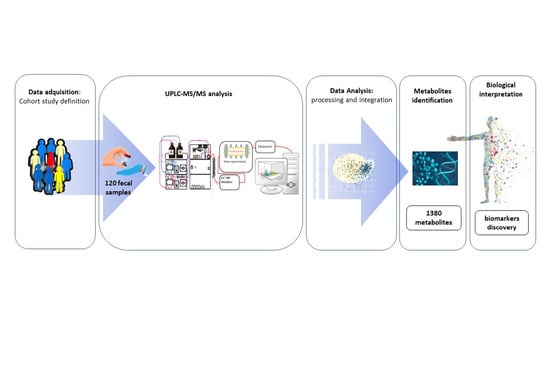
A Comprehensive Metabolomics Analysis of Fecal Samples from Advanced Adenoma and Colorectal Cancer Patients
Abstract
:1. Introduction
2. Results
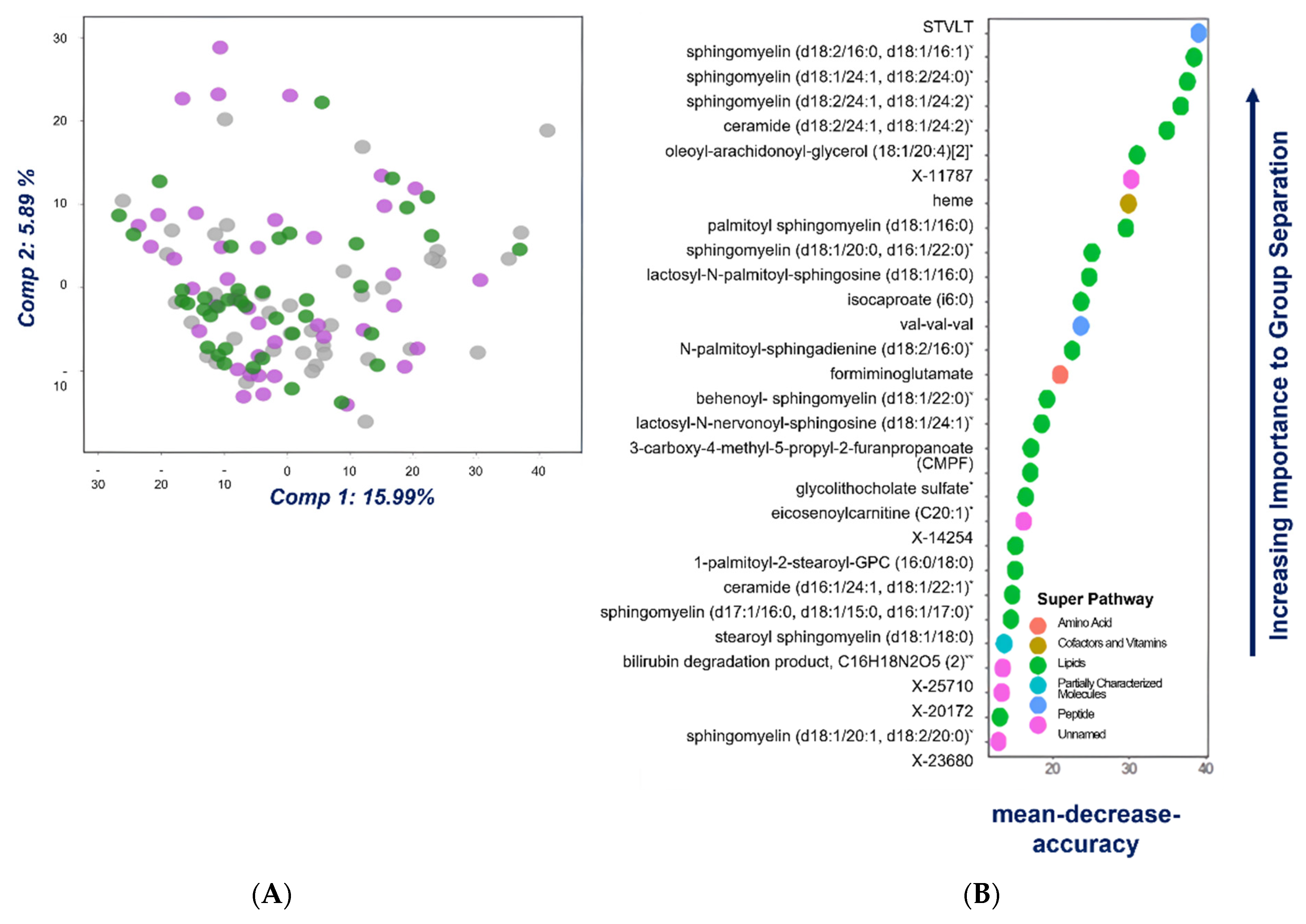
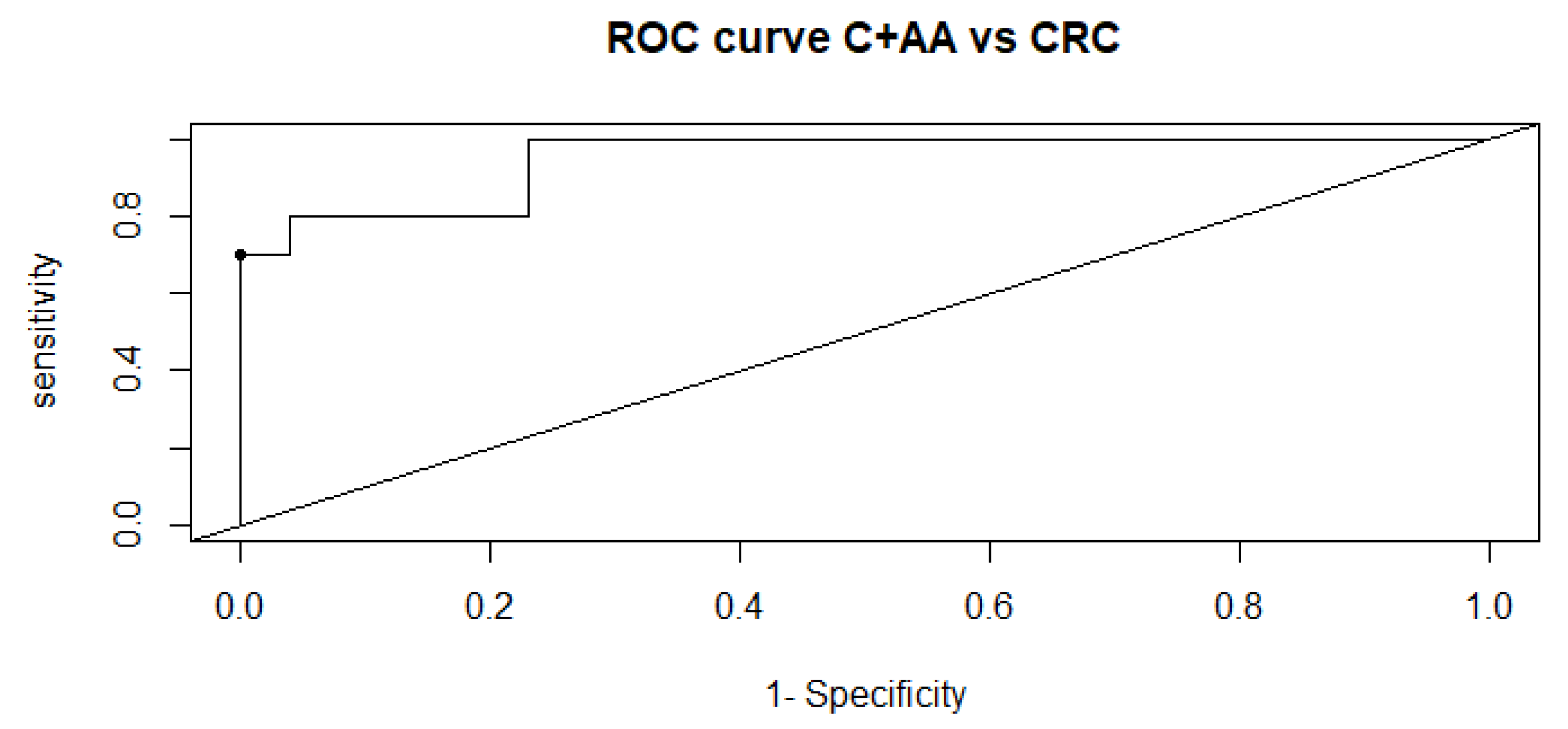
2.1. Univariate, Multivariate and Logistic Regression Analysis
2.2. Comparison of Metabolome of Colorectal Cancer, Advanced Adenoma and Control Groups
3. Discussion
4. Materials and Methods
4.1. Clinical Samples and Study Population
4.2. Sample Preparation and Metabolomics Analysis
4.3. Data Extraction and Compound Identification
4.4. Metabolite Quantification and Data Normalization
4.5. Statistical Analysis for Metabolome and Clinical Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Agency for Research on Cancer. March 2021. Available online: https://gco.iarc.fr/today/data/factsheets/populations/724-spain-fact-sheets.pdf (accessed on 27 October 2021).
