Magnetic Resonance Imaging (MRI) and MR Spectroscopic Methods in Understanding Breast Cancer Biology and Metabolism
Abstract
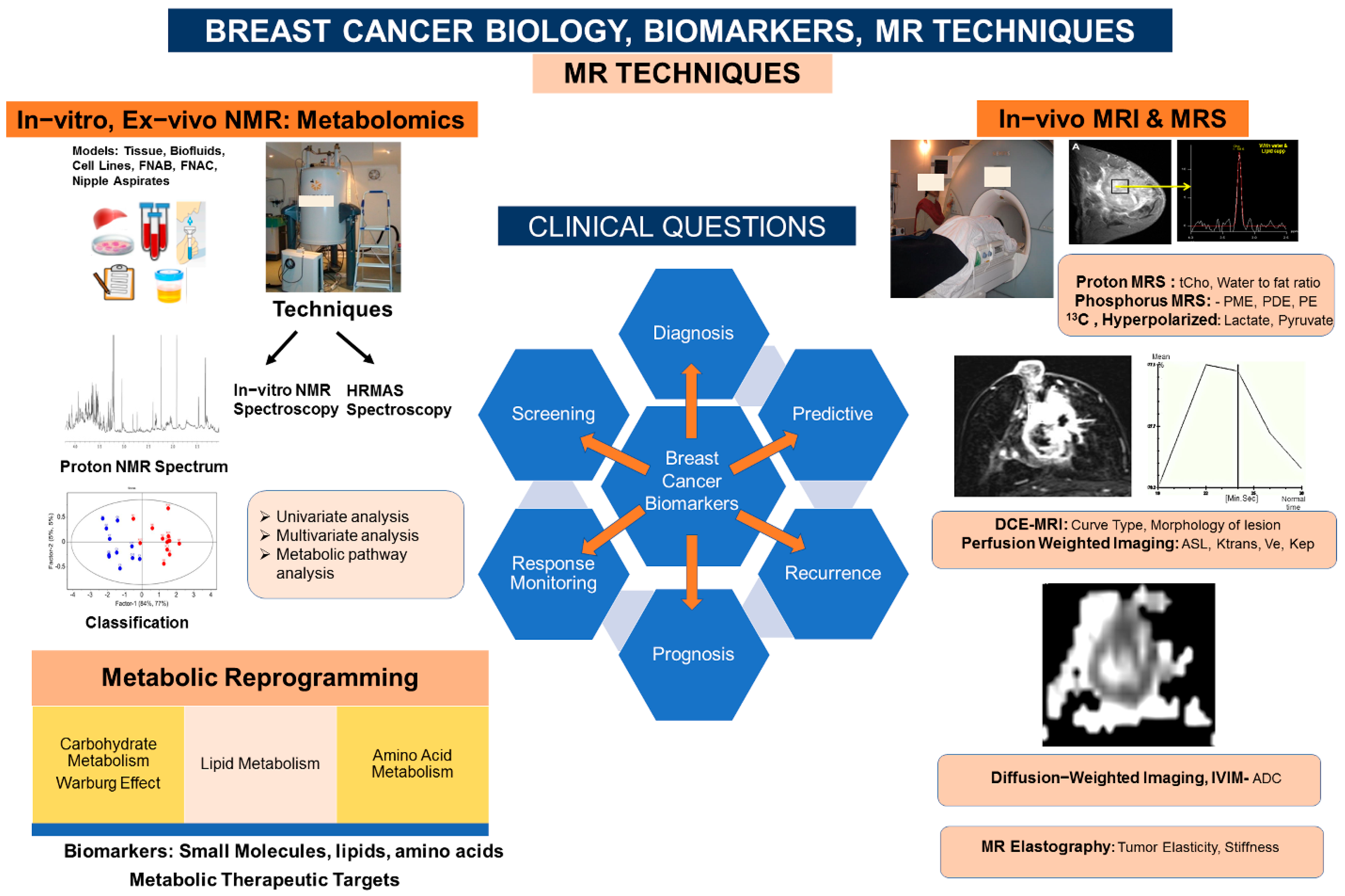
:1. Introduction
2. Breast Cancer Biology: Metabolic Reprogramming
3. Breast Biomarkers: NMR Based Metabolomics, Metabolic Fingerprinting
3.1. Tumor Tissue, Axillary Nodes: HRMAS and In Vitro MRS Studies
3.2. Biofluids
3.3. Aspirates
3.4. Cell-Line Models
3.5. Breast Biomarkers: Living Tissue In Vivo 1H, 31P, and Hyperpolarized 13C MRS
3.6. Breast Biomarkers: Dynamic Contrast-Enhanced MRI (DCE-MRI)
3.7. Breast Biomarkers: Perfusion-Weighted Imaging
3.8. Breast Biomarkers: Diffusion-Weighted Imaging (DWI)
3.9. Breast Biomarkers: MR Elastography (MRE)
3.10. Radiomics
4. Summary, Outlook, and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhl, C. The current status of breast MR imaging. Part I. Choice of technique, image interpretation, diagnostic accuracy, and transfer to clinical practice. Radiology 2007, 244, 356–378. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.M.; Kuhl, C.K.; Moy, L. Contrast-enhanced MRI for breast cancer screening. J. Magn. Reson. Imaging 2019, 50, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, H.F.; Brown, L.F.; Detmar, M.; Dvorak, A.M. Vascular permeability factor/vascular endothelial growth factor, microvascular hyperpermeability, and angiogenesis. Am. J. Pathol. 1995, 146, 1029–1039. [Google Scholar]
- Makkat, S.; Luypaert, R.; Sourbron, S.; Stadnik, T.; De Mey, J. Quantification of perfusion and permeability in breast tumors with a deconvolution-based analysis of second-bolus T1-DCE data. J. Magn. Reson. Imaging 2007, 25, 1159–1167. [Google Scholar] [CrossRef]
- Woodhams, R.; Ramadan, S.; Stanwell, P.; Sakamoto, S.; Hata, H.; Ozaki, M.; Kan, S.; Inoue, Y. Diffusion-weighted imaging of the breast: Principles and clinical applications. Radiographics 2011, 31, 1059–1084. [Google Scholar] [CrossRef] [Green Version]
- Sharma, U.; Sah, R.G.; Agarwal, K.; Parshad, R.; Seenu, V.; Mathur, S.R.; Hari, S.; Jagannathan, N.R. Potential of Diffusion-Weighted Imaging in the Characterization of Malignant, Benign, and Healthy Breast Tissues and Molecular Subtypes of Breast Cancer. Front. Oncol. 2016, 6, 126. [Google Scholar] [CrossRef] [Green Version]
- Sharma, U.; Jagannathan, N.R. Characterization of breast tissues by diffusion weighted MR imaging. Biomed. Spectrosc. Imaging 2014, 3, 1–13. [Google Scholar] [CrossRef]
- Anderson, A.W.; Xie, J.; Pizzonia, J.; Bronen, R.A.; Spencer, D.D.; Gore, J.C. Effects of cell volume fraction changes on apparent diffusion in human cells. Magn. Reson. Imaging 2000, 18, 689–695. [Google Scholar] [CrossRef]
- Guo, Y.; Cai, Y.Q.; Cai, Z.L.; Gao, Y.G.; An, N.Y.; Ma, L.; Mahankali, S.; Gao, J.H. Differentiation of clinically benign and malignant breast lesions using diffusion-weighted imaging. Magn. Reson. Imaging 2002, 16, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Rahbar, H.; Partridge, S.C. Multiparametric MR Imaging of Breast Cancer. Magn. Reson. Imaging Clin. N. Am. 2016, 24, 223–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Li, W.L.; Zhang, Y.L.; Wu, Q.; Guo, Y.M.; Bai, Z.L. Meta-analysis of quantitative diffusion-weighted MR imaging in the differential diagnosis of breast lesions. BMC Cancer 2010, 29, 693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Tang, M.; Min, Z.; Lu, J.; Lei, X.; Zhang, X. Accuracy of combined dynamic contrast-enhanced magnetic resonance imaging and diffusion-weighted imaging for breast cancer detection: A meta-analysis. Acta Radiol. 2016, 57, 651–660. [Google Scholar] [CrossRef]
- Oliphant, T.E.; Manduca, A.; Ehman, R.L.; Greenleaf, J.F. Complex-valued stiffness reconstruction for magnetic resonance elastography by algebraic inversion of the differential equation. Magn. Reson. Med. 2001, 45, 299–310. [Google Scholar] [CrossRef]
- Sinkus, R.; Tanter, M.; Xydeas, T.; Catheline, S.; Bercoff, J.; Fink, M. Viscoelastic shear properties of in vivo breast lesions measured by MR elastography. Magn. Reson. Imaging 2005, 23, 159–165. [Google Scholar] [CrossRef]
- Sinkus, R.; Siegmann, K.; Xydeas, T.; Tanter, M.; Claussen, C.; Fink, M. MR elastography of breast lesions: Understanding the solid/liquid duality can improve the specificity of contrast-enhanced MR mammography. Magn. Reson. Med. 2007, 58, 1135–1144. [Google Scholar] [CrossRef]
- Romero-Garcia, S.; Lopez-Gonzalez, J.S.; Báez-Viveros, J.L.; Aguilar-Cazares, D.; Prado-Garcia, H. Tumor cell metabolism: An integral view. Cancer Biol. Ther. 2011, 12, 939–948. [Google Scholar] [CrossRef] [Green Version]
- Gribbestad, I.S.; Petersen, S.B.; Fjøsne, H.E.; Kvinnsland, S.; Krane, J. 1H NMR spectroscopic characterization of perchloric acid extracts from breast carcinomas and non-involved breast tissue. NMR Biomed. 1994, 7, 181–194. [Google Scholar] [CrossRef]
- Mackinnon, W.B.; Barry, P.A.; Malycha, P.L.; Gillett, D.J.; Russell, P.; Lean, C.L.; Doran, S.T.; Barraclough, B.H.; Bilous, M.; Mountford, C.E. Fine-needle biopsy specimens of benign breast lesions distinguished from invasive cancer ex vivo with proton MR spectroscopy. Radiology 1997, 204, 661–666. [Google Scholar] [CrossRef]
- Kumar, M.; Seenu, V.; Julka, P.K.; Srivastava, A.; Kapila, K.; Rath, G.K.; Jagannathan, N.R. Proton MR Spectroscopy of Human Breast Cancer. In Recent Advances in MR Imaging and Spectroscopy; Jagannathan, N.R., Ed.; Jaypee: New Delhi, India, 2005; pp. 312–344. [Google Scholar]
- Malycha, P.; Mountford, C. Magnetic resonance spectroscopy and breast cancer. Aust. N. Z. J. Surg. 1998, 68, 859–862. [Google Scholar] [CrossRef] [PubMed]
