Flavin-Containing Monooxygenase 3 (FMO3) Is Critical for Dioxin-Induced Reorganization of the Gut Microbiome and Host Insulin Sensitivity
Abstract
:1. Introduction
2. Results
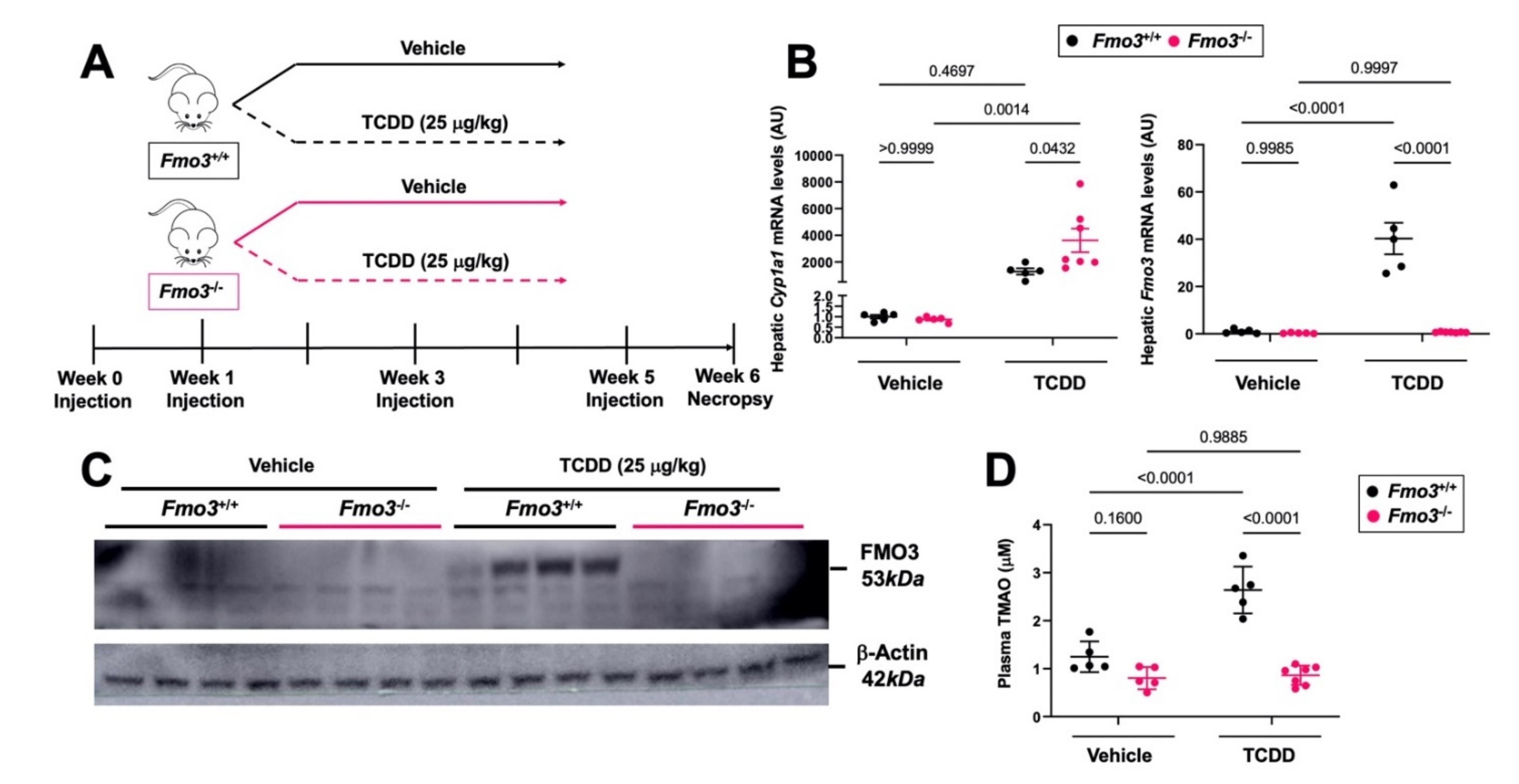
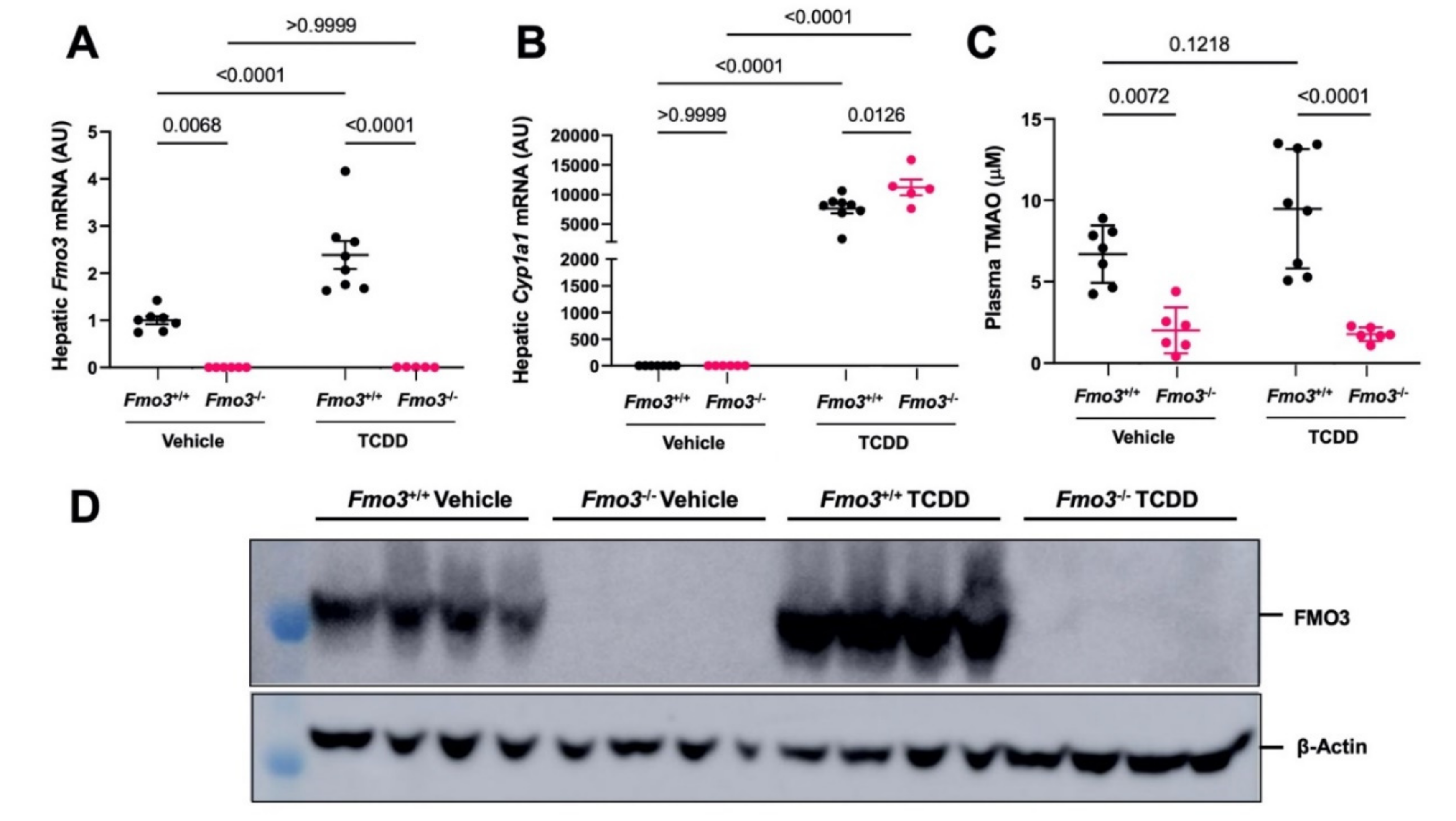
2.1. TCDD Exposure Stimulates the Expression and TMAO-Producing Function of FMO3
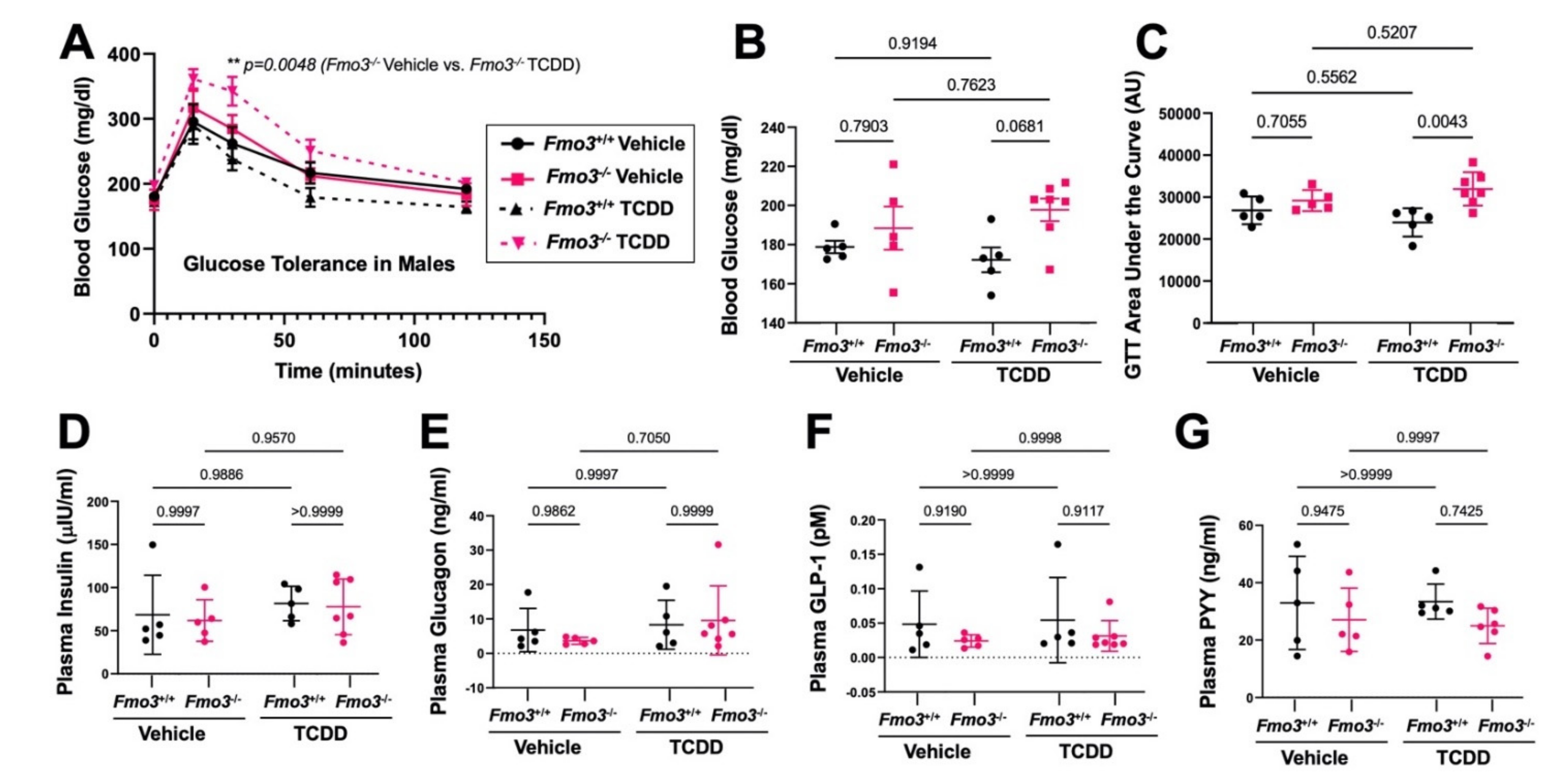
2.2. Genetic Deficiency of Fmo3 Results in Glucose Intolerance in TCDD-Treated Mice
2.3. TCDD-Induced Gene Expression Is Significantly Altered in the Liver of Male Fmo3−/− Mice
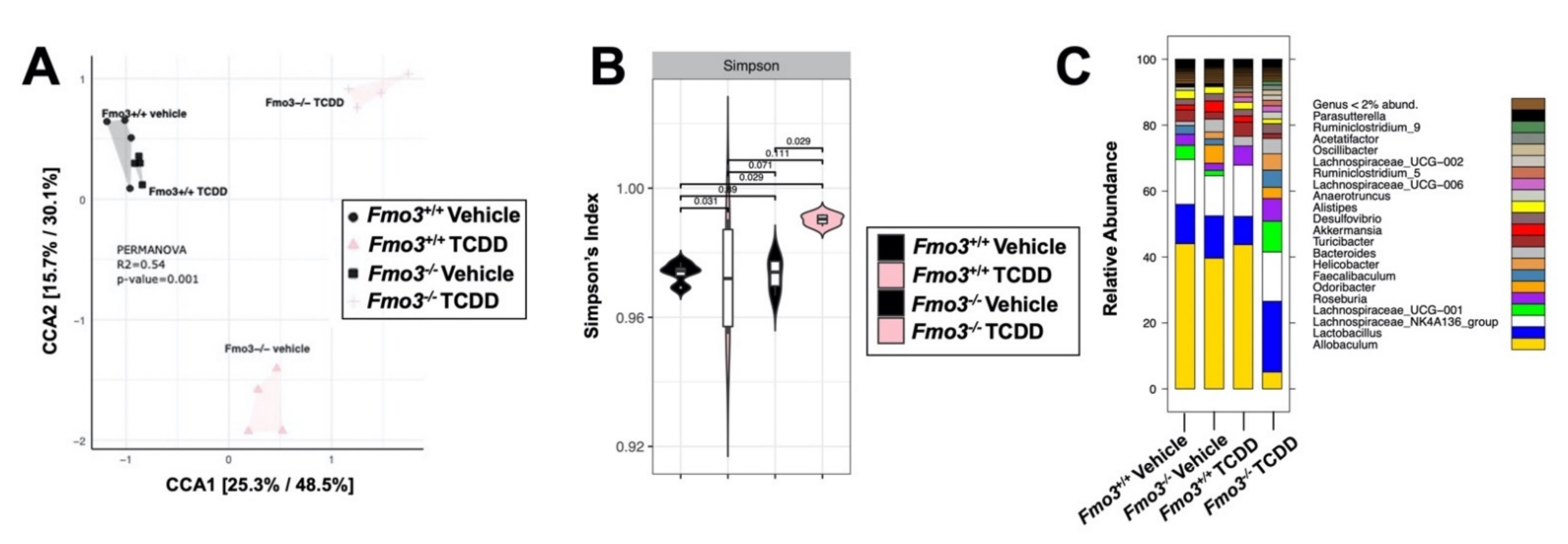
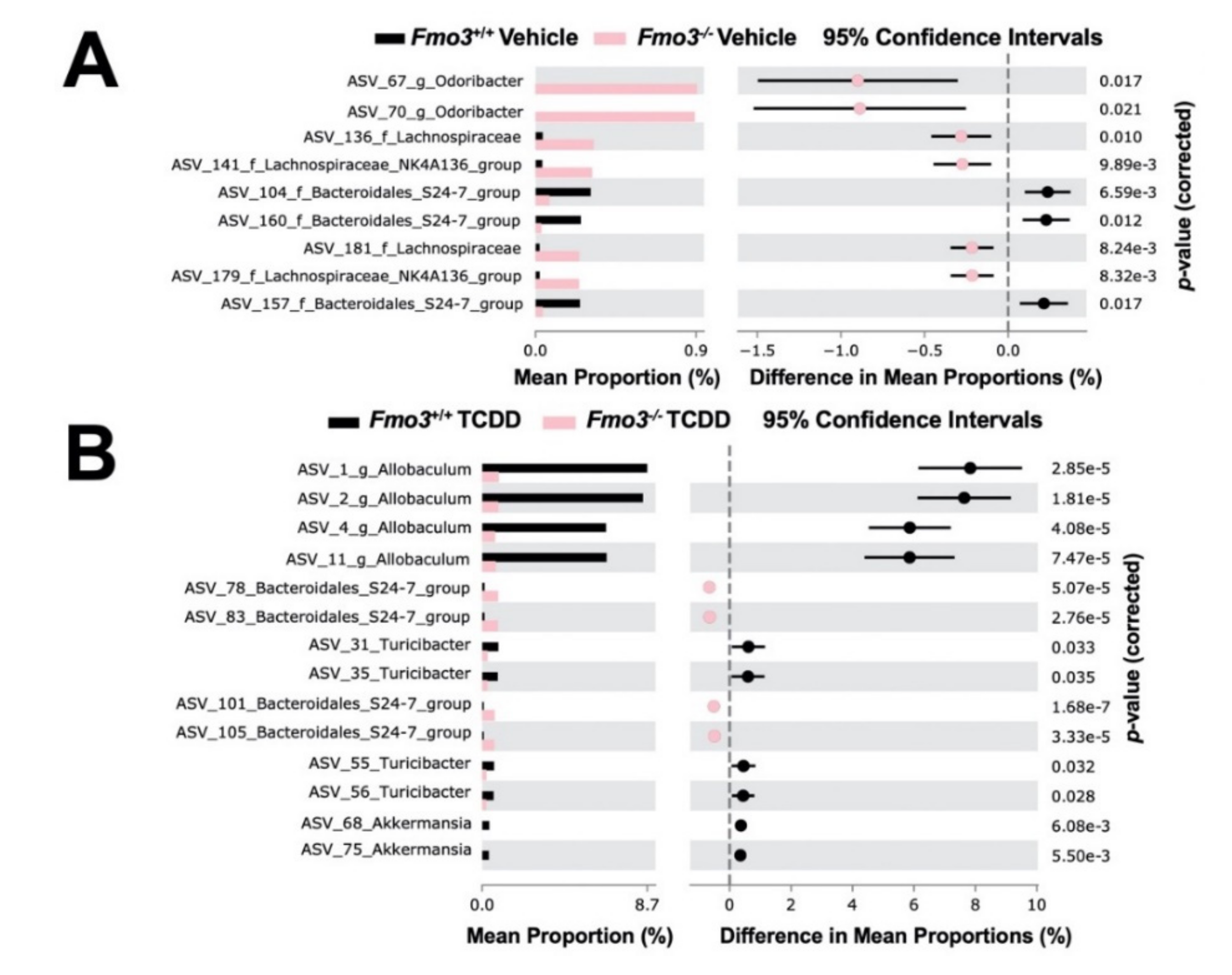
2.4. TCDD-Driven Reorganization of the Gut Microbiome Is Altered in Male Fmo3−/− Mice
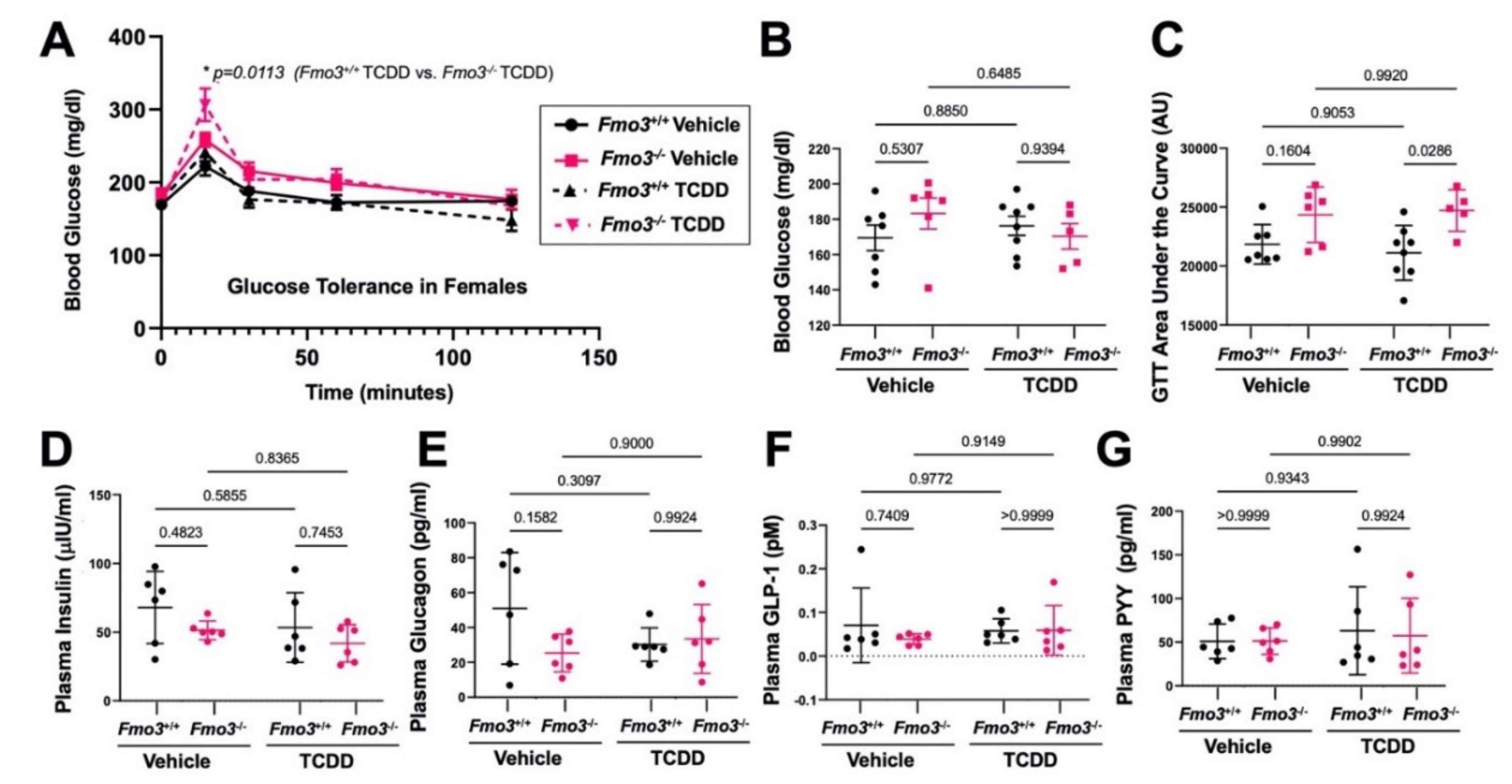
2.5. The Effects of TCDD Are Also Altered in Female Fmo3−/− Mice
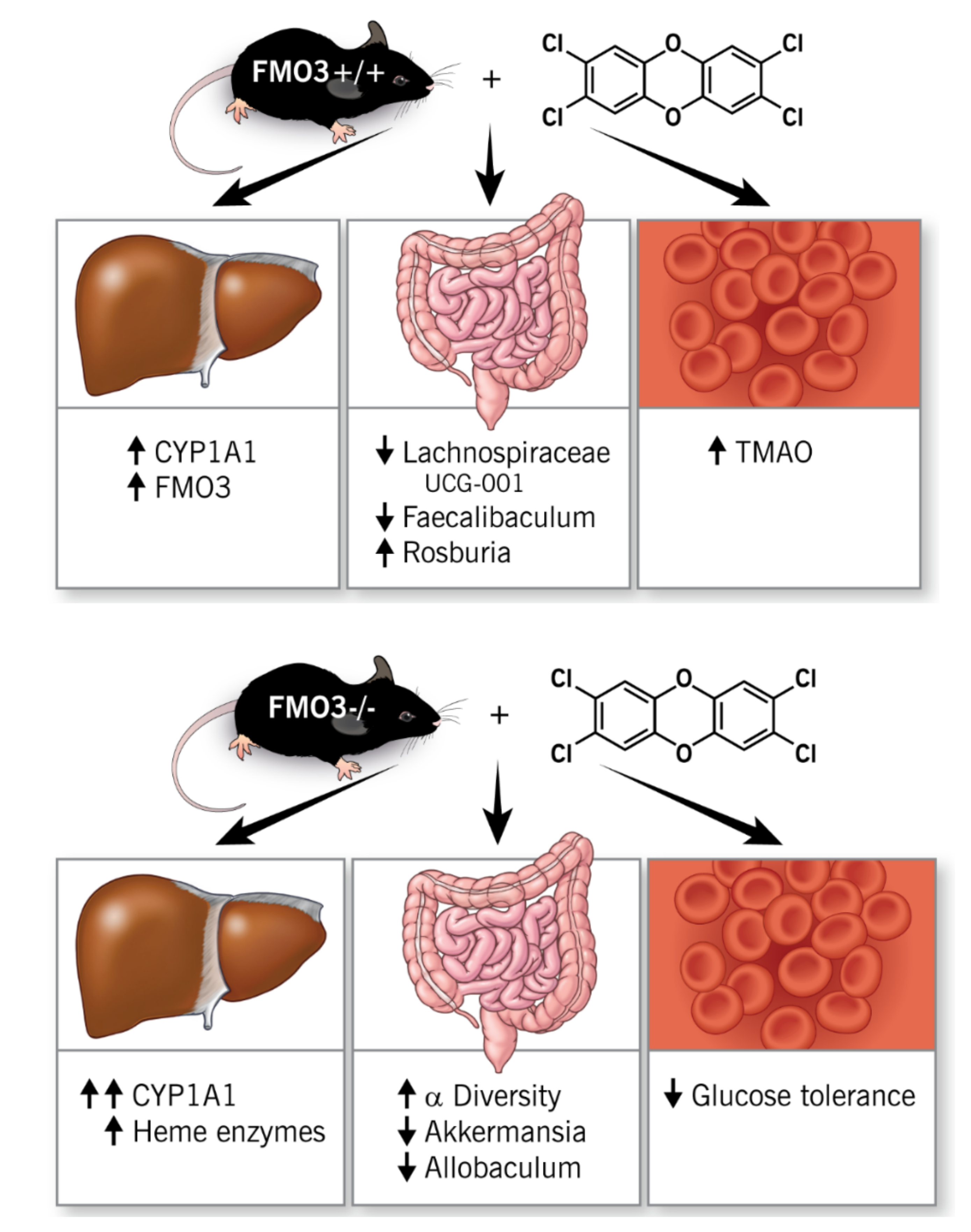
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Animals
4.3. RNA Isolation, RNA Sequencing, and Quantitative PCR (qPCR) Analyses
4.4. Quantification of Plasma Hormone Levels
4.5. Western Blotting
4.6. Quantification of Plasma TMAO Levels
4.7. Gut Microbiome Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kim, Y.A.; Park, J.B.; Woo, M.S.; Lee, S.Y.; Kim, H.Y.; Yoo, Y.H. Persistent Organic Pollutant-Mediated Insulin Resistance. Int. J. Environ. Res. Public Health 2019, 16, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cranmer, M.; Louie, S.; Kennedy, R.H.; Kern, P.A.; Fonseca, V.A. Exposure to 2,3,7,8-Tetrachlorodibenzo-P-Dioxin (Tcdd) Is Associated with Hyperinsulinemia and Insulin Resistance. Toxicol. Sci. 2000, 56, 431–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoyeck, M.P.; Blair, H.; Ibrahim, M.; Solanki, S.; Elsawy, M.; Prakash, A.; Rick, K.R.; Matteo, G.; O’Dwyer, S.; Bruin, J.E. Long-Term Metabolic Consequences of Acute Dioxin Exposure Differ between Male and Female Mice. Sci. Rep. 2020, 10, 1448. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.Y.; Wu, C.L.; Yang, Y.C.; Chang, J.W.; Kuo, Y.C.; Cheng, Y.Y.; Wu, J.S.; Lee, C.C.; Guo, H.R. Association between Dioxin and Diabetes Mellitus in an Endemic Area of Exposure in Taiwan: A Population-Based Study. Medicine 2015, 94, e1730. [Google Scholar] [CrossRef] [PubMed]
- Petriello, M.C.; Hoffman, J.B.; Sunkara, M.; Wahlang, B.; Perkins, J.T.; Morris, A.J.; Hennig, B. Dioxin-Like Pollutants Increase Hepatic Flavin Containing Monooxygenase (Fmo3) Expression to Promote Synthesis of the Pro-Atherogenic Nutrient Biomarker Trimethylamine N-Oxide from Dietary Precursors. J. Nutr. Biochem. 2016, 33, 145–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lind, P.M.; Van Bavel, B.; Salihovic, S.; Lind, L. Circulating Levels of Persistent Organic Pollutants (Pops) and Carotid Atherosclerosis in the Elderly. Environ. Health Perspect. 2012, 120, 38–43. [Google Scholar] [CrossRef]
- Ennour-Idrissi, K.; Ayotte, P.; Diorio, C. Persistent Organic Pollutants and Breast Cancer: A Systematic Review and Critical Appraisal of the Literature. Cancers 2019, 11, 1063. [Google Scholar] [CrossRef] [Green Version]
- Han, M.A.; Kim, J.H.; Song, H.S. Persistent Organic Pollutants, Pesticides, and the Risk of Thyroid Cancer: Systematic Review and Meta-Analysis. Eur. J. Cancer Prev. 2019, 28, 344–349. [Google Scholar] [CrossRef]