- Kolligs, F.T. Diagnostics and Epidemiology of Colorectal Cancer. Visc. Med. 2016, 32, 158–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portillo, I.; Arana-Arri, E.; Idigoras, I.; Bilbao, I.; Martínez-Indart, L.; Bujanda, L.; Gutierrez-Ibarluzea, I. Colorectal and interval cancers of the Colorectal Cancer Screening Program in the Basque Country (Spain). World J. Gastroenterol. 2017, 23, 2731–2742. [Google Scholar] [CrossRef] [PubMed]
- Meklin, J.; SyrjÄnen, K.; Eskelinen, M. Fecal Occult Blood Tests in Colorectal Cancer Screening: Systematic Review and Meta-analysis of Traditional and New-generation Fecal Immunochemical Tests. Anticancer Res. 2020, 40, 3591–3604. [Google Scholar] [CrossRef] [PubMed]
- Elsafi, S.H.; Alqahtani, N.I.; Zakary, N.Y.; Al Zahrani, E.M. The sensitivity, specificity, predictive values, and likelihood ratios of fecal occult blood test for the detection of colorectal cancer in hospital settings. Clin. Exp. Gastroenterol. 2015, 9, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Martín-Blázquez, A.; Díaz, C.; González-Flores, E.; Franco-Rivas, D.; Jiménez-Luna, C.; Melguizo, C.; Prados, J.; Genilloud, O.; Vicente, F.; Caba, O.; et al. Untargeted LC-HRMS-based metabolomics to identify novel biomarkers of metastatic colorectal cancer. Sci. Rep. 2019, 9, 20198. [Google Scholar] [CrossRef]
- Cubiella, J.; Clos-Garcia, M.; Alonso, C.; Martinez-Arranz, I.; Perez-Cormenzana, M.; Barrenetxea, Z.; Berganza, J.; Rodríguez-Llopis, I.; D’Amato, M.; Bujanda, L.; et al. Targeted UPLC-MS Metabolic Analysis of Human faeces Reveals Novel Low-Invasive Candidate Markers for Colorectal Cancer. Cancers 2018, 10, 300. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.; Xue, W.; Yin, H.; Zhang, N.; Zhou, J.; Long, Z.; Wu, C.; Liang, Z.; Xie, K.; Li, S.; et al. Differential Metabolic Alterations and Biomarkers Between Gastric Cancer and Colorectal Cancer: A Systematic Review and Meta-Analysis. OncoTargets Ther. 2020, 13, 6093–6108. [Google Scholar] [CrossRef]
- Hardiman, G. An Introduction to Systems Analytics and Integration of Big Omics Data. Genes 2020, 11, 245. [Google Scholar] [CrossRef] [Green Version]
- Lockhart, D.J.; Winzeler, E.A. Genomics, Gene Expression and DNA Arrays. Nature 2000, 405, 827–836. [Google Scholar] [CrossRef]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-Omics Approaches to Disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Armitage, E.G.; Barbas, C. Metabolomics in cancer biomarker discovery: Current trends and future perspectives. J. Pharm. Biomed. Anal. 2014, 87, 1–11. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, Y.; Zhao, W.; Deng, K.; Wang, Z.; Yang, C.; Ma, L.; Openkova, M.S.; Hou, Y.; Li, K. Metabolomics for biomarker discovery in the diagnosis, prognosis, survival and recurrence of colorectal cancer: A systematic review. Oncotarget 2017, 8, 35460–35472. [Google Scholar] [CrossRef] [Green Version]
- Arana-Arri, E.; Imaz-Ayo, N.; Fernández, M.J.; Idigoras, I.; Bilbao, I.; Bujanda, L.; Bao, F.; Ojembarrena, E.; Gil, I.; Gutiérrez-Ibarluzea, I.; et al. Screening colonoscopy and risk of adverse events among individuals undergoing fecal immunochemical testing in a population-based program: A nested case-control study. United Eur. Gastroenterol. J. 2018, 6, 755–764. [Google Scholar] [CrossRef]
- Bathe, O.F.; Farshidfar, F. From genotype to functional phenotype: Unraveling the metabolomic features of colorectal cancer. Genes 2014, 5, 536–560. [Google Scholar] [CrossRef] [Green Version]
- Griffin, J.L.; Shockcor, J.P. Metabolic profiles of cancer cells. Nat. Rev. Cancer 2004, 4, 551–561. [Google Scholar] [CrossRef]
- Ni, Y.; Xie, G.; Jia, W. Metabonomics of human colorectal cancer: New approaches for early diagnosis and biomarker discovery. J. Proteome Res. 2014, 13, 3857–3870. [Google Scholar] [CrossRef]
- Spratlin, J.L.; Serkova, N.J.; Eckhardt, S.G. Clinical applications of metabolomics in oncology: A review. Clin. Cancer Res. 2009, 15, 431–440. [Google Scholar] [CrossRef] [Green Version]
- Jacob, M.; Lopata, A.L.; Dasouki, M.; Abdel Rahman, A.M. Metabolomics toward Personalized. Mass Spectrom. Rev. 2019, 38, 221–238. [Google Scholar] [CrossRef]
- Wishart, D.S.; Mandal, R.; Stanislaus, A.; Ramirez-Gaona, M. Cancer Metabolomics and the Human Metabolome Database. Metabolites 2016, 6, 10. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Metabolomics for Biomarker Discovery: Moving to the Clinic. Biomed. Res. Int. 2015, 2015, 354671. [Google Scholar] [CrossRef]
- Mamas, M.; Dunn, W.B.; Neyses, L.; Goodacre, R. The role of metabolites and metabolomics in clinically applicable biomarkers of disease. Arch. Toxicol. 2011, 85, 5–17. [Google Scholar] [CrossRef]
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M.; et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009, 69, 4918–4925. [Google Scholar] [CrossRef] [Green Version]
