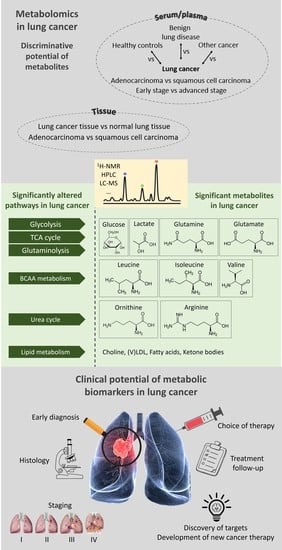
Changes in Metabolism as a Diagnostic Tool for Lung Cancer: Systematic Review
Abstract
:1. Introduction
2. Methods
3. Results
3.1. Metabolic Differentiation between Lung Cancer Patients and Healthy Controls
3.2. Metabolic Differentiation between Lung Cancer Patients and Other Cancer Patients
3.3. Metabolic Differentiation between Lung Cancer and Benign Lung Disease
3.4. Metabolic Differentiation between Early-Stage and Advanced-Stage Lung Cancer
3.5. Metabolic Differentiation between Lung Cancer Tissue and Normal Lung Tissue
3.6. Metabolic Differentiation between Different Histologies of Lung Cancer
| Involved Pathway | Metabolite | Plasma/Serum | Tissue | |||||
|---|---|---|---|---|---|---|---|---|
| Healthy | BC | BPD | Early LC | AC | NLT | AC | ||
| LC | LC | LC | Advanced LC | SCC | LCT | SCC | ||
| Glycolysis | Glucose | ↓ [38,39] ↑ [41,42] | ↑ [41] | ↓ [39] | ↓ [39] | ↓ [71,72] | ||
Lactate | ↑ [38,39,40] ↓ [41,42] | ↓ [41] | ↑ [67] ↓ [39] | ↑ [39] ↓ [40,67] | ↓ [40] | ↑ [71,72] | ↑ [71] | |
Pyruvate | ↑ [71,72] | ↑ [71] | ||||||
| Glutaminolysis | Glutamine | ↑ [38,41] ↓ [40] | ↓ [67] | ↓ [39,40] | ↑ [40] | ↑ [40] | ||
Glutamate | ↑ [38,39] ↓ [40] | ↓ [6] | ↑ [39] | ↑ [40] | ↑ [40] | |||
| BCAA metabolism | Leucine | ↑ [38,39,41,42] | ↑ [67] | ↑ [39] | ||||
Isoleucine | ↑ [38,39,41,42] | ↑ [67] | ↑ [39] ↓ [67] | |||||
Valine | ↑ [41] | ↑ [67] | ↓ [67] | ↑ [40] | ||||
| TCA cycle | Citrate | ↓ [44] | ↓ [67] | |||||
Acetate | ↓ [67] | |||||||
Fumarate | ↑ [44,45] | ↑ [71] | ||||||
| Metabolism involving other amino acids | Tyrosine | ↑ [38,41,52] | ↓ [6] | |||||
Histidine | ↑ [38] | |||||||
| Urea cycle | Ornithine | ↓ [38,50,51,52] | ↓ [71,72] | |||||
Arginine | ↓ [38,52,53] | ↓ [71,72] | ||||||
Creatinine | ↑ [67] | ↓ [67] | ↑ [71,72] | |||||
| Lipid metabolism | Choline | ↓ [38,39] | ↑ [39,67] | ↑ [40] | ||||
| (V)LDL | ↓ [38] | ↑ [39] | ↑ [40] | |||||
| Fatty acids | ↓ [38] | ↑ [41] | ↑ [71,72] | ↑ [40] | ||||
Glycerol | ↑ [39] | ↑ [67] | ↑ [67] | ↑ [71,72] | ||||
| Ketone bodies β-hydroxybutyrate  Acetoacetate  | ↑ [38,39,44] | ↑ [67] | ↓ [67] | |||||
| Reference | Sample Type | Study Population | Measurement Technique | Statistical Analysis | Discriminative Capacity |
|---|---|---|---|---|---|