- Needham, L.L.; Gerthoux, P.M.; Patterson, D.G., Jr.; Brambilla, P.; Turner, W.E.; Beretta, C.; Pirkle, J.L.; Colombo, L.; Sampson, E.J.; Tramacere, P.L.; et al. Serum Dioxin Levels in Seveso, Italy, Population in 1976. Teratog. Carcinog. Mutagenesis 1997, 17, 225–240. [Google Scholar] [CrossRef]
- Park, E.Y.; Park, E.; Kim, J.; Oh, J.-K.; Kim, B.; Hong, Y.-C.; Lim, M.K. Impact of Environmental Exposure to Persistent Organic Pollutants on Lung Cancer Risk. Environ. Int. 2020, 143, 105925. [Google Scholar] [CrossRef]
- Kataria, A.; Trasande, L.; Trachtman, H. The Effects of Environmental Chemicals on Renal Function. Nat. Rev. Nephrol. 2015, 11, 610–625. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.-J.; Wu, P.-Y.; Huang, J.-C.; Chen, S.-C. Environmental Pollution and Chronic Kidney Disease. Int. J. Med. Sci. 2021, 18, 1121–1129. [Google Scholar] [CrossRef] [PubMed]
- Valcke, M.; Levasseur, M.-E.; Da Silva, A.S.; Wesseling, C. Pesticide exposures and chronic kidney disease of unknown etiology: An epidemiologic review. Environ. Health 2017, 16, 49. [Google Scholar] [CrossRef] [Green Version]
- Celius, T.; Roblin, S.; Harper, P.A.; Matthews, J.; Boutros, P.C.; Pohjanvirta, R.; Okey, A.B. Aryl Hydrocarbon Receptor-Dependent Induction of Flavin-Containing Monooxygenase Mrnas in Mouse Liver. Drug Metab. Dispos. 2008, 36, 2499–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrill, J.A.; Layko, D.; Nyska, A.; Hukkanen, R.R.; Manno, R.A.; Grassetti, A.; Lawson, M.; Martin, G.; Budinsky, R.A.; Rowlands, J.C.; et al. Aryl Hydrocarbon Receptor Knockout Rats Are Insensitive to the Pathological Effects of Repeated Oral Exposure to 2,3,7,8-Tetrachlorodibenzo-P-Dioxin. J. Appl. Toxicol. 2016, 36, 802–814. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, A.M.; Dang, C.H. Basic Review of the Cytochrome P450 System. J. Adv. Pract. Oncol. 2013, 4, 263–268. [Google Scholar]
- Michalex, J.; Robinson, J.; Fox, K.; Elequin, V.; Ketchum, N.; Jackson, W.; Pavuk, M.; Grubbs, W.; Cooper, B.; Johnson, P. Pharmacokinetics of Tcdd in Veterans of Operation Ranch Hand: 15-Year Follow-Up. J. Toxicol. Environ. Health Part A 1999, 57, 369–378. [Google Scholar]
- Lee, J.; Prokopec, S.D.; Watson, J.D.; Sun, R.X.; Pohjanvirta, R.; Boutros, P.C. Male and Female Mice Show Significant Differences in Hepatic Transcriptomic Response to 2,3,7,8-Tetrachlorodibenzo-P-Dioxin. BMC Genom. 2015, 16, 625. [Google Scholar] [CrossRef] [Green Version]
- Catucci, G.; Querio, G.; Sadeghi, S.J.; Gilardi, G.; Levi, R. Enzymatically Produced Trimethylamine N-Oxide: Conserving It or Eliminating It. Catalysts 2019, 9, 1028. [Google Scholar] [CrossRef] [Green Version]
- Miao, J.; Ling, A.V.; Manthena, P.V.; Gearing, M.E.; Graham, M.J.; Crooke, R.M.; Croce, K.J.; Esquejo, R.M.; Clish, C.B.; Vicent, D.; et al. Flavin-Containing Monooxygenase 3 as a Potential Player in Diabetes-Associated Atherosclerosis. Nat. Commun. 2015, 6, 6498. [Google Scholar] [CrossRef]