- Sheridan, M.; Ogretmen, B. The Role of Ceramide Metabolism and Signaling in the Regulation of Mitophagy and Cancer Therapy. Cancers 2021, 13, 2475. [Google Scholar] [CrossRef]
- Adje, E.Y.; Balti, R.; Lecouturier, D.; Kouach, M.; Dhulster, P.; Guillochon, D.; Nedjar-Arroume, N. Controlled Enzymatic Hydrolysis: A New Strategy for the Discovery of Antimicrobial Peptides. Probiotics Antimicrob. Proteins 2013, 5, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Dai, X.; Zhou, C.C.; Li, K.X.; Zhang, Y.J.; Lou, X.Y.; Zhu, Y.M.; Sun, Y.L.; Peng, B.X.; Cui, W. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut 2021, 71, 1315–1325. [Google Scholar] [CrossRef]
- Nishiumi, S.; Kobayashi, T.; Ikeda, A.; Yoshie, T.; Kibi, M.; Izumi, Y.; Okuno, T.; Hayashi, N.; Kawano, S.; Takenawa, T.; et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS ONE 2012, 7, e40459. [Google Scholar] [CrossRef]
- Mayerle, J.; Kalthoff, H.; Reszka, R.; Kamlage, B.; Peter, E.; Schniewind, B.; González Maldonado, S.; Pilarsky, C.; Heidecke, C.D.; Schatz, P.; et al. Metabolic biomarker signature to differentiate pancreatic ductal adenocarcinoma from chronic pancreatitis. Gut 2018, 67, 128–137. [Google Scholar] [CrossRef] [Green Version]
- Yuan, B.; Schafferer, S.; Tang, Q.; Scheffler, M.; Nees, J.; Heil, J.; Schott, S.; Golatta, M.; Wallwiener, M.; Sohn, C.; et al. A plasma metabolite panel as biomarkers for early primary breast cancer detection. Int. J. Cancer 2019, 144, 2833–2842. [Google Scholar] [CrossRef]
- Ishikawa, S.; Tamaki, S.; Ohata, M.; Arihara, K.; Itoh, M. Heme induces DNA damage and hyperproliferation of colonic epithelial cells via hydrogen peroxide produced by heme oxygenase: A possible mechanism of heme-induced colon cancer. Mol. Nutr. Food Res. 2010, 54, 1182–1191. [Google Scholar] [CrossRef]
- Lee, D.H.; Anderson, K.E.; Harnack, L.J.; Folsom, A.R.; Jacobs, D.R., Jr. Heme iron, zinc, alcohol consumption, and colon cancer: Iowa Women’s Health Study. J. Natl. Cancer Inst. 2004, 96, 403–407. [Google Scholar] [CrossRef] [Green Version]
- Larsson, S.C.; Adami, H.O.; Giovannucci, E.; Wolk, A. Re: Heme iron, zinc, alcohol consumption, and risk of colon cancer. J. Natl. Cancer Inst. 2005, 97, 232–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balder, H.F.; Vogel, J.; Jansen, M.C.; Weijenberg, M.P.; van den Brandt, P.A.; Westenbrink, S.; van der Meer, R.; Goldbohm, R.A. Heme and chlorophyll intake and risk of colorectal cancer in the Netherlands cohort study. Cancer Epidemiol. Biomark. Prev. 2006, 15, 717–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferris, C.D.; Jaffrey, S.R.; Sawa, A.; Takahashi, M.; Brady, S.D.; Barrow, R.K.; Tysoe, S.A.; Wolosker, H.; Barañano, D.E.; Doré, S.; et al. Haem oxygenase-1 prevents cell death by regulating cellular iron. Nat. Cell. Biol. 1999, 1, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A.I.; Choudhury, M.; da Silva, J.L.; Schwartzman, M.L.; Abraham, N.G. Overexpression of the heme oxygenase gene in renal cell carcinoma. Proc. Soc. Exp. Biol. Med. 1997, 214, 54–61. [Google Scholar] [CrossRef]
- Doi, K.; Akaike, T.; Fujii, S.; Tanaka, S.; Ikebe, N.; Beppu, T.; Shibahara, S.; Ogawa, M.; Maeda, H. Induction of haem oxygenase-1 nitric oxide and ischaemia in experimental solid tumours and implications for tumour growth. Br. J. Cancer 1999, 80, 1945–1954. [Google Scholar] [CrossRef] [Green Version]
- Liepke, C.; Baxmann, S.; Heine, C.; Breithaupt, N.; Ständker, L.; Forssmann, W.G. Human hemoglobin-derived peptides exhibit antimicrobial activity: A class of host defense peptides. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2003, 791, 345–356. [Google Scholar] [CrossRef]
- .Kościuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzałkowska, N.; Jóźwik, A.; Horbańczuk, J.; Krzyżewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: Family of antimicrobial peptides. A review. Mol. Biol. Rep. 2012, 39, 10957–10970. [Google Scholar] [CrossRef] [Green Version]
- Starr, C.G.; Maderdrut, J.L.; He, J.; Coy, D.H.; Wimley, W.C. Pituitary adenylate cyclase-activating polypeptide is a potent broad-spectrum antimicrobial peptide: Structure-activity relationships. Peptides 2018, 104, 35–40. [Google Scholar] [CrossRef]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [Green Version]
- Lau, Q.Y.; Li, J.; Sani, M.A.; Sinha, S.; Li, Y.; Kang, C.; Bhattacharjya, S.; Separovic, F.; Verma, C.; Chia, C.S.B. Elucidating the bactericidal mechanism of action of the linear antimicrobial tetrapeptide BRBR-NH2. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1517–1527. [Google Scholar] [CrossRef]
- Rathinakumar, R.; Walkenhorst, W.F.; Wimley, W.C. Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: The importance of interfacial activity. J. Am. Chem. Soc. 2009, 131, 7609–7617. [Google Scholar] [CrossRef] [Green Version]
- Goedert, J.J.; Sinha, R. Detection of Colorectal Cancer with Two Novel Heme-Related Molecules in Human Feces. International PCT Application No. PCT/US2015/038299, 29 June 2015. 29 June.