- Mountford, C.E.; Somorjai, R.L.; Malycha, P.; Gluch, L.; Lean, C.; Russell, P.; Barraclough, B.; Gillett, D.; Himmelreich, U.; Dolenko, B.; et al. Diagnosis and prognosis of breast cancer by magnetic resonance spectroscopy of fine-needle aspirates analyzed using a statistical classification strategy. Br. J. Surg. 2001, 88, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Mehta, A.; Seenu, V.; Jagannathan, N.R. Biochemical characterization of metastatic lymph nodes of breast cancer patients by in vitro 1H magnetic resonance spectroscopy: A pilot study. Magn. Reson. Imaging 2004, 22, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, N.R.; Sharma, U. Breast Tissue Metabolism by Magnetic Resonance Spectroscopy. Metabolites 2017, 7, 25. [Google Scholar] [CrossRef] [Green Version]
- Silva, C.; Perestrelo, R.; Silva, P.; Tomás, H.; Câmara, J.S. Breast cancer metabolomics: From analytical platforms to multivariate data analysis. A Review. Metabolites 2019, 9, 102. [Google Scholar] [CrossRef] [Green Version]
- Mountford, C.; Ramadan, S.; Stanwell, P.; Malycha, P. Proton MRS of the breast in the clinical setting. NMR Biomed. 2009, 22, 54–64. [Google Scholar] [CrossRef]
- Jobard, E.; Pontoizeau, C.; Blaise, B.J.; Bachelot, T.; Elena-Herrmann, B.; Trédan, O. A serum nuclear magnetic resonance-based metabolomic signature of advanced metastatic human breast cancer. Cancer Lett. 2014, 343, 33–41. [Google Scholar] [CrossRef]
- Suman, S.; Sharma, R.K.; Kumar, V.; Sinha, N.; Shukla, Y. Metabolic fingerprinting in breast cancer stages through 1H NMR spectroscopy-based metabolomic analysis of plasma. J. Pharm. Biomed. Anal. 2018, 160, 38–45. [Google Scholar] [CrossRef]
- Tredwell, G.D.; Miller, J.A.; Chow, H.H.; Thompson, P.A.; Keun, H.C. Metabolomic characterization of nipple aspirate fluid by 1H NMR spectroscopy and GC-MS. J. Proteome Res. 2014, 13, 883–889. [Google Scholar] [CrossRef] [Green Version]
- Nagana Gowda, G.A.; Barding, G.A., Jr.; Dai, J.; Gu, H.; Margineantu, D.H.; Hockenbery, D.M.; Raftery, D. A Metabolomics Study of BPTES Altered Metabolism in Human Breast Cancer Cell Lines. Front. Mol. Biosci. 2018, 5, 49. [Google Scholar] [CrossRef]
- Cheng, L.L.; Chang, I.W.; Smith, B.L.; Gonzalez, R.G. Evaluating Human Breast Ductal Carcinomas with High-Resolution Magic-Angle Spinning Proton Magnetic Resonance Spectroscopy. J. Magn. Reson. 1998, 135, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Sitter, B.; Lundgren, S.; Bathen, T.F.; Halgunset, J.; Fjosne, H.E.; Gribbestad, I.S. Comparison of HR MAS MR spectroscopic profiles of breast cancer tissue with clinical parameters. NMR Biomed. 2006, 19, 30–40. [Google Scholar] [CrossRef]
- Sitter, B.; Sonnewald, U.; Spraul, M.; Fjösne, H.E.; Gribbestad, I.S. High-resolution magic angle spinning MRS of breast cancer tissue. NMR Biomed. 2002, 15, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Bathen, T.F.; Jensen, L.R.; Sitter, B.; Fjösne, H.E.; Halgunset, J.; Axelson, D.E.; Gribbestad, I.S.; Lundgren, S. MR-determined metabolic phenotype of breast cancer in prediction of lymphatic spread, grade, and hormone status. Breast Cancer Res. Treat. 2007, 104, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Giskeødegård, G.F.; Grinde, M.T.; Sitter, B.; Axelson, D.E.; Lundgren, S.; Fjøsne, H.E.; Dahl, S.; Gribbestad, I.S.; Bathen, T.F. Multivariate modeling and prediction of breast cancer prognostic factors using MR metabolomics. J. Proteome Res. 2010, 9, 972–979. [Google Scholar] [CrossRef]
- Choi, J.; Baek, H.-M.; Kim, S.; Kim, M.; Youk, J.; Moon, H.; Kim, E.-K.; Han, K.; Kim, D.-H.; Kim, S. HR-MAS MR spectroscopy of breast cancer tissue obtained with core needle biopsy: Correlation with prognostic factors. PLoS ONE 2012, 7, e51712. [Google Scholar] [CrossRef]
- Maria, R.M.; Altei, W.F.; Selistre-de-Araujo, H.S.; Colnago, L.A. Impact of chemotherapy on metabolic reprogramming: Characterization of the metabolic profile of breast cancer MDA-MB-231 cells using 1H HR-MAS NMR spectroscopy. J. Pharmaceut. Biomed. Anal. 2017, 146, 324–328. [Google Scholar] [CrossRef] [Green Version]
- Fuss, T.L.; Cheng, L.L. Evaluation of Cancer Metabolomics Using ex vivo high resolution magic angle spinning (HRMAS) magnetic resonance spectroscopy (MRS). Metabolites 2016, 6, 11. [Google Scholar] [CrossRef] [Green Version]
- Roebuck, J.R.; Cecil, K.M.; Schnall, M.D.; Lenkinski, R.E. Human breast lesions: Characterization with proton MR spectroscopy. Radiology 1998, 209, 269–275. [Google Scholar] [CrossRef]
- Jagannathan, N.R.; Kumar, M.; Seenu, V.; Coshic, O.; Dwivedi, S.N.; Julka, P.K.; Srivastava, A.; Rath, G.K. Evaluation of total choline from in vivo volume localized proton MR spectroscopy and its response to neoadjuvant chemotherapy in locally advanced breast cancer. Br. J. Cancer 2001, 84, 1016–1022. [Google Scholar] [CrossRef] [Green Version]
- Sah, R.G.; Sharma, U.; Parshad, R.; Seenu, V.; Mathur, S.R.; Jagannathan, N.R. Association of estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 status with total choline concentration and tumor volume in breast cancer patients: An MRI and in vivo proton MRS study. Magn. Reson. Med. 2012, 68, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Katz-Brull, R.; Lavin, P.T.; Lenkinski, R.E. Clinical utility of proton magnetic resonance spectroscopy in characterizing breast lesions. J. Natl. Cancer Inst. 2002, 94, 1197–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagannathan, N.R. Application of in vivo MR methods in the study of breast cancer metabolism. NMR Biomed. 2018, 20, e4032. [Google Scholar] [CrossRef] [PubMed]
- Baltzer, P.A.; Dietzel, M. Breast lesions: Diagnosis by using proton MR spectroscopy at 1.5 and 3.0 T—Systematic review and meta-analysis. Radiology 2013, 267, 735–746. [Google Scholar] [CrossRef]
- Cen, D.; Xu, L. Differential diagnosis between malignant and benign breast lesions using single-voxel proton MRS: A meta-analysis. J. Cancer Res. Clin. Oncol. 2014, 140, 993–1001. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.J.; Song, H.S.; Chen, L.H. 1H-MRS evaluation of breast lesions by using total choline signal-to-noise ratio as an indicator of malignancy: A meta-analysis. Med. Oncol. 2015, 32, 160. [Google Scholar] [CrossRef]