| Zhang et al., 2016 [38] | Serum |
| 1H-NMR RRLC | OPLS-DA | LC vs. healthy: 100% sens, 100% spec |
| Puchades-Carrasco et al., 2016 [39] | Serum |
| 1H-NMR | OPLS-DA | LC vs. healthy based on all metabolites: 92% sens, 95% spec, R² 0.931, Q² 0.873 LC vs. BDP vs. healthy based on 5 metabolites: 77% sens, 77.5% spec E-LC vs. A-LC: R² 0.779, Q² 0.592 |
| Berker et al., 2019 [40] | Serum |
| HRMAS-MRS | LDA CCA | ROC_AUC LC: 0.989 |
| Tissue |
| HRMAS-MRS | LDA CCA | None reported | |
| Louis et al., 2016 [41] | Plasma |
| 1H-NMR | OPLS-DA | Training LC vs. healthy: correct classification of 78% of LC, 92% of controls Validation LC vs. healthy: 71% sens, 81% spec AC vs. SCC: correct classification of 81% of AC, 38% of SCC |
| Derveaux et al., 2021 [42] | Plasma |
| 1H-NMR | OPLS-DA | Training LC vs. healthy: 85% sens, 93% spec Validation LC vs. healthy: 74% sens, 74% spec |
| Maeda et al., 2010 [52] | Plasma |
| LC-MS | Logistic regression | ROC_AUC LC: 0.817 ROC_AUC stage I: 0.796 ROC_AUC AC: 0.795 ROC_AUC SCC: 0.860 |
| Chen et al., 2015 [45] | Serum |
| LC-MS GC-MS | PLS-DA | LC-MS:
|
| Deja et al., 2014 [67] | Serum |
| 1H-NMR | OPLS-DA | COPD vs. LC: R²X 0.682, R²Y 0.762, Q² 0.568, AUC 0.993 COPD vs. E-LC: R²X 0.694, R²Y 0.809, Q² 0.651, AUC: 1 COPD vs. A-LC: R²X 0.663, R²Y 0.909, Q² 0.595, AUC; 1 E-LC vs. A-LC: R²X 0.732, R²Y 0.908, Q² 0.298, AUC: 0.904 |
| Vanhove et al., 2018 [6] | Plasma |
| 1H-NMR | PLS-DA | LC vs. inflammation:
|
| Moreno et al., 2018 [71] | Tissue |
| LC-MS GC-MS | PLS-DA | None reported |
| Zhang et al., 2020 [44] | Plasma |
| LC-MS HPLC-MS/MS | PLS-DA Logistic regression | Stage I/II vs. healthy: 0.919 sens, 0.900 spec, AUC 0.959 |
| Kowalczyk et al., 2021 [72] | Plasma |
| LC-MS: UHPLC combined with QTOF | PLS-DA | None reported |
| Tissue |
| LC-MS: UHPLC combined with QTOF | PLS-DA | RPLC: AC vs. SCC vs. control: R² 0.983, Q² 0.853 HILIC: AC vs. SCC vs. control: R² 0.858, Q² 0.732 | |
| Qi et al., 2021 [50] | Plasma |
| LC-MS | Logistic regressionOPLS-DA | LC vs. healthy all stages
|
4. Discussion
5. Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Lung Cancer Survival Rates. 2021. Available online: https://www.cancer.org/cancer/lung-cancer/detection-diagnosis-staging/survival-rates.html (accessed on 29 January 2021).