- Schugar, R.C.; Shih, D.M.; Warrier, M.; Helsley, R.N.; Burrows, A.; Ferguson, D.; Brown, A.L.; Gromovsky, A.D.; Heine, M.; Chatterjee, A.; et al. The Tmao-Producing Enzyme Flavin-Containing Monooxygenase 3 Regulates Obesity and the Beiging of White Adipose Tissue. Cell Rep. 2017, 19, 2451–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shih, D.M.; Wang, Z.; Lee, R.; Meng, Y.; Che, N.; Charugundla, S.; Qi, H.; Wu, J.; Pan, C.; Brown, J.M.; et al. Flavin Containing Monooxygenase 3 Exerts Broad Effects on Glucose and Lipid Metabolism and Atherosclerosis. J. Lipid Res. 2015, 56, 22–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warrier, M.; Shih, D.M.; Burrows, A.C.; Ferguson, D.; Gromovsky, A.D.; Brown, A.L.; Marshall, S.; McDaniel, A.; Schugar, R.C.; Wang, Z.; et al. The Tmao-Generating Enzyme Flavin Monooxygenase 3 Is a Central Regulator of Cholesterol Balance. Cell Rep. 2015, 10, 326–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petriello, M.C.; Charnigo, R.; Sunkara, M.; Soman, S.; Pavuk, M.; Birnbaum, L.; Morris, A.J.; Hennig, B. Relationship between Serum Trimethylamine N-Oxide and Exposure to Dioxin-Like Pollutants. Environ. Res. 2018, 162, 211–218. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal Microbiota Metabolism of L-Carnitine, a Nutrient in Red Meat, Promotes Atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Henderson, A.; Petriello, M.C.; Romano, K.A.; Gearing, M.; Miao, J.; Schell, M.; Sandoval-Espinola, W.J.; Tao, J.; Sha, B.; et al. Trimethylamine N-Oxide Binds and Activates Perk to Promote Metabolic Dysfunction. Cell Metab. 2019, 30, 1141–1151.e5. [Google Scholar] [CrossRef]
- Gupta, N.; Buffa, J.A.; Roberts, A.B.; Sangwan, N.; Skye, S.M.; Li, L.; Ho, K.J.; Varga, J.; DiDonato, J.A.; Tang, W.W.; et al. Targeted Inhibition of Gut Microbial Trimethylamine N-Oxide Production Reduces Renal Tubulointerstitial Fibrosis and Functional Impairment in a Murine Model of Chronic Kidney Disease. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 1239–1255. [Google Scholar] [CrossRef]
- Johnson, C.; Prokopienko, A.J.; West, R.E.; Nolin, T.D.; Stubbs, J.R. Decreased Kidney Function Is Associated with Enhanced Hepatic Flavin Monooxygenase Activity and Increased Circulating Trimethylamine N-Oxide Concentrations in Mice. Drug Metab. Dispos. 2018, 46, 1304. [Google Scholar]
- Pelletier, C.C.; Croyal, M.; Ene, L.; Aguesse, A.; Billon-Crossouard, S.; Krempf, M.; Lemoine, S.; Guebre-Egziabher, F.; Juillard, L.; Soulage, C.O. Elevation of Trimethylamine-N-Oxide in Chronic Kidney Disease: Contribution of Decreased Glomerular Filtration Rate. Toxins 2019, 11, 635. [Google Scholar] [CrossRef] [Green Version]
- Robinson-Cohen, C.; Newitt, R.; Shen, D.D.; Rettie, A.E.; Kestenbaum, B.R.; Himmelfarb, J.; Yeung, C.K. Association of Fmo3 Variants and Trimethylamine N-Oxide Concentration, Disease Progression, and Mortality in Ckd Patients. PLoS ONE 2016, 11, e0161074. [Google Scholar] [CrossRef]
- Zhang, W.; Miikeda, A.; Zuckerman, J.; Jia, X.; Charugundla, S.; Zhou, Z.; Kaczor-Urbanowicz, K.E.; Magyar, C.; Guo, F.; Wang, Z.; et al. Inhibition of Microbiota-Dependent Tmao Production Attenuates Chronic Kidney Disease in Mice. Sci. Rep. 2021, 11, 518. [Google Scholar] [CrossRef]