- Gallagher, F.A.; Woitek, R.; McLean, M.A.; Gill, A.B.; Manzano Garcia, R.; Provenzano, E.; Riemer, F.; Kaggie, J.; Chhabra, A.; Ursprung, S.; et al. Imaging breast cancer using hyperpolarized carbon-13 MRI. Proc. Natl. Acad. Sci. USA 2020, 117, 2092–2098. [Google Scholar] [CrossRef] [Green Version]
- Racker, E. Why do tumor cells have a high aerobic glycolysis? J. Cell. Physiol. 1976, 89, 697–700. [Google Scholar] [CrossRef]
- Eingenbrodt, E.; Glossmann, H. Glycolysis. One of the keys to cancer? Trends Pharmacol. Sci. 1980, 1, 240–245. [Google Scholar] [CrossRef]
- Leach, M.; Moyec, L.; Podo, F. MRS of tumors: Basic principles. In Magnetic Resonance Spectroscopy in Biology and Medicine; Cetaines, J.D., de Bovee, W.M.M.J., Podo, F., Eds.; Pergamon Press: Oxford, UK, 1992; pp. 295–344. [Google Scholar]
- Vander Heiden, M.G.; Cantley, L.; Thompson, C. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [Green Version]
- Hirschhaeuser, F.; Sattler, U.; Mueller-Klieser, W. Lactate: A metabolic key player in cancer. Cancer Res. 2011, 71, 6921–6925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haukaas, T.H.; Euceda, L.R.; Giskeødegård, G.F.; Lamichhane, S.; Krohn, M.; Jernström, S.; Aure, M.R.; Lingjærde, O.C.; Schlichting, E.; Garred, Ø.; et al. Oslo Breast Cancer Consortium (OSBREAC). Metabolic clusters of breast cancer in relation to gene- and protein expression subtypes. Cancer Metab. 2016, 4, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, N.; Wildes, F.; Takagi, T.; Glunde, K.; Bhujwalla, Z.M. The tumor microenvironment modulates choline and lipid metabolism. Front. Oncol. 2016, 6, 262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montemezzi, S.; Cavedon, C.; Camera, L.; Meliadò, G.; Caumo, F.; Baglio, I.; Sardanelli, F. 1H MR spectroscopy of suspicious breast mass lesions at 3T: A clinical experience. Radiol. Med. 2017, 122, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Ramazan, A.; Demircioglu, O.; Ugurlu, U.; Kaya, H.; Aribal, E. Efficacy of single voxel 1H MR spectroscopic imaging at 3T for the differentiation of benign and malignant breast lesions. Clin. Imaging 2016, 40, 831–836. [Google Scholar] [CrossRef] [PubMed]
- Aribal, E.; Asadov, R.; Ramazan, A.; Ugurlu, M.Ü.; Kaya, H. Multiparametric breast MRI with 3T: Effectivity of combination of contrast enhanced MRI, DWI and 1H single voxel spectroscopy in differentiation of Breast tumors. Eur. J. Radiol. 2016, 85, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Clauser, P.; Marcon, M.; Dietzel, M.; Baltzer, P.A.T. A new method to reduce false positive results in breast MRI by evaluation of multiple spectral regions in proton MR-spectroscopy. Eur. J. Radiol. 2017, 92, 51–57. [Google Scholar] [CrossRef]
- Gillies, R.J.; Barry, J.A.; Ross, B.D. In vitro and in vivo 13C and 31P NMR analyses of phosphocholine metabolism in rat glioma cells. Magn. Reson. Med. 1994, 32, 310–318. [Google Scholar] [CrossRef]
- Katz-Brull, R.; Seger, D.; Rivenson-Segal, D. Metabolic markers of breast cancer: Enhanced choline metabolism and reduced choline-ether-phospholipid synthesis. Cancer Res. 2002, 62, 1966–1970. [Google Scholar]
- Warden, C.H.; Friedkin, M.; Geiger, P.J. Regulation of choline kinase activity and phosphatidyl choline biosynthesis by mitogenic growth factors in 3T3 fibroblasts. J. Biol. Chem. 1985, 260, 6006–6011. [Google Scholar] [CrossRef]
- Noh, D.Y.; Ahn, S.J.; Lee, R.A.; Park, I.A.; Kim, J.H.; Suh, P.G.; Ryu, S.H.; Lee, K.H.; Han, J.S. Overexpression of phospholipase D1 in human breast cancer tissues. Cancer Lett. 2000, 161, 207–214. [Google Scholar] [CrossRef]
- Glunde, K.; Jie, C.; Bhujwalla, Z.M. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004, 64, 4270–4276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogin, L.; Papa, M.Z.; Polak-Charcon, S.; Degani, H. TNF modulations of phospholipid metabolism in human breast cancer cells. Biochim. Biophys. Acta 1998, 1392, 217–232. [Google Scholar] [CrossRef]
- Tozaki, M.; Hoshi, K. 1H MR spectroscopy of invasive ductal carcinoma: Correlations with FDG PET and histologic prognostic factors. AJR Am. J. Roentgenol. 2010, 194, 1384–1390. [Google Scholar] [CrossRef] [PubMed]
- Baio, G.; Rescinito, G.; Rosa, F.; Pace, D.; Boccardo, S.; Basso, L.; Salvi, S.; Calabrese, M.; Truini, M.; Neumaier, C.E. Correlation between choline peak at MR spectroscopy and calcium-sensing receptor expression level in breast cancer: A preliminary clinical study. Mol. Imaging Biol. 2015, 17, 548–556. [Google Scholar] [CrossRef]
- Agarwal, K.; Hariprasad, G.; Rani, K.; Sharma, U.; Mathur, S.R.; Seenu, V.; Parshad, R.; Jagannathan, N.R. Is there an association between enhanced choline and β-catenin pathway in breast cancer? A pilot study by MR Spectroscopy and ELISA. Sci. Rep. 2017, 7, 2221. [Google Scholar] [CrossRef] [Green Version]
- Lukey, M.J.; Katt, W.P.; Cerione, R.A. Targeting amino acid metabolism for cancer therapy. Drug Discov. Today 2016, 6, 281–289. [Google Scholar] [CrossRef] [Green Version]
- Hensley, C.T.; Wasti, A.T.; DeBerardinis, R.J. Glutamine and cancer: Cell biology, physiology, and clinical opportunities. J. Clin. Investig. 2013, 123, 3678–3684. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, R.; Jain, M.; Madhusudhan, N.; Sheppard, N.G.; Strittmatter, L.; Kampf, C.; Huang, J.; Asplund, A.; Mootha, V.K. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014, 5, 3128. [Google Scholar] [CrossRef] [Green Version]