- Lu, T.; Yang, X.; Huang, Y.; Zhao, M.; Li, M.; Ma, K.; Yin, J.; Zhan, C.; Wang, Q. Trends in the incidence, treatment, and survival of patients with lung cancer in the last four decades. Cancer Manag. Res. 2019, 11, 943–953. [Google Scholar] [CrossRef] [Green Version]
- Bade, B.C.; Dela Cruz, C.S. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin. Chest. Med. 2020, 41, 1–24. [Google Scholar] [CrossRef]
- Brown, J.C.; Winters-Stone, K.; Lee, A.; Schmitz, K.H. Cancer, physical activity, and exercise. Compr. Physiol. 2012, 2, 2775–2809. [Google Scholar]
- Vanhove, K.; Giesen, P.; Owokotomo, O.E.; Mesotten, L.; Louis, E.; Shkedy, Z.; Thomeer, M.; Adriaensens, P. The plasma glutamate concentration as a complementary tool to differentiate benign PET-positive lung lesions from lung cancer. BMC Cancer 2018, 18, 868. [Google Scholar] [CrossRef]
- Tanoue, L.T.; Tanner, N.T.; Gould, M.K.; Silvestri, G.A. Lung cancer screening. Am. J. Respir. Crit. Care Med. 2015, 191, 19–33. [Google Scholar] [CrossRef] [Green Version]
- National Lung Screening Trial Research Team; Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef] [Green Version]
- Swensen, S.J.; Jett, J.R.; Hartman, T.E.; Midthun, D.E.; Sloan, J.A.; Sykes, A.M.; Aughenbaugh, G.L.; Clemens, M.A. Lung cancer screening with CT: Mayo Clinic experience. Radiology 2003, 226, 756–761. [Google Scholar] [CrossRef]
- Love, C.; Tomas, M.B.; Tronco, G.G.; Palestro, C.J. FDG PET of infection and inflammation. Radiographics 2005, 25, 1357–1368. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Montalva, A.; Barios, M.; Salvador, F.; Villar, A.; Tortola, T.; Molina-Morant, D.; Lorenzo-Bosquet, C.; Espinosa-Pereiro, J.; Molina, I. Usefulness of FDG PET/CT in the management of tuberculosis. PLoS ONE 2019, 14, e0221516. [Google Scholar] [CrossRef]
- Rosenbaum, S.J.; Lind, T.; Antoch, G.; Bockisch, A. False-positive FDG PET uptake—The role of PET/CT. Eur. Radiol. 2006, 16, 1054–1065. [Google Scholar] [CrossRef]
- Granville, C.A.; Dennis, P.A. An overview of lung cancer genomics and proteomics. Am. J. Respir. Cell Mol. Biol. 2005, 32, 169–176. [Google Scholar] [CrossRef]
- Najafi, Z.; Mohamadnia, A.; Ahmadi, R.; Mahmoudi, M.; Bahrami, N.; Khosravi, A.; Jamaati, H.; Tabarsi, P.; Kazem pour Dizaji, M.; Shirian, S. Proteomic and genomic biomarkers for Non-Small Cell Lung Cancer: Peroxiredoxin, Haptoglobin, and Alpha-1 antitrypsin. Cancer Med. 2020, 9, 3974–3982. [Google Scholar] [CrossRef]
- Wishart, D.S. Is Cancer a Genetic Disease or a Metabolic Disease? EBioMedicine 2015, 2, 478–479. [Google Scholar] [CrossRef] [Green Version]
- Holmes, E.; Wilson, I.D.; Nicholson, J.K. Metabolic phenotyping in health and disease. Cell 2008, 134, 714–717. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Senga, S.S.; Grose, R.P. Hallmarks of cancer—The new testament. Open Biol. 2021, 11, 200358. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Miyamoto, S.; Taylor, S.L.; Barupal, D.K.; Taguchi, A.; Wohlgemuth, G.; Wikoff, W.R.; Yoneda, K.Y.; Gandara, D.R.; Hanash, S.M.; Kim, K.; et al. Systemic Metabolomic Changes in Blood Samples of Lung Cancer Patients Identified by Gas Chromatography Time-of-Flight Mass Spectrometry. Metabolites 2015, 5, 192–210. [Google Scholar] [CrossRef] [Green Version]
- Fouad, Y.A.; Aanei, C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017, 7, 1016–1036. [Google Scholar] [PubMed]