- Goedert, J.J.; Sampson, J.N.; Moore, S.C.; Xiao, Q.; Xiong, X.; Hayes, R.B.; Ahn, J.; Shi, J.; Sinha, R. Fecal metabolomics: Assay performance and association with colorectal cancer. Carcinogenesis 2014, 35, 2089–2096. [Google Scholar] [CrossRef] [Green Version]
- Santos, C.R.; Schulze, A. Lipid metabolism in cancer. FEBS J. 2012, 279, 2610–2623. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, X.; Liu, X.; Sun, H.; Guo, Z.; Zheng, G.; Zhang, Y.; Sun, W. UPLC-MS based urine untargeted metabolomic analyses to differentiate bladder cancer from renal cell carcinoma. BMC Cancer 2019, 19, 1195. [Google Scholar] [CrossRef] [Green Version]
- Amara, C.S.; Vantaku, V.; Lotan, Y.; Putluri, N. Recent advances in the metabolomic study of bladder cancer. Expert Rev. Proteom. 2019, 16, 315–324. [Google Scholar] [CrossRef]
- Ordóñez, Y.F.; González, J.; Bedia, C.; Casas, J.; Abad, J.L.; Delgado, A.; Fabrias, G. 3-Ketosphinganine provokes the accumulation of dihydroshingolipids and induces autophagy in cancer cells. Mol. Biosyst. 2016, 12, 1166–1173. [Google Scholar] [CrossRef] [Green Version]
- Morad, S.A.; Cabot, M.C. Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer 2013, 13, 51–65. [Google Scholar] [CrossRef]
- Machala, M.; Procházková, J.; Hofmanová, J.; Králiková, L.; Slavík, J.; Tylichová, Z.; Ovesná, P.; Kozubík, A.; Vondráček, J. Colon Cancer and Perturbations of the Sphingolipid Metabolism. Int. J. Mol. Sci. 2019, 20, 6051. [Google Scholar] [CrossRef] [Green Version]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- Chen, J.; Hou, H.; Chen, H.; Luo, Y.; Zhang, L.; Zhang, Y.; Liu, H.; Zhang, F.; Liu, Y.; Wang, A.; et al. Urinary metabolomics for discovering metabolic biomarkers of laryngeal cancer using UPLC-QTOF/MS. J. Pharm. Biomed. Anal. 2019, 167, 83–89. [Google Scholar] [CrossRef]
- Kim, M.; Vogtmann, E.; Ahlquist, D.A.; Devens, M.E.; Kisiel, J.B.; Taylor, W.R.; White, B.A.; Hale, V.L.; Sung, J.; Chia, N.; et al. Fecal Metabolomic Signatures in Colorectal Adenoma Patients Are Associated with Gut Microbiota and Early Events of Colorectal Cancer Pathogenesis. mBio 2020, 11, e03186-19. [Google Scholar] [CrossRef] [Green Version]
- Sweeney, E.A.; Sakakura, C.; Shirahama, T.; Masamune, A.; Ohta, H.; Hakomori, S.; Igarashi, Y. Sphingosine and its methylated derivative N,N-dimethylsphingosine (DMS) induce apoptosis in a variety of human cancer cell lines. Int. J. Cancer 1996, 66, 358–366. [Google Scholar] [CrossRef]
- Ahn, E.H.; Schroeder, J.J. Induction of apoptosis by sphingosine, sphinganine, and C(2)-ceramide in human colon cancer cells, but not by C(2)-dihydroceramide. Anticancer Res. 2010, 30, 2881–2884. [Google Scholar]
- Schiffmann, S.; Ziebell, S.; Sandner, J.; Birod, K.; Deckmann, K.; Hartmann, D.; Rode, S.; Schmidt, H.; Angioni, C.; Geisslinger, G.; et al. Activation of ceramide synthase 6 by celecoxib leads to a selective induction of C16:0-ceramide. Biochem. Pharmacol. 2010, 80, 1632–1640. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, S.; Pandey, A. The Yin and Yang of lactosylceramide metabolism: Implications in cell function. Biochim. Biophys. Acta. 2008, 1780, 370–382. [Google Scholar] [CrossRef]
- Kakugawa, Y.; Wada, T.; Yamaguchi, K.; Yamanami, H.; Ouchi, K.; Sato, I.; Miyagi, T. Up-regulation of plasma membrane-associated ganglioside sialidase (Neu3) in human colon cancer and its involvement in apoptosis suppression. Proc. Natl. Acad. Sci. USA 2002, 99, 10718–10723. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Dueck, D.A.; Chan, M.; Tran, K.; Wong, J.T.; Jay, F.T.; Littman, C.; Stimpson, R.; Choy, P.C. The modulation of choline phosphoglyceride metabolism in human colon cancer. Mol. Cell. Biochem. 1996, 162, 97–103. [Google Scholar] [CrossRef]
- Ruiz-Cabello, J.; Cohen, J.S. Phospholipid metabolites as indicators of cancer cell function. NMR Biomed. 1992, 5, 226–233. [Google Scholar] [CrossRef]
- Jackowski, S. Coordination of membrane phospholipid synthesis with the cell cycle. J. Biol. Chem. 1994, 269, 3858–3867. [Google Scholar] [CrossRef]
- Jackowski, S. Cell cycle regulation of membrane phospholipid metabolism. J. Biol. Chem. 1996, 271, 20219–20222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podo, F. Tumour phospholipid metabolism. NMR Biomed. 1999, 12, 413–439. [Google Scholar] [CrossRef]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2014, 6, 6528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Resh, M.D. Cholesterol depletion from the plasma membrane triggers ligand-independent activation of the epidermal growth factor receptor. J. Biol. Chem. 2002, 277, 49631–49637. [Google Scholar] [CrossRef] [Green Version]