- Sitter, B.; Bathen, T.F.; Singstad, T.; Fjøsne, H.E.; Lundgren, S.; Halgunset, J.; Gribbestad, I.S. Quantification of Metabolites in breast cancer patients with different clinical prognosis using HR MAS MR spectroscopy. NMR Biomed. 2010, 23, 424–431. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, H.; Wang, Y.; Liu, C.; Zhu, W.; Zheng, S.; Wan, F. Taurine induces the apoptosis of breast cancer cells by regulating apoptosis-related proteins of mitochondria. Int. J. Mol. Med. 2015, 35, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Perez, I.; Posma, J.M.; Serrano-Contreras, J.I.; Boulangé, C.L.; Chan, Q.; Frost, G.; Stamler, J.; Elliott, P.; Lindon, J.C.; Holmes, E.; et al. Identifying unknown metabolites using NMR-based metabolic profiling techniques. Nat. Protoc. 2020, 15, 2538–2567. [Google Scholar] [CrossRef] [PubMed]
- Stebbing, J.; Sharma, A.; North, B.; Athersuch, T.J.; Zebrowski, A.; Pchejetski, D.; Coombes, R.C.; Nicholson, J.K.; Keun, H.C. A metabolic phenotyping approach to understanding relationships between metabolic syndrome and breast tumour responses to chemotherapy. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2012, 23, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Connelly, J.; Lindon, J.C.; Holmes, E. Metabonomics: A platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov. 2002, 1, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Brindle, J.T.; Antti, H.; Holmes, E.; Tranter, G.; Nicholson, J.K.; Bethell, H.W.; Clarke, S.; Schofield, P.M.; McKilligin, E.; Mosedale, D.E.; et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nat. Med. 2002, 8, 1439–1444. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.M.; Darzi, A.W.; Takats, Z.; Lindon, J.C. Metabolic phenotyping in clinical and surgical environments. Nature 2012, 491, 384–392. [Google Scholar] [CrossRef]
- Sharma, U.; Jagannathan, N.R. Breast Cancer Metabolomics Using NMR. Methods Mol. Biol. 2019, 2037, 195–213. [Google Scholar] [CrossRef]
- Li, M.; Song, Y.; Cho, N.; Chang, J.M.; Koo, H.R.; Yi, A.; Kim, H.; Park, S.; Moon, W.K. An HR-MAS MR metabolomics study on breast tissues obtained with core needle biopsy. PLoS ONE 2011, 6, e25563. [Google Scholar] [CrossRef] [Green Version]
- Bathen, T.F.; Geurts, B.; Sitter, B.; Fjøsne, H.E.; Lundgren, S.; Buydens, L.M.; Gribbestad, I.S.; Postma, G.; Giskeødegård, G.F. Feasibility of MR metabolomics for immediate analysis of resection margins during breast cancer surgery. PLoS ONE 2013, 8, e61578. [Google Scholar] [CrossRef] [Green Version]
- Gogiashvili, M.; Horsch, S.; Marchan, R.; Gianmoena, K.; Cadenas, C.; Tanner, B.; Naumann, S.; Ersova, D.; Lippek, F.; Rahnenführer, J.; et al. Impact of intratumoral heterogeneity of breast cancer tissue on quantitative metabolomics using high-resolution magic angle spinning 1H NMR spectroscopy. NMR Biomed. 2018, 31, e3862. [Google Scholar] [CrossRef]
- Chae, E.Y.; Shin, H.J.; Kim, S.; Baek, H.M.; Yoon, D.; Kim, S.; Shim, Y.E.; Kim, H.H.; Cha, J.H.; Choi, W.J.; et al. The Role of High-Resolution Magic Angle Spinning 1H Nuclear Magnetic Resonance Spectroscopy for Predicting the Invasive Component in Patients with Ductal Carcinoma in Situ Diagnosed on Preoperative Biopsy. PLoS ONE 2016, 11, e0161038. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Yoon, D.; Yun, M.; Choi, J.S.; Park, V.Y.; Kim, E.K.; Jeong, J.; Koo, J.S.; Yoon, J.H.; Moon, H.J.; et al. Metabolomics of Breast Cancer Using High-Resolution Magic Angle Spinning Magnetic Resonance Spectroscopy: Correlations with 18F-FDG Positron Emission Tomography-Computed Tomography, Dynamic Contrast-Enhanced and Diffusion-Weighted Imaging MRI. PLoS ONE 2016, 11, e0159949. [Google Scholar] [CrossRef] [PubMed]
- Borgan, E.; Sitter, B.; Lingjærde, O.C.; Johnsen, H.; Lundgren, S.; Bathen, T.F.; Sørlie, T.; Børresen-Dale, A.L.; Gribbestad, I.S. Merging transcriptomics and metabolomics—Advances in breast cancer profiling. BMC Cancer 2010, 10, 628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giskeødegård, G.F.; Lundgren, S.; Sitter, B.; Fjøsne, H.E.; Postma, G.; Buydens, L.M.; Gribbestad, I.S.; Bathen, T.F. Lactate and glycine-potential MR biomarkers of prognosis in estrogen receptor-positive breast cancers. NMR Biomed. 2012, 25, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.D.; Lamichhane, S.; Lundgren, S.; Bofin, A.; Fjøsne, H.; Giskeødegård, G.F.; Bathen, T.F. Metabolic characterization of triple negative breast cancer. BMC Cancer 2014, 14, 941. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.S.; Baek, H.M.; Kim, S.; Kim, M.J.; Youk, J.H.; Moon, H.J.; Kim, E.K.; Nam, Y.K. Magnetic resonance metabolic profiling of breast cancer tissue obtained with core needle biopsy for predicting pathologic response to neoadjuvant chemotherapy. PLoS ONE 2013, 8, e83866. [Google Scholar] [CrossRef]
- Beckonert, O.; Monnerjahn, J.; Bonk, U.; Leibfritz, D. Visualizing metabolic changes in breast-cancer tissue using 1H-NMR spectroscopy and self-organizing maps. NMR Biomed. 2003, 16, 1–11. [Google Scholar] [CrossRef]
- Seenu, V.; Pavan Kumar, M.N.; Sharma, U.; Gupta, S.D.; Mehta, S.N.; Jagannathan, N.R. Potential of magnetic resonance spectroscopy to detect metastasis in axillary lymph nodes in breast cancer. Magn. Reson. Imaging 2005, 23, 1005–1010. [Google Scholar] [CrossRef]
- Richard, V.; Conotte, R.; Mayne, D.; Colet, J.M. Does the 1H-NMR plasma metabolome reflect the host-tumor interactions in human breast cancer? Oncotarget 2017, 8, 49915–49930. [Google Scholar] [CrossRef]
- Hart, C.D.; Vignoli, A.; Tenori, L.; Uy, G.L.; Van To, T.; Adebamowo, C.; Hossain, S.M.; Biganzoli, L.; Risi, E.; Love, R.R.; et al. Serum metabolomic profiles identify ER-positive early breast cancer patients at increased risk of disease recurrence in a multicenter population. Clin. Cancer Res. 2017, 23, 1422–1431. [Google Scholar] [CrossRef] [Green Version]
- Tenori, L.; Oakman, C.; Morris, P.G.; Gralka, E.; Turner, N.; Cappadona, S.; Fornier, M.; Hudis, C.; Norton, L.; Luchinat, C.; et al. Serum metabolomic profiles evaluated after surgery may identify patients with oestrogen receptor negative early breast cancer at increased risk of disease recurrence. Results from a retrospective study. Mol. Oncol. 2015, 9, 128–139. [Google Scholar] [CrossRef]