- Elia, I.; Haigis, M.C. Metabolites and the tumour microenvironment: From cellular mechanisms to systemic metabolism. Nat. Metab. 2021, 3, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Venneti, S.; Nagrath, D. Glutaminolysis: A Hallmark of Cancer Metabolism. Annu. Rev. Biomed. Eng. 2017, 19, 163–194. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Dang, C.V. Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res. 2006, 66, 8927–8930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanhove, K.; Graulus, G.-J.; Mesotten, L.; Thomeer, M.; Derveaux, E.; Noben, J.-P.; Guedens, W.; Adriaensens, P. The Metabolic Landscape of Lung Cancer: New Insights in a Disturbed Glucose Metabolism. Front. Oncol. 2019, 9, 1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, K.; Hoffmann, P.; Voelkl, S.; Meidenbauer, N.; Ammer, J.; Edinger, M.; Gottfried, E.; Schwarz, S.; Rothe, G.; Hoves, S.; et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 2007, 109, 3812–3819. [Google Scholar] [CrossRef]
- Sonveaux, P.; Copetti, T.; De Saedeleer, C.J.; Vegran, F.; Verrax, J.; Kennedy, K.M.; Moon, E.J.; Dhup, S.; Danhier, P.; Frérart, F.; et al. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PLoS ONE 2012, 7, e33418. [Google Scholar] [CrossRef]
- Jin, L.; Alesi, G.N.; Kang, S. Glutaminolysis as a target for cancer therapy. Oncogene 2016, 35, 3619–3625. [Google Scholar] [CrossRef] [Green Version]
- Dang, C.V. Glutaminolysis: Supplying carbon or nitrogen or both for cancer cells? Cell Cycle 2010, 9, 3884–3886. [Google Scholar] [CrossRef] [Green Version]
- Vanhove, K.; Derveaux, E.; Graulus, G.J.; Mesotten, L.; Thomeer, M.; Noben, J.P.; Guedens, W.; Adriaensens, P. Glutamine Addiction and Therapeutic Strategies in Lung Cancer. Int. J. Mol. Sci. 2019, 20, 252. [Google Scholar] [CrossRef] [Green Version]
- Vyas, S.; Zaganjor, E.; Haigis, M.C. Mitochondria and Cancer. Cell 2016, 166, 555–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, L.; Zhou, Y. Crucial role of the pentose phosphate pathway in malignant tumors (Review). Oncol. Lett. 2019, 17, 4213–4221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gotzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. Ann. Intern. Med. 2009, 151, W65–W94. [Google Scholar] [CrossRef] [Green Version]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, X.; Wang, C.; Zhang, H.; Cai, Z. Non-targeted and targeted metabolomics approaches to diagnosing lung cancer and predicting patient prognosis. Oncotarget 2016, 7, 63437–63448. [Google Scholar] [CrossRef] [Green Version]
- Puchades-Carrasco, L.; Jantus-Lewintre, E.; Perez-Rambla, C.; Garcia-Garcia, F.; Lucas, R.; Calabuig, S.; Blasco, A.; Dopazo, J.; Camps, C.; Pineda-Lucena, A. Serum metabolomic profiling facilitates the non-invasive identification of metabolic biomarkers associated with the onset and progression of non-small cell lung cancer. Oncotarget 2016, 7, 12904–12916. [Google Scholar] [CrossRef] [Green Version]
- Berker, Y.; Vandergrift, L.A.; Wagner, I.; Su, L.; Kurth, J.; Schuler, A.; Dinges, S.S.; Habbel, P.; Nowak, J.; Mark, E.; et al. Magnetic Resonance Spectroscopy-based Metabolomic Biomarkers for Typing, Staging, and Survival Estimation of Early-Stage Human Lung Cancer. Sci. Rep. 2019, 9, 10319. [Google Scholar] [CrossRef] [Green Version]
- Louis, E.; Adriaensens, P.; Guedens, W.; Bigirumurame, T.; Baeten, K.; Vanhove, K.; Vandeurzen, K.; Darquennes, K.; Vansteenkiste, J.; Dooms, C.; et al. Detection of Lung Cancer through Metabolic Changes Measured in Blood Plasma. J. Thorac. Oncol. 2016, 11, 516–523. [Google Scholar] [CrossRef] [Green Version]
- Derveaux, E.; Thomeer, M.; Mesotten, L.; Reekmans, G.; Adriaensens, P. Detection of Lung Cancer via Blood Plasma and (1) H-NMR Metabolomics: Validation by a Semi-Targeted and Quantitative Approach Using a Protein-Binding Competitor. Metabolites 2021, 11, 537. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Chang, N.C.; Rudnicki, M.A. Skeletal Muscle Remodeling and Regeneration. Pathobiol. Hum. Dis. 2014, 567–579. [Google Scholar]