- Sagawa, H.; Tazuma, S.; Kajiyama, G. Protection against hydrophobic bile salt-induced cell membrane damage by liposomes and hydrophilic bile salts. Am. J. Physiol. 1993, 264, 835–839. [Google Scholar] [CrossRef]
- Kandell, R.L.; Bernstein, C. Bile salt/acid induction of DNA damage in bacterial and mammalian cells: Implications for colon cancer. Nutr. Cancer 1991, 16, 227–238. [Google Scholar] [CrossRef]
- Reddy, B.S.; Watanabe, K.; Weisburger, J.H.; Wynder, E.L. Promoting effect of bile acids in colon carcinogenesis in germ-free and conventional F344 rats. Cancer Res. 1977, 37, 3238–3242. [Google Scholar]
- McGarr, S.E.; Ridlon, J.M.; Hylemon, P.B. Diet, anaerobic bacterial metabolism, and colon cancer: A review of the literature. J. Clin. Gastroenterol. 2005, 39, 98–109. [Google Scholar]
- Johnson, C.H.; Santidrian, A.F.; LeBoeuf, S.E.; Kurczy, M.E.; Rattray, N.J.W.; Rattray, Z.; Warth, B.; Ritland, M.; Hoang, L.T.; Loriot, C.; et al. Metabolomics guided pathway analysis reveals link between cancer metastasis, cholesterol sulfate, and phospholipids. Cancer Metab. 2017, 5, 9. [Google Scholar] [CrossRef] [Green Version]
- Kuroki, T.; Ikuta, T.; Kashiwagi, M.; Kawabe, S.; Ohba, M.; Huh, N.; Mizuno, K.; Ohno, S.; Yamada, E.; Chida, K. Cholesterol sulfate. An activator of protein kinase C mediating squamous cell differentiation: A review. Mutat. Res. 2000, 462, 189–195. [Google Scholar] [CrossRef]
- Anderson, N.M.; Mucka, P.; Kern, J.G.; Feng, H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell 2018, 9, 216–237. [Google Scholar] [CrossRef]
- Field, M.S.; Anguera, M.C.; Page, R.; Stover, P.J. 5,10-Methenyltetrahydrofolate synthetase activity is increased in tumors and modifies the efficacy of antipurine LY309887. Arch. Biochem. Biophys. 2009, 481, 145–150. [Google Scholar] [CrossRef] [Green Version]
- Cubiella, J.; Vega, P.; Salve, M.; Díaz-Ondina, M.; Alves, M.T.; Quintero, E.; Álvarez-Sánchez, V.; Fernández-Bañares, F.; Boadas, J.; Campo, R.; et al. Development and external validation of a faecal immunochemical test-based prediction model for colorectal cancer detection in symptomatic patients. BMC Med. 2016, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Montrose, D.C.; Zhou, X.K.; Kopelovich, L.; Yantiss, R.K.; Karoly, E.D.; Subbaramaiah, K.; Dannenberg, A.J. Metabolic profiling, a noninvasive approach for the detection of experimental colorectal neoplasia. Cancer Prev. Res. 2012, 5, 1358–1367. [Google Scholar] [CrossRef] [Green Version]
- Goufo, P.; Cortez, I. Metabolite profile data of grapevine plants with brown wood streaking and grapevine leaf stripe (esca complex disease) symptoms. Data Brief 2021, 39, 107623. [Google Scholar] [CrossRef]
- Ford, L.; Kennedy, A.D.; Goodman, K.D.; Pappan, K.L.; Evans, A.M.; Miller, L.A.D.; Wulff, J.E.; Wiggs, B.R.; Lennon, J.J.; Elsea, S.; et al. Precision of a Clinical Metabolomics Profiling Platform for Use in the Identification of Inborn Errors of Metabolism. J. Appl. Lab. Med. 2020, 5, 342–356. [Google Scholar] [CrossRef]
- Goodman, K.; Mitchell, M.; Evans, A.M.; Miller, L.A.D.; Ford, L.; Wittmann, B.; Kennedy, A.D.; Toal, D. Assessment of the effects of repeated freeze thawing and extended bench top processing of plasma samples using untargeted metabolomics. Metabolomics 2021, 17, 31. [Google Scholar] [CrossRef]
- Evans, A.M.; Bridgewater, B.; Liu, Q.; Mitchell, M.W.; Robinson, R.J.; Dai, H.; Steward, S.J.; DeHaven, C.D.; Miller, L.A.D. High resolution mass spectrometry improves data quality and quality as compared to unit mass resolution mass spectrometry in high throughput profiling metabolomics. Metabolomics 2014, 4, 1. [Google Scholar]
- DeHaven, C.D.; Evans, A.M.; Dai, H.; Lawton, K.A. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J. Cheminform. 2010, 2, 9. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, A.D.; Wittmann, B.M.; Evans, A.M.; Miller, L.A.D.; Toal, D.R.; Lonergan, S.; Elsea, S.H.; Pappan, K.L. Metabolomics in the clinic: A review of the shared and unique features of untargeted metabolomics for clinical research and clinical testing. J. Mass Spectrom. 2018, 53, 1143–1154. [Google Scholar] [CrossRef]
- DeHaven, C.D.; Evans, A.M.; Dai, H.; Lawton, K.A. Software Techniques for Enabling High-Throughput Analysis of Metabolomics Datasets. Available online: https://www.intechopen.com/chapters/28007 (accessed on 26 December 2021).