- Chen, L.; Chen, Y.; Zhao, M.; Zheng, L.; Fan, D. Changes in the Concentrations of Trimethylamine N-Oxide (Tmao) and Its Precursors in Patients with Amyotrophic Lateral Sclerosis. Sci. Rep. 2020, 10, 15198. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, S.; Ogliari, P.; Davin, A.; Corato, M.; Cova, E.; Abel, K.; Cashman, J.R.; Ceroni, M.; Cereda, C. Flavin-Containing Monooxygenase Mrna Levels Are up-Regulated in Als Brain Areas in Sod1-Mutant Mice. Neurotox. Res. 2011, 20, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Govindarajulu, M.; Pinky, P.D.; Steinke, I.; Bloemer, J.; Ramesh, S.; Kariharan, T.; Rella, R.T.; Bhattacharya, S.; Dhanasekaran, M.; Suppiramaniam, V.; et al. Gut Metabolite Tmao Induces Synaptic Plasticity Deficits by Promoting Endoplasmic Reticulum Stress. Front. Mol. Neurosci. 2020, 13, 138. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Ke, Y.; Zhan, R.; Liu, C.; Zhao, M.; Zeng, A.; Shi, X.; Ji, L.; Cheng, S.; Pan, B.; et al. Trimethylamine-N-Oxide Promotes Brain Aging and Cognitive Impairment in Mice. Aging Cell 2018, 17, e12768. [Google Scholar] [CrossRef]
- Vogt, N.M.; Romano, K.A.; Darst, B.F.; Engelman, C.D.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Blennow, K.; Zetterberg, H.; Bendlin, B.B.; et al. The Gut Microbiota-Derived Metabolite Trimethylamine N-Oxide Is Elevated in Alzheimer’s Disease. Alzheimer’s Res. Ther. 2018, 10, 124. [Google Scholar] [CrossRef] [Green Version]
- Bennett, B.J.; de Aguiar Vallim, T.Q.; Wang, Z.; Shih, D.M.; Meng, Y.; Gregory, J.; Allayee, H.; Lee, R.; Graham, M.; Crooke, R.; et al. Trimethylamine-N-Oxide, a Metabolite Associated with Atherosclerosis, Exhibits Complex Genetic and Dietary Regulation. Cell Metab. 2013, 17, 49–60. [Google Scholar] [CrossRef] [Green Version]
- Schugar, R.C.; Gliniak, C.M.; Osborn, L.J.; Massey, W.; Sangwan, N.; Horak, A.; Banerjee, R.; Orabi, D.; Helsley, R.N.; Brown, A.L.; et al. Gut Microbe-Targeted Choline Trimethylamine Lyase Inhibition Improves Obesity Via Rewiring of Host Circadian Rhythms. eLife 2022, 11, e63998. [Google Scholar] [CrossRef]
- Sundberg, C.D.; Hankinson, O. A Crispr/Cas9 Whole-Genome Screen Identifies Genes Required for Aryl Hydrocarbon Receptor-Dependent Induction of Functional Cyp1a1. Toxicol. Sci. 2019, 170, 310–319. [Google Scholar] [CrossRef]
- Biljes, D.; Hammerschmidt-Kamper, C.; Kadow, S.; Diel, P.; Weigt, C.; Burkart, V.; Esser, C. Impaired Glucose and Lipid Metabolism in Ageing Aryl Hydrocarbon Receptor Deficient Mice. EXCLI J. 2015, 14, 1153–1163. [Google Scholar]
- Fernandez-Salguero, P.M.; Hilbert, D.M.; Rudikoff, S.; Ward, J.M.; Gonzalez, F.J. Aryl-Hydrocarbon Receptor-Deficient Mice Are Resistant to 2,3,7,8-Tetrachlorodibenzo-P-Dioxin-Induced Toxicity. Toxicol. Appl. Pharmacol. 1996, 140, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Natividad, J.M.; Agus, A.; Planchais, J.; Lamas, B.; Jarry, A.C.; Martin, R.; Michel, M.-L.; Chong-Nguyen, C.; Roussel, R.; Straube, M.; et al. Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab. 2018, 28, 737–749.