- Asiago, V.M.; Alvarado, L.Z.; Shanaiah, N.; Gowda, G.A.; Owusu-Sarfo, K.; Ballas, R.A.; Raftery, D. Early detection of recurrent breast cancer using metabolite profiling. Cancer Res. 2010, 70, 8309–8318. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Sharma, R.K.; Chagtoo, M.; Agarwal, G.; George, N.; Sinha, N.; Godbole, M.M. 1H NMR metabolomics reveals association of high expression of inositol 1, 4, 5 trisphosphate receptor and Metabolites in breast cancer patients. PLoS ONE 2017, 12, e0169330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flote, V.G.; Vettukattil, R.; Bathen, T.F.; Egeland, T.; McTiernan, A.; Frydenberg, H.; Husøy, A.; Finstad, S.E.; Lømo, J.; Garred, Ø.; et al. Lipoprotein subfractions by nuclear magnetic resonance are associated with tumor characteristics in breast cancer. Lipids Health Dis. 2016, 15, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tenori, L.; Oakman, C.; Claudino, W.M.; Bernini, P.; Cappadona, S.; Nepi, S.; Biganzoli, L.; Arbushites, M.C.; Luchinat, C.; Bertini, I.; et al. Exploration of serum metabolomic profiles and outcomes in women with metastatic breast cancer: A pilot study. Mol. Oncol. 2012, 6, 437–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, S.; Liu, L.; Zhang, J.; Bowers, J.; Gowda, G.A.; Seeger, H.; Fehm, T.; Neubauer, H.J.; Vogel, U.; Clare, S.E.; et al. Metabolomics approach for predicting response to neoadjuvant chemotherapy for breast cancer. Mol. Oncol. 2013, 7, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Cao, M.D.; Cheng, M.; Rizwan, A.; Jiang, L.; Krishnamachary, B.; Bhujwalla, Z.M.; Bathen, T.F.; Glunde, K. Targeting choline phospholipid metabolism: GDPD5 and GDPD6 silencing decrease breast cancer cell proliferation, migration, and invasion. NMR Biomed. 2016, 29, 1098–1107. [Google Scholar] [CrossRef]
- Jagannathan, N.R.; Singh, M.; Govindaraju, V.; Raghunathan, P.; Coshic, O.; Julka, P.K.; Rath, G.K. Volume localized in vivo proton MR spectroscopy of breast carcinoma: Variation of water-fat ratio in patients receiving chemotherapy. NMR Biomed. 1998, 11, 414–422. [Google Scholar] [CrossRef]
- Kumar, M.; Jagannathan, N.R.; Seenu, V.; Dwivedi, S.N.; Julka, P.K.; Rath, G.K. Monitoring the therapeutic response of locally advanced breast cancer patients: Sequential in vivo proton MR spectroscopy study. J. Magn. Reson. Imaging 2006, 24, 325–332. [Google Scholar] [CrossRef]
- Manton, D.J.; Chaturvedi, A.; Hubbard, A.; Lind, M.J.; Lowry, M.; Maraveyas, A.; Pickles, M.D.; Tozer, D.J.; Turnbull, L.W. Neoadjuvant chemotherapy in breast cancer: Early response prediction with quantitative MR imaging and spectroscopy. Br. J. Cancer 2006, 94, 427–435. [Google Scholar] [CrossRef]
- Wang, J.; Wang, M.Y.; Kuo, W.H.; Chen, K.L.; Shih, T.T. Proton MR spectroscopy of normal breasts: Association of risk factors for breast cancer with water and lipid composition of the breast. Magn. Reson. Imaging 2016, 34, 524–528. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, K.; Sharma, U.; Mathur, S.; Seenu, V.; Parshad, R.; Jagannathan, N.R. Study of lipid metabolism by estimating the fat fraction in different breast tissues and in various breast tumor sub-types by in vivo (1) H MR spectroscopy. Magn. Reson. Imaging 2018, 49, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.B.; Horvat, J.V.; Hancu, I.; Sutton, O.M.; Bernard-Davila, B.; Weber, M.; Oh, J.H.; Marino, M.A.; Avendano, D.; Leithner, D.; et al. Quantitative in vivo proton MR spectroscopic assessment of lipid metabolism: Value for breast cancer diagnosis and prognosis. J. Magn. Reson. Imaging 2019, 50, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.B.; Brennan, S.B.; Ishill, N.M.; Morris, E.A.; Liberman, L.; Dershaw, D.D.; Bartella, L.; Koutcher, J.A.; Huang, W. Diagnostic usefulness of water-to-fat ratio and choline concentration in malignant and benign breast lesions and normal breast parenchyma: An in vivo 1H MRS study. J. Magn. Reson. Imaging 2011, 33, 855–863. [Google Scholar] [CrossRef]
- Drisis, S.; Flamen, P.; Ignatiadis, M.; Metens, T.; Chao, S.L.; Chintinne, M.; Lemort, M. Total choline quantification measured by 1H MR spectroscopy as early predictor of response after neoadjuvant treatment for locally advanced breast cancer: The impact of immunohistochemical status. J. Magn. Reson. Imaging 2018, 48, 982–993. [Google Scholar] [CrossRef]
- Sharma, U.; Baek, H.M.; Su, M.Y.; Jagannathan, N.R. In vivo 1H MRS in the assessment of the therapeutic response of breast cancer patients. NMR Biomed. 2011, 24, 700–711. [Google Scholar] [CrossRef] [Green Version]
- Bolan, P.J.; Kim, E.; Herman, B.A.; Newstead, G.M.; Rosen, M.A.; Schnall, M.D.; Pisano, E.D.; Weatherall, P.T.; Morris, E.A.; Lehman, C.D.; et al. ACRIN Trial team ISPY-1 Investigators. MR spectroscopy of breast cancer for assessing early treatment response: Results from the ACRIN 6657 MRS trial. J. Magn. Reson. Imaging 2017, 46, 290–302. [Google Scholar] [CrossRef]
- Sharma, U.; Agarwal, K.; Sah, R.G.; Parshad, R.; Seenu, V.; Mathur, S.; Gupta, S.D.; Jagannathan, N.R. Can multi-parametric MR based approach improve the predictive value of pathological and clinical therapeutic response in breast cancer patients? Front. Oncol. 2018, 8, 319. [Google Scholar] [CrossRef]
- Sharma, U.; Jagannathan, N.R. In vivo magnetic resonance spectroscopy in breast cancer. In Modern Magnetic Resonance; Webb, G.A., Ed.; Springer: Dordrecht, The Netherlands, 2008. [Google Scholar] [CrossRef]
- Schmitz, A.M.; Veldhuis, W.B.; Menke-Pluijmers, M.B.; van der Kemp, W.J.; van der Velden, T.A.; Kock, M.C.; Westenend, P.J.; Klomp, D.W.; Gilhuijs, K.G. Multiparametric MRI with dynamic contrast enhancement, diffusion-weighted imaging, and 31-Phosphorus spectroscopy at 7 T for characterization of breast cancer. Investig. Radiol. 2015, 50, 766–771. [Google Scholar] [CrossRef]
