Liu, C.; Wang, C.; Yao, H. Comprehensive resource utilization of waste using the black soldier fly (Hermetia illucens (L.)) (Diptera: Stratiomyidae). Animals 2019, 9, 349. [Google Scholar]
- Batish, I.; Brits, D.; Valencia, P.; Miyai, C.; Rafeeq, S.; Xu, Y.; Galanopoulos, M.; Sismour, E.; Ovissipour, R. Effects of enzymatic hydrolysis on the functional properties, antioxidant activity and protein structure of black soldier fly (Hermetia illucens) protein. Insects 2020, 11, 876. [Google Scholar]
- Cullere, M.; Tasoniero, G.; Giaccone, V.; Acuti, G.; Marangon, A.; Dalle Zotte, A. Black soldier fly as dietary protein source for broiler quails: Meat proximate composition, fatty acid and amino acid profile, oxidative status and sensory traits. Animal 2018, 12, 640–647. [Google Scholar]
- Pervin, M.A.; Jahan, H.; Akter, R.; Omri, A.; Hossain, Z. Appraisal of different levels of soybean meal in diets on growth, digestive enzyme activity, antioxidation, and gut histology of tilapia (Oreochromis niloticus). Fish Physiol. Biochem. 2020, 46, 1397–1407. [Google Scholar] [PubMed]
- Yan, Y.; Wang, J.; Dong, X.; Cai, Y.; Wang, Y.; Ren, L.; Zhang, C.; Tao, M.; Luo, K.; Zeng, Y.; et al. Quantitative proteomic analysis of hepatic tissue in allotetraploid hybridized from red crucian carp and common carp identified crucial proteins and pathways associated with metabolism and growth rate. Proteomics 2021, 22, e2100115. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.B.; Feng, L.; Jiang, W.D.; Liu, Y.; Wu, P.; Jiang, J.; Kuang, S.Y.; Tang, L.; Tang, W.N.; Zhang, Y.A.; et al. Physical and flavor characteristics, fatty acid profile, antioxidant status and Nrf2-dependent antioxidant enzyme gene expression changes in young grass carp (Ctenopharyngodon idella) fillets fed dietary valine. PLoS ONE 2017, 12, e0169270. [Google Scholar]
- Arshad, M.S.; Sohaib, M.; Ahmad, R.S.; Nadeem, M.T.; Imran, A.; Arshad, M.U.; Kwon, J.H.; Amjad, Z. Ruminant meat flavor influenced by different factors with special reference to fatty acids. Lipids Health Dis. 2018, 17, 223. [Google Scholar] [PubMed] [Green Version]
- Radzikowska, U.; Rinaldi, A.O.; Çelebi Sözener, Z.; Karaguzel, D.; Wojcik, M.; Cypryk, K.; Akdis, M.; Akdis, C.A.; Sokolowska, M. The influence of dietary fatty acids on immune responses. Nutrients 2019, 11, 2990. [Google Scholar]
- Nemeth, M.; Wallner, B.; Schuster, D.; Siutz, C.; Quint, R.; Wagner, K.H.; Millesi, E. Effects of dietary fatty acids on the social life of male guinea pigs from adolescence to adulthood. Horm. Behav. 2020, 124, 104784. [Google Scholar]
- Stejskal, V.; Tran, H.Q.; Prokesova, M.; Gebauer, T.; Giang, P.T.; Gai, F.; Gasco, L. Partially defatted Hermetia illucens larva meal in diet of Eurasian perch (Perca fluviatilis) juveniles. Animals 2020, 10, 1876. [Google Scholar]
- Bruni, L.; Belghit, I.; Lock, E.J.; Secci, G.; Taiti, C.; Parisi, G. Total replacement of dietary fish meal with black soldier fly (Hermetia illucens) larvae does not impair physical, chemical or volatile composition of farmed Atlantic salmon (Salmo salar L.). J. Sci. Food Agric. 2020, 100, 1038–1047. [Google Scholar]
- Mancini, S.; Medina, I.; Iaconisi, V.; Gai, F.; Basto, A.; Parisi, G. Impact of black soldier fly larvae meal on the chemical and nutritional characteristics of rainbow trout fillets. Animal 2018, 12, 1672–1681. [Google Scholar]
- Caimi, C.; Gasco, L.; Biasato, I.; Malfatto, V.; Varello, K.; Prearo, M.; Pastorino, P.; Bona, M.C.; Francese, D.R.; Schiavone, A.; et al. Could dietary black soldier fly meal inclusion affect the liver and intestinal histological traits and the oxidative stress biomarkers of Siberian sturgeon (Acipenser baerii) juveniles. Animals 2020, 10, 155. [Google Scholar]
- Rimoldi, S.; Antonini, M.; Gasco, L.; Moroni, F.; Terova, G. Intestinal microbial communities of rainbow trout (Oncorhynchus mykiss) may be improved by feeding a Hermetia illucens meal/low-fishmeal diet. Fish Physiol. Biochem. 2021, 47, 365–380. [Google Scholar] [PubMed]
- Zhang, P.; Fu, L.; Liu, H.; Huda, N.U.; Zhu, X.; Han, D.; Jin, J.; Yang, Y.; Kim, Y.S.; Xie, S. Effects of inosine 5′-monophosphate supplementation in high fishmeal and high soybean diets on growth, immune-related gene expression in gibel carp (Carassius auratus gibelio var. CAS Ⅲ), and its challenge against Aeromonas hydrophila infection. Fish Shellfish. Immunol. 2019, 86, 913–921. [Google Scholar] [PubMed]