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierre, S.; Chevallier, A.; Teixeira-Clerc, F.; Ambolet-Camoit, A.; Bui, L.C.; Bats, A.S.; Fournet, J.C.; Fernandez-Salguero, P.; Aggerbeck, M.; Lotersztajn, S.; et al. Aryl Hydrocarbon Receptor-Dependent Induction of Liver Fibrosis by Dioxin. Toxicol. Sci. 2014, 137, 114–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, D.; Dunaief, J.L. Retinal Iron Homeostasis in Health and Disease. Front. Aging Neurosci. 2013, 5, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esmith, A.; McCulloh, R.J. Hemopexin and Haptoglobin: Allies against Heme Toxicity from Hemoglobin Not Contenders. Front. Physiol. 2015, 6, 187. [Google Scholar]
- Vinchi, F.; Gastaldi, S.; Silengo, L.; Altruda, F.; Tolosano, E. Hemopexin Prevents Endothelial Damage and Liver Congestion in a Mouse Model of Heme Overload. Am. J. Pathol. 2008, 173, 289–299. [Google Scholar] [CrossRef] [Green Version]
- Scheja, L.; Heese, B.; Zitzer, H.; Michael, M.; Siesky, A.M.; Pospisil, H.; Beisiegel, U.; Seedorf, K. Acute-Phase Serum Amyloid a as a Marker of Insulin Resistance in Mice. Exp. Diabetes Res. 2008, 2008, 230837. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Yang, Y.; Qin, Z.; Cai, J.; Guo, X.; Tang, Y.; Wan, J.; Su, D.-F.; Liu, X. The Acute-Phase Protein Orosomucoid Regulates Food Intake and Energy Homeostasis Via Leptin Receptor Signaling Pathway. Diabetes 2016, 65, 1630. [Google Scholar] [CrossRef] [Green Version]
- Komori, H.; Nishi, K.; Uehara, N.; Watanabe, H.; Shuto, T.; Suenaga, A.; Maruyama, T.; Otagiri, M. Characterization of Hepatic Cellular Uptake of A1-Acid Glycoprotein (Agp), Part 2: Involvement of Hemoglobin Β-Chain on Plasma Membranes in the Uptake of Human Agp by Liver Parenchymal Cells. J. Pharm. Sci. 2012, 101, 1607–1615. [Google Scholar] [CrossRef]
- Chen, G.I.; Tisayakorn, S.; Jorgensen, C.; D’Ambrosio, L.M.; Goudreault, M.; Gingras, A.C. Pp4r4/Kiaa1622 Forms a Novel Stable Cytosolic Complex with Phosphoprotein Phosphatase 4. J. Biol. Chem. 2008, 283, 29273–29284. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.C.-T.; Tang-Oxley, Q.; Qiu, W.R.; Wang, Y.-P.; Mihindukulasuriya, K.A.; Afshar, R.; Tan, T.-H. Protein Phosphatase X Interacts with C-Rel and Stimulates C-Rel/Nuclear Factor Κb Activity. J. Biol. Chem. 1998, 273, 33561–33565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Dong, W.; Zou, T.; Ouyang, L.; He, G.; Liu, Y.; Qi, Y. Protein Phosphatase 4 Negatively Regulates Lps Cascade by Inhibiting Ubiquitination of Traf6. FEBS Lett. 2008, 582, 2843–2849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Huang, X.; Jiao, J.; Zhang, H.; Liu, J.; Qin, W.; Meng, X.; Shen, T.; Lin, Y.; Chu, J.; et al. Protein Phosphatase 4 (Pp4) Functions as a Critical Regulator in Tumor Necrosis Factor (Tnf)-A-Induced Hepatic Insulin Resistance. Sci. Rep. 2015, 5, 18093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Gui, W.; Rimal, B.; Koo, I.; Smith, P.B.; Nichols, R.G.; Cai, J.; Liu, Q.; Patterson, A.D. Metabolic Impact of Persistent Organic Pollutants on Gut Microbiota. Gut Microbes 2020, 12, 1848209. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Zhu, J.; Le Roy, C.I.; Mompeo, O.; Young, K.; Rebholz, C.M.; Selvin, E.; North, K.E.; Mohney, R.P.; Bell, J.T.; et al. Serum Metabolites Reflecting Gut Microbiome Alpha Diversity Predict Type 2 Diabetes. Gut Microbes 2020, 11, 1632–1642. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, C.C.; Monteil, M.A.; Davis, E.M. Overweight and Obesity in Children Are Associated with an Abundance of Firmicutes and Reduction of Bifidobacterium in Their Gastrointestinal Microbiota. Child. Obes. 2020, 16, 204–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig, H.; Hausmann, B.; Schreder, M.; Pönisch, W.; Zojer, N.; Knop, S.; Gunsilius, E.; Egle, A.; Petzer, A.; Einsele, H.; et al. Reduced Alpha Diversity of the Oral Microbiome Correlates with Short Progression-Free Survival in Patients with Relapsed/Refractory Multiple Myeloma Treated with Ixazomib-Based Therapy (Agmt Mm 1, Phase Ii Trial). eJHaem 2021, 2, 102–106. [Google Scholar] [CrossRef]
- Li, C.; You, Z.; Lin, Y.; Liu, H.; Su, J. Skin Microbiome Differences Relate to the Grade of Acne Vulgaris. J. Dermatol. 2019, 46, 787–790. [Google Scholar] [CrossRef]
- Vogtmann, E.; Flores, R.; Yu, G.; Freedman, N.D.; Shi, J.; Gail, M.H.; Dye, B.A.; Wang, G.-Q.; Klepac-Ceraj, V.; Paster, B.J.; et al. Association between Tobacco Use and the Upper Gastrointestinal Microbiome among Chinese Men. Cancer Causes Control 2015, 26, 581–588. [Google Scholar] [CrossRef] [Green Version]
- Greer, R.L.; Dong, X.; de Moraes, A.; Zielke, R.A.; Fernandes, G.R.; Peremyslova, E.; Vasquez-Perez, S.; Schoenborn, A.A.; Gomes, E.P.; Pereira, A.C.; et al. Akkermansia Muciniphila Mediates Negative Effects of Ifnγ on Glucose Metabolism. Nat. Commun. 2016, 7, 13329. [Google Scholar] [CrossRef]
- Van Hul, M.; Geurts, L.; Plovier, H.; Druart, C.; Everard, A.; Ståhlman, M.; Rhimi, M.; Chira, K.; Teissedre, P.-L.; Delzenne, N.M.; et al. Reduced Obesity, Diabetes, and Steatosis Upon Cinnamon and Grape Pomace Are Associated with Changes in Gut Microbiota and Markers of Gut Barrier. Am. J. Physiol. Endocrinol. Metab. 2018, 314, E334–E352. [Google Scholar] [CrossRef] [PubMed]
- Dopkins, N.; Neameh, W.H.; Hall, A.; Lai, Y.; Rutkovsky, A.; Gandy, A.O.; Lu, K.; Nagarkatti, P.S.; Nagarkatti, M. Effects of Acute 2,3,7,8-Tetrachlorodibenzo-P-Dioxin Exposure on the Circulating and Cecal Metabolome Profile. Int. J. Mol. Sci. 2021, 22, 11801. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, Y.; Dong, Z.; Xu, M.; Gao, M.; Cong, M.; Liu, S. Tcdd Promotes Liver Fibrosis through Disordering Systemic and Hepatic Iron Homeostasis. J. Hazard. Mater. 2020, 395, 122588. [Google Scholar] [CrossRef]
- Catucci, G.; Aramini, D.; Sadeghi, S.J.; Gilardi, G. Ligand Stabilization and Effect on Unfolding by Polymorphism in Human Flavin-Containing Monooxygenase 3. Int. J. Biol. Macromol. 2020, 162, 1484–1493. [Google Scholar] [CrossRef] [PubMed]
- Catucci, G.; Bortolussi, S.; Rampolla, G.; Cusumano, D.; Gilardi, G.; Sadeghi, S.J. Flavin-Containing Monooxygenase 3 Polymorphic Variants Significantly Affect Clearance of Tamoxifen and Clomiphene. Basic Clin. Pharmacol. Toxicol. 2018, 123, 687–691. [Google Scholar] [CrossRef] [PubMed]