- Harris, T.; Degani, H.; Frydman, L. Hyperpolarized 13C NMR studies of glucose metabolism in living breast cancer cell cultures. NMR Biomed. 2013, 26, 1831–1843. [Google Scholar] [CrossRef]
- Nelson, S.J.; Kurhanewicz, J.; Vigneron, D.B.; Larson, P.E.; Harzstark, A.L.; Ferrone, M.; van Criekinge, M.; Chang, J.W.; Bok, R.; Park, I.; et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-¹³C] pyruvate. Sci. Transl. Med. 2013, 5, 198ra108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golman, K.; in ’t Zandt, R.; Thaning, M. Real-time metabolic imaging. Proc. Natl. Acad. Sci. USA 2006, 103, 11270–11275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albers, M.J.; Bok, R.; Chen, A.P.; Cunningham, C.H.; Zierhut, M.L.; Zhang, V.Y.; Kohler, S.J.; Tropp, J.; Hurd, R.E.; Yen, Y.F.; et al. Hyperpolarized 13C lactate, pyruvate, and alanine: Noninvasive biomarkers for prostate cancer detection and grading. Cancer Res. 2008, 68, 8607–8615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woitek, R.; McLean, M.A.; Gill, A.B.; Grist, J.T.; Provenzano, E.; Patterson, A.J.; Ursprung, S.; Torheim, T.; Zaccagna, F.; Locke, M.; et al. Hyperpolarized 13C MRI of Tumor Metabolism Demonstrates Early Metabolic Response to Neoadjuvant Chemotherapy in Breast Cancer. Radiol. Imaging Cancer 2020, 2, e200017. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.M.; Cho, N.; Moy, L. Breast MRI: State of the Art. Radiology 2019, 292, 520–536. [Google Scholar] [CrossRef]
- Sharma, U.; Agarwal, K.; Hari, S.; Mathur, S.R.; Seenu, V.; Parshad, R.; Jagannathan, N.R. Role of diffusion weighted imaging and magnetic resonance spectroscopy in breast cancer patients with indeterminate dynamic contrast enhanced magnetic resonance imaging findings. Magn. Reson. Imaging 2019, 61, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.K.; Schrading, S.; Strobel, K.; Schild, H.H.; Hilgers, R.D.; Bieling, H.B. Abbreviated breast magnetic resonance imaging (MRI): First postcontrast subtracted images and maximum-intensity projection-a novel approach to breast cancer screening with MRI. Am. J. Clin. Oncol. 2014, 32, 2304–2310. [Google Scholar] [CrossRef]
- Leithner, D.; Moy, L.; Morris, E.A.; Marino, M.A.; Helbich, T.H.; Pinker, K. Abbreviated MRI of the Breast: Does It Provide Value? J. Magn. Reson Imaging 2019, 49, e85–e100. [Google Scholar] [CrossRef]
- Onishi, N.; Sadinski, M.; Hughes, M.C.; Ko, E.S.; Gibbs, P.; Gallagher, K.M.; Fung, M.M.; Hunt, T.J.; Martinez, D.F.; Shukla-Dave, A.; et al. Ultrafast dynamic contrast-enhanced breast MRI may generate prognostic imaging markers of breast cancer. Breast Cancer Res. 2020, 22, 58. [Google Scholar] [CrossRef]
- Franklin, S.L.; Voormolen, N.; Bones, I.K.; Korteweg, T.; Wasser, M.; Dankers, H.G.; Cohen, D.; van Stralen, M.; Bos, C.; van Osch, M. Feasibility of velocity-selective arterial spin labeling in breast cancer patients for noncontrast-enhanced perfusion imaging. J. Magn. Reson. Imaging 2021, 54, 1282–1291. [Google Scholar] [CrossRef]
- Kawashima, M.; Katada, Y.; Shukuya, T.; Kojima, M.; Nozaki, M. MR perfusion imaging using the arterial spin labeling technique for breast cancer. J. Magn. Reson. Imaging 2012, 35, 436–440. [Google Scholar] [CrossRef]
- Fischer, U.; Korthauer, A.; Baum, F.; Luftner-Nagel, S.; Heyden, D.; Marten-Engelke, K. Short first-pass MRI of the breast. Acta Radiol. 2012, 53, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.K.; Bieling, H.; Gieseke, J.; Ebel, T.; Mielcarek, P.; Far, F.; Folkers, P.; Elevelt, A.; Schild, H.H. Breast neoplasms: T2* susceptibility-contrast, first-pass perfusion MR imaging. Radiology 1997, 202, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Delille, J.P.; Slanetz, P.J.; Yeh, E.D.; Kopans, D.B.; Garrido, L. Breast cancer: Regional blood flow and blood volume measured with magnetic susceptibility-based MR imaging-initial results. Radiology 2002, 223, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Delille, J.P.; Slanetz, P.J.; Yeh, E.D.; Kopans, D.B.; Garrido, L. Physiologic changes in breast magnetic resonance imaging during the menstrual cycle: Perfusion imaging, signal enhancement, and influence of the T1 relaxation time of breast tissue. Breast J 2005, 11, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Park, V.Y.; Kim, E.K.; Kim, M.J.; Yoon, J.H.; Moon, H.J. Perfusion parameters on breast dynamic contrast-enhanced MRI are associated with disease-specific survival in patients with triple-negative breast cancer. AJR. Am. J. Roentgenol. 2017, 208, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Tofts, P.S.; Brix, G.; Buckley, D.L.; Evelhoch, J.L.; Henderson, E.; Knopp, M.V.; Larsson, H.B.; Lee, T.Y.; Mayr, N.A.; Parker, G.J.; et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: Standardized quantities and symbols. J. Magn. Reson. Imaging 1999, 10, 223–232. [Google Scholar] [CrossRef]
- Yim, H.; Kang, D.K.; Jung, Y.S.; Jeon, G.S.; Kim, T.H. Analysis of kinetic curve and model-based perfusion parameters on dynamic contrast-enhanced MRI in breast cancer patients: Correlations with dominant stroma type. Magn. Reson. Imaging 2016, 34, 60–65. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, S.H.; Kim, Y.J.; Kang, B.J.; An, Y.Y.; Lee, A.W.; Song, B.J.; Park, Y.S.; Lee, H.B. Enhancement parameters on dynamic contrast enhanced breast MRI: Do they correlate with prognostic factors and subtypes of breast cancers? Magn. Reson. Imaging 2015, 33, 72–80. [Google Scholar] [CrossRef]
- Koo, H.R.; Cho, N.; Song, I.C.; Kim, H.; Chang, J.M.; Yi, A.; Yun, B.L.; Moon, W.K. Correlation of perfusion parameters on dynamic contrast-enhanced MRI with prognostic factors and subtypes of breast cancers. J. Magn. Reson. Imaging 2012, 36, 145–151. [Google Scholar] [CrossRef]