- https://cran.r-project.org/. Available online: https://cran.r-project.org/web/packages/randomForest/randomForest.pdf (accessed on 26 December 2021).
- https://cran.r-project.org/. Available online: https://cran.r-project.org/web/packages/RRreg/RRreg.pdf (accessed on 26 December 2021).
- Domínguez-Almendros, S.; Benítez-Parejo, N.; Gonzalez-Ramirez, A.R. Logistic regression models. Allergol. Immunopathol. 2011, 39, 295–305. [Google Scholar] [CrossRef]
- https://cran.r-project.org/. Available online: https://cran.r-project.org/web/packages/caret/caret.pdf (accessed on 26 December 2021).
- https://cran.r-project.org/. Available online: https://cran.r-project.org/web/packages/pROC/pROC.pdf (accessed on 26 December 2021).
- https://cran.r-project.org/. Available online: https://cran.r-project.org/web/packages/performance/performance.pdf (accessed on 26 December 2021).
- https://cran.r-project.org/. Available online: https://cran.r-project.org/web/packages/ROCR/ROCR.pdf (accessed on 26 December 2021).
- Puchades-Carrasco, L.; Pineda-Lucena, A. Metabolomics Applications in Precision Medicine: An Oncological Perspective. Curr. Top. Med. Chem. 2017, 17, 2740–2751. [Google Scholar] [CrossRef] [Green Version]
- RStudio Team. RStudio: Integrated Development for R. RStudio; PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 26 December 2021).
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [Green Version]
| Pathway | Biochemical Name | AA vs. C | CRC vs. C | AA + CRC vs. C | CRC vs. AA | C + AA vs. CRC | MSI | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold Change | q-Value | Fold Change | q-Value | Fold Change | q-Value | Fold Change | q-Value | Fold Change | q-Value | |||
| AMINO ACID | ||||||||||||
| Histidine Metabolism | formiminoglutamate | 0.92 | 0.709 | 2.02 | 0.0817 | 1.47 | 0.5331 | 2.21 | 0.0089 | 2.11 | 0.0064 | 1 |
| PEPTIDE | ||||||||||||
| Polypeptide | val-val-ala | 0.51 | 0.731 | 2.02 | 0.0705 | 1.27 | 0.4626 | 3.96 | 0.0076 | 2.68 | 0.0064 | 1 |
| STVLT | 0.46 | 0.8245 | 11.83 | 0.0065 | 6.14 | 0.241 | 25.98 | 0.0019 | 16.26 | 0.0022 | 1 | |
| LIPID | ||||||||||||
| Fatty Acid, Dicarboxylate | 3-carboxy-4-methyl-5-propyl-2-furanpropanoate | 1.3 | 0.7987 | 2.78 | 0.0339 | 2.04 | 0.2488 | 2.13 | 0.1472 | 2.42 | 0.0233 | 1 |
| Fatty Acid Metabolism | eicosenoylcarnitine (C20:1) | 0.73 | 0.6723 | 0.41 | 0.0063 | 0.57 | 0.241 | 0.57 | 0.7251 | 0.48 | 0.0274 | 2 |
| Diacylglycerol | oleoyl-arachidonoyl-glycerol (18:1/20:4) [2] (DAG 38:5) | 0.75 | 0.8245 | 4.11 | 0.0065 | 2.43 | 0.241 | 5.5 | 0.0017 | 4.7 | 0.0015 | 2 |
| Ceramide | ceramide (d18:2/24:1, d18:1/24:2) | 0.65 | 0.7016 | 1.92 | 0.0771 | 1.28 | 0.5331 | 2.94 | 0.0004 | 2.32 | 0.001 | 2 |
| LacCer | lactosyl-N-palmitoyl-sphingosine (d18:1/16:0) (LacCer 34:1) | 0.53 | 0.731 | 3.06 | 0.0631 | 1.8 | 0.4776 | 5.78 | 0.0013 | 4,00 | 0.0016 | 1 |
| lactosyl-N-nervonoyl-sphingosine (d18:1/24:1) (LacCer 42:3) | 0.42 | 0.6788 | 3.13 | 0.1213 | 1.78 | 0.5463 | 7.39 | 0.0016 | 4.4 | 0.0041 | 2 | |