- Zhang, L.; Zheng, J.; Ahmed, R.; Huang, G.; Reid, J.; Mandal, R.; Maksymuik, A.; Sitar, D.S.; Tappia, P.S.; Ramjiawan, B.; et al. A High-Performing Plasma Metabolite Panel for Early-Stage Lung Cancer Detection. Cancers 2020, 12, 622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Ma, Z.; Li, A.; Li, H.; Wang, B.; Zhong, J.; Min, L.; Dai, L. Metabolomic profiling of human serum in lung cancer patients using liquid chromatography/hybrid quadrupole time-of-flight mass spectrometry and gas chromatography/mass spectrometry. J. Cancer Res. Clin. Oncol. 2015, 141, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Haferkamp, S.; Drexler, K.; Federlin, M.; Schlitt, H.J.; Berneburg, M.; Adamski, J.; Gaumann, A.; Geissler, E.K.; Ganapathy, V.; Parkinson, E.K.; et al. Extracellular Citrate Fuels Cancer Cell Metabolism and Growth. Front. Cell Dev. Biol. 2020, 8, 602476. [Google Scholar] [CrossRef]
- King, A.; Selak, M.A.; Gottlieb, E. Succinate dehydrogenase and fumarate hydratase: Linking mitochondrial dysfunction and cancer. Oncogene 2006, 25, 4675–4682. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, T.M. The complex role of branched chain amino acids in diabetes and cancer. Metabolites 2013, 3, 931–945. [Google Scholar] [CrossRef] [Green Version]
- Garlick, P.J. The role of leucine in the regulation of protein metabolism. J. Nutr. 2005, 135, S1553–S1556. [Google Scholar] [CrossRef] [Green Version]
- Qi, S.A.; Wu, Q.; Chen, Z.; Zhang, W.; Zhou, Y.; Mao, K.; Li, J.; Li, Y.; Chen, J.; Huang, Y.; et al. High-resolution metabolomic biomarkers for lung cancer diagnosis and prognosis. Sci. Rep. 2021, 11, 11805. [Google Scholar] [CrossRef]
- Proenza, A.M.; Oliver, J.; Palou, A.; Roca, P. Breast and lung cancer are associated with a decrease in blood cell amino acid content. J. Nutr. Biochem. 2003, 14, 133–138. [Google Scholar] [CrossRef]
- Maeda, J.; Higashiyama, M.; Imaizumi, A.; Nakayama, T.; Yamamoto, H.; Daimon, T.; Yamakado, M.; Imamura, F.; Kodama, K. Possibility of multivariate function composed of plasma amino acid profiles as a novel screening index for non-small cell lung cancer: A case control study. BMC Cancer 2010, 10, 690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, J.; Xu, L.; Li, W.; Zheng, C.; Wu, L. Targeted metabolomics for serum amino acids and acylcarnitines in patients with lung cancer. Exp. Ther. Med. 2019, 18, 188–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Wu, L.; Li, K.; Liu, F.; Wang, L.; Zhang, D.; Zhou, J.; Ma, X.; Wang, S.; Yang, S. Ornithine aminotransferase promoted the proliferation and metastasis of non-small cell lung cancer via upregulation of miR-21. J. Cell. Physiol. 2019, 234, 12828–12838. [Google Scholar] [CrossRef] [PubMed]
- Albaugh, V.L.; Pinzon-Guzman, C.; Barbul, A. Arginine-Dual roles as an onconutrient and immunonutrient. J. Surg. Oncol. 2017, 115, 273–280. [Google Scholar] [CrossRef]
- Zhao, M.; Jung, Y.; Jiang, Z.; Svensson, K.J. Regulation of Energy Metabolism by Receptor Tyrosine Kinase Ligands. Front. Physiol. 2020, 11, 354. [Google Scholar] [CrossRef] [Green Version]
- Jutel, M.; Blaser, K.; Akdis, C.A. The role of histamine in regulation of immune responses. Chem. Immunol. Allergy 2006, 91, 174–187. [Google Scholar]
- Reddy, A.V.; Killampalli, L.K.; Prakash, A.R.; Naag, S.; Sreenath, G.; Biraggari, S.K. Analysis of lipid profile in cancer patients, smokers, and nonsmokers. Dent. Res. J. 2016, 13, 494–499. [Google Scholar]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Du, G. Dysregulated lipid metabolism in cancer. World J. Biol. Chem. 2012, 3, 167–174. [Google Scholar] [CrossRef]