- Bae, J.; Song, Y.; Moniruzzaman, M.; Hamidoghli, A.; Lee, S.; Je, H.; Choi, W.; Min, T.; Bai, S.C. Evaluation of dietary soluble extract hydrolysates with or without supplementation of inosine monophosphate based on growth, hematology, non-specific immune responses and disease resistance in juvenile Nile Tilapia Oreochromis niloticus. Animals 2021, 11, 1107. [Google Scholar] [PubMed]
- Hong, H.; Regenstein, J.M.; Luo, Y. The importance of ATP-related compounds for the freshness and flavor of post-mortem fish and shellfish muscle: A review. Crit. Rev. Food Sci. Nutr. 2017, 57, 1787–1798. [Google Scholar]
- Li, X.; Amadou, I.; Zhou, G.Y.; Qian, L.Y.; Zhang, J.L.; Wang, D.L.; Cheng, X.R. Flavor components comparison between the neck meat of donkey, swine, bovine, and sheep. Food Sci. Anim. Resour. 2020, 40, 527–540. [Google Scholar]
- Xu, Y.; Wang, R.; Zhao, H.; Yin, Y.; Li, X.; Yi, S.; Li, J. Effect of heat treatment duration on the interaction between fish myosin and selected flavor compounds. J. Sci. Food Agric. 2020, 100, 4457–4463. [Google Scholar]
- Bezman, Y.; Rouseff, R.L.; Naim, M. 2-Methyl-3-furanthiol and methional are possible off-flavors in stored orange juice: Aroma-similarity, NIF/SNIF GC-O, and GC analyses. J. Agric. Food Chem. 2001, 49, 5425–5432. [Google Scholar]
- Ganeko, N.; Shoda, M.; Hirohara, I.; Bhadra, A.; Ishida, T.; Matsuda, H.; Takamura, H.; Matoba, T. Analysis of volatile flavor compounds of sardine (Sardinops melanostica) by solid phase microextraction. J. Food Sci. 2008, 73, S83–S88. [Google Scholar]
- Zheng, A.; Chang, W.; Liu, G.; Yue, Y.; Li, J.; Zhang, S.; Cai, H.; Yang, A.; Chen, Z. Molecular differences in hepatic metabolism between AA broiler and big bone chickens: A proteomic study. PLoS ONE 2016, 11, e0164702. [Google Scholar]
- Zhao, X.L.; Han, Y.; Ren, S.T.; Ma, Y.M.; Li, H.; Peng, X.X. L-proline increases survival of tilapias infected by Streptococcus agalactiae in higher water temperature. Fish Shellfish. Immunol. 2005, 44, 33–42. [Google Scholar]
- Pezzatti, J.; González-Ruiz, V.; Codesido, S.; Gagnebin, Y.; Joshi, A.; Guillarme, D.; Schappler, J.; Picard, D.; Boccard, J.; Rudaz, S. A scoring approach for multi-platform acquisition in metabolomics. J. Chromatogr. A 2019, 1592, 47–54. [Google Scholar] [PubMed]
- Zhu, W.; Zhang, H.; Li, X.; Meng, Q.; Shu, R.; Wang, M.; Zhou, G.; Wang, H.; Miao, L.; Zhang, J.; et al. Cold adaptation mechanisms in the ghost moth Hepialus xiaojinensis: Metabolic regulation and thermal compensation. J. Insect Physiol. 2016, 85, 76–85. [Google Scholar] [PubMed] [Green Version]
- Stacklies, W.; Redestig, H.; Scholz, M.; Walther, D.; Selbig, J. Pcamethods—A bioconductor package providing PCA methods for incomplete data. Bioinformatics 2007, 23, 1164–1167. [Google Scholar] [PubMed]
- Hackstadt, A.J.; Hess, A.M. Filtering for increased power for microarray data analysis. BMC Bioinform. 2009, 10, 11. [Google Scholar]
- Jonsson, P.; Johansson, A.I.; Gullberg, J.; Trygg, J.; Grung, B.; Marklund, S.; Sjöström, M.; Antti, H.; Moritz, T. High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal. Chem. 2005, 77, 5635–5642. [Google Scholar]
- Chong, J.; Xia, J. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar]
- Pu, D.; Duan, W.; Huang, Y.; Zhang, L.; Zhang, Y.; Sun, B.; Ren, F.; Zhang, H.; Tang, Y. Characterization of the dynamic texture perception and the impact factors on the bolus texture changes during oral processing. Food Chem. 2021, 339, 128078. [Google Scholar]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, B.; Li, J.; Xu, L.; Liu, H.; Yang, M. Metabolomic Effects of the Dietary Inclusion of Hermetia illucens Larva Meal in Tilapia. Metabolites 2022, 12, 286. https://doi.org/10.3390/metabo12040286
Ye B, Li J, Xu L, Liu H, Yang M. Metabolomic Effects of the Dietary Inclusion of Hermetia illucens Larva Meal in Tilapia. Metabolites. 2022; 12(4):286. https://doi.org/10.3390/metabo12040286
Chicago/Turabian StyleYe, Bo, Jian Li, Lijun Xu, Hui Liu, and Manjun Yang. 2022. "Metabolomic Effects of the Dietary Inclusion of Hermetia illucens Larva Meal in Tilapia" Metabolites 12, no. 4: 286. https://doi.org/10.3390/metabo12040286
APA StyleYe, B., Li, J., Xu, L., Liu, H., & Yang, M. (2022). Metabolomic Effects of the Dietary Inclusion of Hermetia illucens Larva Meal in Tilapia. Metabolites, 12(4), 286. https://doi.org/10.3390/metabo12040286