| Sphingomyelin (SM) | palmitoyl sphingomyelin (d18:1/16:0) (SM 34:1) | 0.59 | 0.7225 | 2.3 | 0.0309 | 1.45 | 0.4556 | 3.89 | 0.001 | 2.9 | 0.001 | 1 |
| behenoyl sphingomyelin (d18:1/22:0) (SM 40:1) | 0.56 | 0.6788 | 2.04 | 0.1068 | 1.3 | 0.5332 | 3.64 | 0.0021 | 2.61 | 0.005 | 2 | |
| SM (d17:1/16:0, d18:1/15:0, d16:1/17:0) | 0.52 | 0.6965 | 1.89 | 0.1643 | 1.2 | 0.5332 | 3.67 | 0.008 | 2.49 | 0.0233 | 2 | |
| SM (d18:2/16:0, d18:1/16:1) (SM 34:2) | 0.64 | 0.7359 | 5.16 | 0.0017 | 2.9 | 0.241 | 8.01 | 0.0001 | 6.28 | 0.0002 | 2 | |
| SM (d18:1/20:0, d16:1/22:0) (SM 38:1) | 0.45 | 0.7339 | 1.63 | 0.0779 | 1.04 | 0.461 | 3.61 | 0.0033 | 2.25 | 0.0064 | 2 | |
| SM (d18:1/24:1, d18:2/24:0) (SM 42:2) | 0.5 | 0.7186 | 4.1 | 0.0039 | 2.3 | 0.3232 | 8.19 | 0.00007 | 5.46 | 0.00008 | 2 | |
| SM (d18:2/24:1, d18:1/24:2) (SM 42:3) | 0.59 | 0.7359 | 6.55 | 0.0017 | 3.57 | 0.241 | 11.19 | 0.0001 | 8.26 | 0.0002 | 2 | |
| Secondary Bile Acid Metabolism | glycolithocholate sulfate | 2.05 | 0.731 | 0.28 | 0.1213 | 1.17 | 0.5439 | 0.14 | 0.0332 | 0.19 | 0.0071 | 2 |
| glycocholenate sulfate | 0.4 | 0.8598 | 0.1 | 0.1643 | 0.25 | 0.472 | 0.24 | 0.2052 | 0.14 | 0.0398 | 2 | |
| NUCLEOTIDE | ||||||||||||
| Pyrimidine Metabolism | cytidine | 0.93 | 0.7359 | 0.46 | 0.0417 | 0.7 | 0.2488 | 0.5 | 0.3399 | 0.48 | 0.0398 | 1 |
| COFACTOR AND VITAMINS | ||||||||||||
| Hemoglobin and Porphyrin Metabolism | heme | 0.33 | 0.7604 | 8.44 | 0.0088 | 4.38 | 0.2885 | 25.62 | 0.0008 | 12.69 | 0.0011 | 1 |
| bilirubin (Z,Z) | 0.52 | 0.7484 | 0.16 | 0.0813 | 0.34 | 0.3114 | 0.31 | 0.3557 | 0.21 | 0.0457 | 1 | |
| bilirubin (E,E) | 0.77 | 0.7849 | 0.19 | 0.1589 | 0.48 | 0.5331 | 0.25 | 0.0497 | 0.21 | 0.0105 | 2 | |
| XENOBIOTICS | ||||||||||||
| Xanthine Metabolism | 3,7-dimethylurate | 1.18 | 0.8245 | 0.42 | 0.125 | 0.8 | 0.461 | 0.36 | 0.1135 | 0.39 | 0.0398 | 1 |
| PARTIALLY CHARACTERIZED MOLECULES (PCM) | ||||||||||||
| PCM | bilirubin degradation product, C16H18N2O5 (2) | 0.91 | 0.7329 | 0.31 | 0.0219 | 0.61 | 0.2488 | 0.34 | 0.2451 | 0.32 | 0.0064 | 3 |
| UN NAMED | ||||||||||||
| N/A | X-11787 | 1.28 | 0.8318 | 3.57 | 0.0065 | 2.43 | 0.241 | 2.78 | 0.0127 | 3.13 | 0.0027 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Telleria, O.; Alboniga, O.E.; Clos-Garcia, M.; Nafría-Jimenez, B.; Cubiella, J.; Bujanda, L.; Falcón-Pérez, J.M. A Comprehensive Metabolomics Analysis of Fecal Samples from Advanced Adenoma and Colorectal Cancer Patients. Metabolites 2022, 12, 550. https://doi.org/10.3390/metabo12060550
Telleria O, Alboniga OE, Clos-Garcia M, Nafría-Jimenez B, Cubiella J, Bujanda L, Falcón-Pérez JM. A Comprehensive Metabolomics Analysis of Fecal Samples from Advanced Adenoma and Colorectal Cancer Patients. Metabolites. 2022; 12(6):550. https://doi.org/10.3390/metabo12060550
Chicago/Turabian StyleTelleria, Oiana, Oihane E. Alboniga, Marc Clos-Garcia, Beatriz Nafría-Jimenez, Joaquin Cubiella, Luis Bujanda, and Juan Manuel Falcón-Pérez. 2022. "A Comprehensive Metabolomics Analysis of Fecal Samples from Advanced Adenoma and Colorectal Cancer Patients" Metabolites 12, no. 6: 550. https://doi.org/10.3390/metabo12060550