- de Kruijf, E.M.; van Nes, J.G.; van de Velde, C.J.; Putter, H.; Smit, V.T.; Liefers, G.J.; Kuppen, P.J.; Tollenaar, R.A.; Mesker, W.E. Tumor-stroma ratio in the primary tumor is a prognostic factor in early breast cancer patients, especially in triple-negative carcinoma patients. Breast Cancer Res. Treat. 2011, 125, 687–696. [Google Scholar] [CrossRef]
- Dekker, T.J.; van de Velde, C.J.; van Pelt, G.W.; Kroep, J.R.; Julien, J.P.; Smit, V.T.; Tollenaar, R.A.; Mesker, W.E. Prognostic significance of the tumor-stroma ratio: Validation study in node-negative premenopausal breast cancer patients from the EORTC perioperative chemotherapy (POP) trial (10854). Breast Cancer Res. Treat. 2013, 139, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Moorman, A.M.; Vink, R.; Heijmans, H.J.; van der Palen, J.; Kouwenhoven, E.A. The prognostic value of tumour-stroma ratio in triple-negative breast cancer. ELSO 2012, 38, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Mei, N.; Yin, B.; Peng, W. Correlation of DCE-MRI Perfusion Parameters and Molecular Biology of Breast Infiltrating Ductal Carcinoma. Front. Oncol. 2021, 11, 561735. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.C. An introduction to ASL labeling techniques. J. Magn. Reson. Imaging 2014, 40, 1–10. [Google Scholar] [CrossRef]
- Dai, W.; Garcia, D.; de Bazelaire, C.; Alsop, D.C. Continuous flow-driven inversion for arterial spin labeling using pulsed radio frequency and gradient fields. Magn. Reson. Med. 2008, 60, 1488–1497. [Google Scholar] [CrossRef] [Green Version]
- Alsop, D.C.; Detre, J.A.; Golay, X.; Günther, M.; Hendrikse, J.; Hernandez-Garcia, L.; Lu, H.; MacIntosh, B.J.; Parkes, L.M.; Smits, M.; et al. Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn. Reson. Med. 2015, 73, 102–116. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.G. Quantification of relative cerebral blood flow change by flow-sensitive alternating inversion recovery (FAIR) technique: Application to functional mapping. Magn. Reson. Med. 1995, 34, 293–301. [Google Scholar] [CrossRef]
- Nery, F.; Buchanan, C.E.; Harteveld, A.A.; Odudu, A.; Bane, O.; Cox, E.F.; Derlin, K.; Gach, H.M.; Golay, X.; Gutberlet, M.; et al. Consensus-based technical recommendations for clinical translation of renal ASL MRI. Magn. Reson. Mater. Phys. Biol. Med. 2020, 33, 141–161. [Google Scholar] [CrossRef] [Green Version]
- Wong, E.C.; Cronin, M.; Wu, W.C.; Inglis, B.; Frank, L.R.; Liu, T.T. Velocity-selective arterial spin labeling. Magn. Reson. Med. 2006, 55, 1334–1341. [Google Scholar] [CrossRef] [Green Version]
- Yabuuchi, H.; Matsuo, Y.; Sunami, S.; Kamitani, T.; Kawanami, S.; Setoguchi, T.; Sakai, S.; Hatakenaka, M.; Kubo, M.; Tokunaga, E.; et al. Detection of non-palpable breast cancer in asymptomatic women by using unenhanced diffusion-weighted and T2-weighted MR imaging: Comparison with mammography and dynamic contrast-enhanced MR imaging. Eur. Radiol. 2011, 21, 11–17. [Google Scholar] [CrossRef]
- Richard, R.; Thomassin, I.; Chapellier, M.; Scemama, A.; de Cremoux, P.; Varna, M.; Giacchetti, S.; Espié, M.; de Kerviler, E.; de Bazelaire, C. Diffusion-weighted MRI in pretreatment prediction of response to neoadjuvant chemotherapy in patients with breast cancer. Eur. Radiol. 2013, 23, 2420–2431. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Danishad, K.K.; Seenu, V.; Jagannathan, N.R. Longitudinal study of the assessment by MRI and diffusion-weighted imaging of tumor response in patients with locally advanced breast cancer undergoing neoadjuvant chemotherapy. NMR Biomed. 2009, 22, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Bokacheva, L.; Kaplan, J.B.; Giri, D.D.; Patil, S.; Gnanasigamani, M.; Nyman, C.G.; Deasy, J.O.; Morris, E.A.; Thakur, S.B. Intravoxel incoherent motion diffusion-weighted MRI at 3.0 T differentiates malignant breast lesions from benign lesions and breast parenchyma. J. Magn. Reson. Imaging 2014, 40, 813–823. [Google Scholar] [CrossRef] [Green Version]
- Iima, M.; Yano, K.; Kataoka, M.; Umehana, M.; Murata, K.; Kanao, S.; Togashi, K.; Le Bihan, D. Quantitative non-Gaussian diffusion and intravoxel incoherent motion magnetic resonance imaging: Differentiation of malignant and benign breast lesions. Investig. Radiol. 2015, 50, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Liang, C.; Liu, Z.; Zhang, S.; Huang, B. Intravoxel incoherent motion (IVIM) in evaluation of breast lesions: Comparison with conventional DWI. Eur. J. Radiol. 2013, 82, e782–e789. [Google Scholar] [CrossRef]
- Jensen, J.H.; Helpern, J.A. MRI quantification of non-Gaussian water diffusion by kurtosis analysis. NMR Biomed. 2010, 23, 698–710. [Google Scholar] [CrossRef]
- Partridge, S.C.; Ziadloo, A.; Murthy, R.; White, S.W.; Peacock, S.; Eby, P.R.; DeMartini, W.B.; Lehman, C.D. Diffusion tensor MRI: Preliminary anisotropy measures and mapping of breast tumors. J. Magn. Reson. Imaging 2010, 31, 339–347. [Google Scholar] [CrossRef]
- Baltzer, P.A.; Schäfer, A.; Dietzel, M.; Grässel, D.; Gajda, M.; Camara, O.; Kaiser, W.A. Diffusion tensor magnetic resonance imaging of the breast: A pilot study. Eur. Radiol. 2011, 21, 1–10. [Google Scholar] [CrossRef]
- Meng, L.; Zhou, J.; Sasano, H.; Suzuki, T.; Zeitoun, K.M.; Bulun, S.E. Tumor necrosis factor alpha and interleukin 11 secreted by malignant breast epithelial cells inhibit adipocyte differentiation by selectively down-regulating CCAAT/enhancer binding protein alpha and peroxisome proliferator-activated receptor gamma: Mechanism of desmoplastic reaction. Cancer Res. 2001, 61, 2250–2255. [Google Scholar]
- Yang, J.Y.; Qiu, B.S. The Advance of Magnetic Resonance Elastography in Tumor Diagnosis. Front. Oncol. 2021, 11, 722703. [Google Scholar] [CrossRef] [PubMed]