- Catucci, G.; Occhipinti, A.; Maffei, M.; Gilardi, G.; Sadeghi, S.J. Effect of Human Flavin-Containing Monooxygenase 3 Polymorphism on the Metabolism of Aurora Kinase Inhibitors. Int. J. Mol. Sci. 2013, 14, 2707–2716. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Catucci, G.; Castrignanò, S.; Gilardi, G.; Sadeghi, S.J. Inactivation Mechanism of N61s Mutant of Human Fmo3 Towards Trimethylamine. Sci. Rep. 2017, 7, 14668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, C.; Catucci, G.; di Nardo, G.; Gilardi, G.; Sadeghi, S.J. Human Flavin-Containing Monooxygenase 3: Structural Mapping of Gene Polymorphisms and Insights into Molecular Basis of Drug Binding. Gene 2016, 593, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Helsley, R.N.; Miyata, T.; Kadam, A.; Varadharajan, V.; Sangwan, N.; Huang, E.C.; Banerjee, R.; Brown, A.L.; Fung, K.K.; Massey, W.J.; et al. Gut Microbial Trimethylamine Is Elevated in Alcohol-Associated Hepatitis and Contributes to Ethanol-Induced Liver Injury in Mice. eLife 2022, 11, e76554. [Google Scholar] [CrossRef]
- Pathak, P.; Helsley, R.N.; Brown, A.L.; Buffa, J.A.; Choucair, I.; Nemet, I.; Gogonea, C.B.; Gogonea, V.; Wang, Z.; Garcia-Garcia, J.C.; et al. Small Molecule Inhibition of Gut Microbial Choline Trimethylamine Lyase Activity Alters Host Cholesterol and Bile Acid Metabolism. Am. J. Physiol. Heart Circ. Physiol. 2020, 318, H1474–H1486. [Google Scholar] [CrossRef]
- Roberts, A.B.; Gu, X.; Buffa, J.A.; Hurd, A.G.; Wang, Z.; Zhu, W.; Gupta, N.; Skye, S.M.; Cody, D.B.; Levison, B.S.; et al. Development of a Gut Microbe-Targeted Nonlethal Therapeutic to Inhibit Thrombosis Potential. Nat. Med. 2018, 24, 1407–1417. [Google Scholar] [CrossRef]
- Wang, Z.; Roberts, A.B.; Buffa, J.A.; Levison, B.S.; Zhu, W.; Org, E.; Gu, X.; Huang, Y.; Zamanian-Daryoush, M.; Culley, M.K.; et al. Non-Lethal Inhibition of Gut Microbial Trimethylamine Production for the Treatment of Atherosclerosis. Cell 2015, 163, 1585–1595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Xu, C.-X.; Krager, S.L.; Bottum, K.M.; Liao, D.-F.; Tischkau, S.A. Aryl Hydrocarbon Receptor Deficiency Enhances Insulin Sensitivity and Reduces Ppar-A Pathway Activity in Mice. Environ. Health Perspect. 2011, 119, 1739–1744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, X.; Liu, Y.; Long, J.; Chen, S.; Liao, G.; Wu, S.; Li, C.; Wang, L.; Ling, W.; Zhu, H. Trimethylamine N-Oxide Aggravates Liver Steatosis through Modulation of Bile Acid Metabolism and Inhibition of Farnesoid X Receptor Signaling in Nonalcoholic Fatty Liver Disease. Mol. Nutr. Food Res. 2019, 63, 1900257. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Star: Ultrafast Universal Rna-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript Assembly and Quantification by Rna-Seq Reveals Unannotated Transcripts and Isoform Switching During Cell Differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.M.; Boysen, M.S.; Chung, S.; Fabiyi, O.; Morrison, R.F.; Mandrup, S.; McIntosh, M.K. Conjugated Linoleic Acid Induces Human Adipocyte Delipidation: Autocrine/Paracrine Regulation of Mek/Erk Signaling by Adipocytokines. J. Biol. Chem. 2004, 279, 26735–26747. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut Flora Metabolism of Phosphatidylcholine Promotes Cardiovascular Disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Levison, B.S.; Hazen, J.E.; Donahue, L.; Li, X.M.; Hazen, S.L. Measurement of Trimethylamine-N-Oxide by Stable Isotope Dilution Liquid Chromatography Tandem Mass Spectrometry. Anal. Biochem. 