- Musharraf, S.G.; Mazhar, S.; Choudhary, M.I.; Rizi, N.; Atta-ur, R. Plasma metabolite profiling and chemometric analyses of lung cancer along with three controls through gas chromatography-mass spectrometry. Sci. Rep. 2015, 5, 8607. [Google Scholar] [CrossRef] [Green Version]
- Feng, S.; Wang, H.; Liu, J.; Aa, J.; Zhou, F.; Wang, G. Multi-dimensional roles of ketone bodies in cancer biology: Opportunities for cancer therapy. Pharmacol. Res. 2019, 150, 104500. [Google Scholar] [CrossRef] [PubMed]
- Louis, E.; Adriaensens, P.; Guedens, W.; Vanhove, K.; Vandeurzen, K.; Darquennes, K.; Vansteenkiste, J.; Dooms, C.; de Jonge, E.; Thomeer, M.; et al. Metabolic phenotyping of human blood plasma: A powerful tool to discriminate between cancer types? Ann. Oncol. 2016, 27, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Rysman, E.; Brusselmans, K.; Scheys, K.; Timmermans, L.; Derua, R.; Munck, S.; Van Veldhoven, P.P.; Waltregny, D.; Daniëls, V.W.; Machiels, J.; et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010, 70, 8117–8126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, R.; Louis, E.; Stinkens, K.; Mesotten, L.; Jonge, E.; Thomeer, M.; Caenepeel, P.; Adriaensens, P. Metabolic Phenotyping of Blood Plasma by Proton Nuclear Magnetic Resonance to Discriminate between Colorectal Cancer, Breast Cancer and Lung Cancer. Metab. Open Access 2016, 6, 3. [Google Scholar]
- Christen, S.; Lorendeau, D.; Schmieder, R.; Broekaert, D.; Metzger, K.; Veys, K.; Elia, I.; Buescher, J.M.; Orth, M.F.; Davidson, S.M.; et al. Breast Cancer-Derived Lung Metastases Show Increased Pyruvate Carboxylase-Dependent Anaplerosis. Cell Rep. 2016, 17, 837–848. [Google Scholar] [CrossRef] [Green Version]
- Deja, S.; Porebska, I.; Kowal, A.; Zabek, A.; Barg, W.; Pawelczyk, K.; Stanimirova, I.; Daszykowski, M.; Korzeniewska, A.; Jankowska, R.; et al. Metabolomics provide new insights on lung cancer staging and discrimination from chronic obstructive pulmonary disease. J. Pharm. Biomed. Anal. 2014, 100, 369–380. [Google Scholar] [CrossRef]
- Kuo, W.K.; Liu, Y.C.; Chu, C.M.; Hua, C.C.; Huang, C.Y.; Liu, M.H.; Wang, C.H. Amino Acid-Based Metabolic Indexes Identify Patients With Chronic Obstructive Pulmonary Disease And Further Discriminates Patients In Advanced BODE Stages. Int. J. Chron. Obstruct. Pulmon. Dis. 2019, 14, 2257–2266. [Google Scholar] [CrossRef] [Green Version]
- Monleon, D.; Wang, L.; Tang, Y.; Liu, S.; Mao, S.; Ling, Y.; Liu, D.; He, X.; Wang, X. Metabonomic Profiling of Serum and Urine by 1H NMR-Based Spectroscopy Discriminates Patients with Chronic Obstructive Pulmonary Disease and Healthy Individuals. PLoS ONE 2013, 8, e65675. [Google Scholar]
- Saoi, M.; Britz-McKibbin, P. New Advances in Tissue Metabolomics: A Review. Metabolites 2021, 11, 672. [Google Scholar] [CrossRef]
- Moreno, P.; Jimenez-Jimenez, C.; Garrido-Rodriguez, M.; Calderon-Santiago, M.; Molina, S.; Lara-Chica, M.; Priego-Capote, F.; Salvatierra, Á.; Muñoz, E.; Calzado, M.A. Metabolomic profiling of human lung tumor tissues—Nucleotide metabolism as a candidate for therapeutic interventions and biomarkers. Mol. Oncol. 2018, 12, 1778–1796. [Google Scholar] [CrossRef] [Green Version]
- Kowalczyk, T.; Kisluk, J.; Pietrowska, K.; Godzien, J.; Kozlowski, M.; Reszec, J.; Sierko, E.; Naumnik, W.; Mróz, R.; Moniuszko, M.; et al. The Ability of Metabolomics to Discriminate Non-Small-Cell Lung Cancer Subtypes Depends on the Stage of the Disease and the Type of Material Studied. Cancers 2021, 13, 3314. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, A.; Sugimoto, M.; Suzuki, A.; Hatakeyama, Y.; Enomoto, A.; Harada, S.; Soga, T.; Tomita, M.; Takebayashi, T. Effects of processing and storage conditions on charged metabolomic profiles in blood. Electrophoresis 2015, 36, 2148–2155. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.H. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol. Biol. 2015, 1277, 161–193. [Google Scholar] [PubMed]