- McKnight, A.L.; Kugel, J.L.; Rossman, P.J.; Manduca, A.; Hartmann, L.C.; Ehman, R.L. MR Elastography of Breast Cancer: Preliminary Results. Am. J. Roentgenol. 2002, 178, 1411–1417. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, J.; Sinkus, R.; Lorenzen, M.; Dargatz, M.; Leussler, C.; Röschmann, P.; Adam, G. MR elastography of the breast:preliminary clinical results. In RoFo: Fortschritte auf dem Gebiete der Rontgenstrahlen und der Nuklearmedizin; Georg Thieme Verlag Stuttgart: New York, NY, USA, 2002; Volume 174, pp. 830–834. [Google Scholar] [CrossRef]
- Xydeas, T.; Siegmann, K.; Sinkus, R.; Krainick-Strobel, U.; Miller, S.; Claussen, C.D. Magnetic resonance elastography of the breast: Correlation of signal intensity data with viscoelastic properties. Investig. Radiol. 2005, 40, 412–420. [Google Scholar] [CrossRef]
- Balleyguier, C.; Lakhdar, A.B.; Dunant, A.; Mathieu, M.C.; Delaloge, S.; Sinkus, R. Value of Whole Breast Magnetic Resonance Elastography Added to MRI for Lesion Characterization. NMR Biomed. 2018, 31, e3795. [Google Scholar] [CrossRef] [PubMed]
- Siegmann, K.C.; Xydeas, T.; Sinkus, R.; Kraemer, B.; Vogel, U.; Claussen, C.D. Diagnostic Value of MR Elastography in Addition to Contrast-Enhanced MR Imaging of the Breast-Initial Clinical Results. Eur. Radiol. 2010, 20, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Tagliafico, A.S.; Piana, M.; Schenone, D.; Lai, R.; Massone, A.M.; Houssami, N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast 2020, 49, 74–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Crivelli, P.; Ledda, R.E.; Parascandolo, N.; Fara, A.; Soro, D.; Conti, M. A new challenge for radiologists: Radiomics in breast cancer. BioMed Res. Int. 2018, 2018, 6120703. [Google Scholar] [CrossRef] [Green Version]
- Parekh, V.S.; Jacobs, M.A. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ Breast Cancer 2017, 3, 43. [Google Scholar] [CrossRef] [Green Version]
- Whitney, H.M.; Taylor, N.S.; Drukker, K.; Edwards, A.V.; Papaioannou, J.; Schacht, D.; Giger, M.L. Additive Benefit of Radiomics Over Size Alone in the Distinction Between Benign Lesions and Luminal A Cancers on a Large Clinical Breast MRI Dataset. Acad. Rdaiol. 2019, 26, 202–209. [Google Scholar] [CrossRef]
- Dong, Y.; Feng, Q.; Yang, W.; Lu, Z.; Deng, C.; Zhang, L.; Lian, Z.; Liu, J.; Luo, X.; Pei, S.; et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur. Radiol. 2018, 28, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhu, Y.; Liu, Z.; Yu, T.; He, C.; Jiang, W.; Kan, Y.; Dong, D.; Tian, J.; Luo, Y. Radiomic nomogram for prediction of axillary lymph node metastasis in breast cancer. Eur. Radiol. 2019, 29, 3820–3829. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Ji, Y.; Qi, L.; Guo, X.; Jian, X.; Liu, P. Breast cancer Ki67 expression prediction by DCE-MRI radiomics features. Clin. Radiol. 2018, 73, 909. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Bignotti, B.; Rossi, F.; Matos, J.; Calabrese, M.; Valdora, F.; Houssami, N. Breast cancer Ki-67 expression prediction by digital breast tomosynthesis radiomics features. Eur. Radiol. Exp. 2019, 3, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Characteristics | Magnetic Resonance Spectroscopy | Magnetic Resonance Imaging | ||
|---|---|---|---|---|
| In Vitro | Ex Vivo | In Vivo | ||
| Information | Biochemical composition (metabolite detection) | Biochemical composition (metabolite detection) | Biochemical composition (metabolite detection) | Anatomic (structure and morphology), functional |
| Sample/Subject | Tissue extract, biofluids, cell lines, aspirates | Excised tissues/biopsies | Living humans/organisms | Living humans/organisms |
| Equipment | NMR Spectrometer | NMR Spectrometer with accessories for HRMAS | Human MRI Scanner | Human MRI Scanner |
| Field Strength | High field strength 9.4 T–21.1 T | High field strength 9.4 T–18.8 T | 1.5 T–7 T | 1.5 T–7 T |
| Nuclei of interest | 1H, 13C, 31P, 23Na, 19F | 1H, 13C | 1H, 31P, 23Na, 19F 13C- hyperpolarized | 1H from fat and water |
| Data | 1D/2D spectra | 1D/2D spectra | SVS 1D, SVS-2D, CSI (MRSI) | Conventional T1, T2-weighted, DCE-MRI, Diffusion-weighted, Perfusion weighted, MR Elastography, fMRI |
| Advantages | High sensitivity and resolution, detection of a large number of metabolites, easy quantification, easy experimentation | High sensitivity and resolution, detection of a large number of metabolites, quantification not that easy, special experimentation | Organ-specific metabolite composition, and longitudinal studies. | Organ-specific structural and functional studies, longitudinal studies possible. |
| Limitations | Tissue excision is invasive | Tissue excision is invasive | Low sensitivity and resolution, detection of a small number of metabolites, Claustrophobia of patients | Claustrophobia of patients, contrast required in some studies |
| Reproducibility | Lesser than in vivo | Lesser than in vivo | High | High |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, U.; Jagannathan, N.R. Magnetic Resonance Imaging (MRI) and MR Spectroscopic Methods in Understanding Breast Cancer Biology and Metabolism. Metabolites 2022, 12, 295. https://doi.org/10.3390/metabo12040295
Sharma U, Jagannathan NR. Magnetic Resonance Imaging (MRI) and MR Spectroscopic Methods in Understanding Breast Cancer Biology and Metabolism. Metabolites. 2022; 12(4):295. https://doi.org/10.3390/metabo12040295
Chicago/Turabian StyleSharma, Uma, and Naranamangalam R. Jagannathan. 2022. "Magnetic Resonance Imaging (MRI) and MR Spectroscopic Methods in Understanding Breast Cancer Biology and Metabolism" Metabolites 12, no. 4: 295. https://doi.org/10.3390/metabo12040295
APA StyleSharma, U., & Jagannathan, N. R. (2022). Magnetic Resonance Imaging (MRI) and MR Spectroscopic Methods in Understanding Breast Cancer Biology and Metabolism. Metabolites, 12(4), 295. https://doi.org/10.3390/metabo12040295