2014, 455, 35–40. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. Qiime Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. Dada2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMurdie, P.J.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer Publishing Company: New York, NY, USA, 2009. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Hollander, M.; Wolfe, D.A. Nonparametric Statistical Methods, 2nd ed.; Wiley-Interscience: Hoboken, NJ, USA, 1999; Volume 19, pp. 1386–1388. ISBN 0-471-19045-4. [Google Scholar]
- Witkowski, M.; Witkowski, M.; Friebel, J.; Buffa, J.A.; Li, X.S.; Wang, Z.; Sangwan, N.; Li, L.; DiDonato, J.A.; Tizian, C.; et al. Endothelial Tissue Factor Contributes to Trimethylamine N-Oxide-Enhanced Arterial Thrombosis. Cardiovasc. Res. 2021, cvab263. [Google Scholar] [CrossRef]



| Primer | Sequence (5′–3′) |
|---|---|
| Fmo3 F | CCCACATGCTTTGAGAGGAG |
| Fmo3 R | GGAAGAGTTGGTGAAGACCG |
| Cyp1a1 F | CTGAAGGTGGTAGTTCTTGGAG |
| Cyp1a1 R | CCATACATGGAAGGCATGATCTA |
| CycloA F | GCGGCAGGTCCATCTACG |
| CycloA R | GCCATCCAGCCATTCAGTC |
| Alas1 F | CACTGTCCGAGTCACATCATC |
| Alas1 R | TGATGGCCTGGACGTAGATA |
| Col1a1 F | ATGTTCAGCTTTGTGGACCTC |
| Col1a1 R | CAGAAAGCACAGCACTCGC |
| IL-1b F | AGTTGACGGACCCCAAAAG |
| IL-1b R | AGCTGGATGCTCTCATCAGG |
| TNFa F | CCACCACGCTCTTCTGTCTAC |
| TNFa R | AGGGTCTGGGCCATAGAACT |
| IL-6 F | GCTACCAAACTGGATATAATCAGGA |
| IL-6 R | CCAGGTAGCTATGGTACTCCAGAA |
| Antibody | Vendor | Product # |
|---|---|---|
| FMO3 | Abcam | ab126790 |
| HPX | R&D Systems | AF7007 |
| HP | Abcam | ab256454 |
| β-Actin HRP | Cell Signaling | 4970S |
| Rabbit HRP | Cell Signaling | 7074S |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massey, W.; Osborn, L.J.; Banerjee, R.; Horak, A.; Fung, K.K.; Orabi, D.; Chan, E.R.; Sangwan, N.; Wang, Z.; Brown, J.M. Flavin-Containing Monooxygenase 3 (FMO3) Is Critical for Dioxin-Induced Reorganization of the Gut Microbiome and Host Insulin Sensitivity. Metabolites 2022, 12, 364. https://doi.org/10.3390/metabo12040364
Massey W, Osborn LJ, Banerjee R, Horak A, Fung KK, Orabi D, Chan ER, Sangwan N, Wang Z, Brown JM. Flavin-Containing Monooxygenase 3 (FMO3) Is Critical for Dioxin-Induced Reorganization of the Gut Microbiome and Host Insulin Sensitivity. Metabolites. 2022; 12(4):364. https://doi.org/10.3390/metabo12040364
Chicago/Turabian StyleMassey, William, Lucas J. Osborn, Rakhee Banerjee, Anthony Horak, Kevin K. Fung, Danny Orabi, E. Ricky Chan, Naseer Sangwan, Zeneng Wang, and J. Mark Brown. 2022. "Flavin-Containing Monooxygenase 3 (FMO3) Is Critical for Dioxin-Induced Reorganization of the Gut Microbiome and Host Insulin Sensitivity" Metabolites 12, no. 4: 364. https://doi.org/10.3390/metabo12040364
APA StyleMassey, W., Osborn, L. J., Banerjee, R., Horak, A., Fung, K. K., Orabi, D., Chan, E. R., Sangwan, N., Wang, Z., & Brown, J. M. (2022). Flavin-Containing Monooxygenase 3 (FMO3) Is Critical for Dioxin-Induced Reorganization of the Gut Microbiome and Host Insulin Sensitivity. Metabolites, 12(4), 364. https://doi.org/10.3390/metabo12040364