- Moyer, V.A.; US Preventive Services Task Force. Screening for lung cancer: U.S. Preventive Services Task Force recommendation statement. Ann. Intern. Med. 2014, 160, 330–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Koning, H.J.; van der Aalst, C.M.; de Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.W.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N.; et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef]
- Kuhn, T.; Floegel, A.; Sookthai, D.; Johnson, T.; Rolle-Kampczyk, U.; Otto, W.; von Bergen, M.; Boeing, H.; Kaaks, R. Higher plasma levels of lysophosphatidylcholine 18:0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med. 2016, 14, 13. [Google Scholar] [CrossRef] [Green Version]
- His, M.; Viallon, V.; Dossus, L.; Gicquiau, A.; Achaintre, D.; Scalbert, A.; Ferrari, P.; Romieu, I.; Onland-Moret, N.C.; Weiderpass, E.; et al. Prospective analysis of circulating metabolites and breast cancer in EPIC. BMC Med. 2019, 17, 178. [Google Scholar] [CrossRef]
- Guertin, K.A.; Loftfield, E.; Boca, S.M.; Sampson, J.N.; Moore, S.C.; Xiao, Q.; Huang, W.Y.; Xiong, X.; Freedman, N.D.; Cross, A.J.; et al. Serum biomarkers of habitual coffee consumption may provide insight into the mechanism underlying the association between coffee consumption and colorectal cancer. Am. J. Clin. Nutr. 2015, 101, 1000–1011. [Google Scholar] [CrossRef]
- Udo, R.; Katsumata, K.; Kuwabara, H.; Enomoto, M.; Ishizaki, T.; Sunamura, M.; Nagakawa, Y.; Soya, R.; Sugimoto, M.; Tsuchida, A. Urinary charged metabolite profiling of colorectal cancer using capillary electrophoresis-mass spectrometry. Sci. Rep. 2020, 10, 21057. [Google Scholar] [CrossRef]


| 1H-NMR | HPLC | (LC/GC)-MS | |
|---|---|---|---|
| Sensitivity | Low | Higher | Highest |
| Sample preparation | Minimal sample preparation required | Extra sample preparation steps required: e.g., derivatization, solvent extraction | Extra sample preparation steps required: e.g., derivatization, solvent extraction |
| Number of detectable metabolites | 30–100 | 300–1000+ | 300–1000+ |
| Number of samples in one run | Analysis of 1 sample in 1 run | Analysis of more samples in 1 run | Analysis of more samples in 1 run |
| Cost per sample | Low | High | High |
| Reproducibility | High | Average | Average |
| Tissue samples | Can be analyzed directly | Requires tissue extraction | Requires tissue extraction |
| Speed | Fast | Slower | Slower |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mariën, H.; Derveaux, E.; Vanhove, K.; Adriaensens, P.; Thomeer, M.; Mesotten, L. Changes in Metabolism as a Diagnostic Tool for Lung Cancer: Systematic Review. Metabolites 2022, 12, 545. https://doi.org/10.3390/metabo12060545
Mariën H, Derveaux E, Vanhove K, Adriaensens P, Thomeer M, Mesotten L. Changes in Metabolism as a Diagnostic Tool for Lung Cancer: Systematic Review. Metabolites. 2022; 12(6):545. https://doi.org/10.3390/metabo12060545
Chicago/Turabian StyleMariën, Hanne, Elien Derveaux, Karolien Vanhove, Peter Adriaensens, Michiel Thomeer, and Liesbet Mesotten. 2022. "Changes in Metabolism as a Diagnostic Tool for Lung Cancer: Systematic Review" Metabolites 12, no. 6: 545. https://doi.org/10.3390/metabo12060545
APA StyleMariën, H., Derveaux, E., Vanhove, K., Adriaensens, P., Thomeer, M., & Mesotten, L. (2022). Changes in Metabolism as a Diagnostic Tool for Lung Cancer: Systematic Review. Metabolites, 12(6), 545. https://doi.org/10.3390/metabo12060545