Differential Protein Expression among Two Different Ovine ARDS Phenotypes—A Preclinical Randomized Study
Abstract
1. Introduction
2. Results
2.1. Studied Population
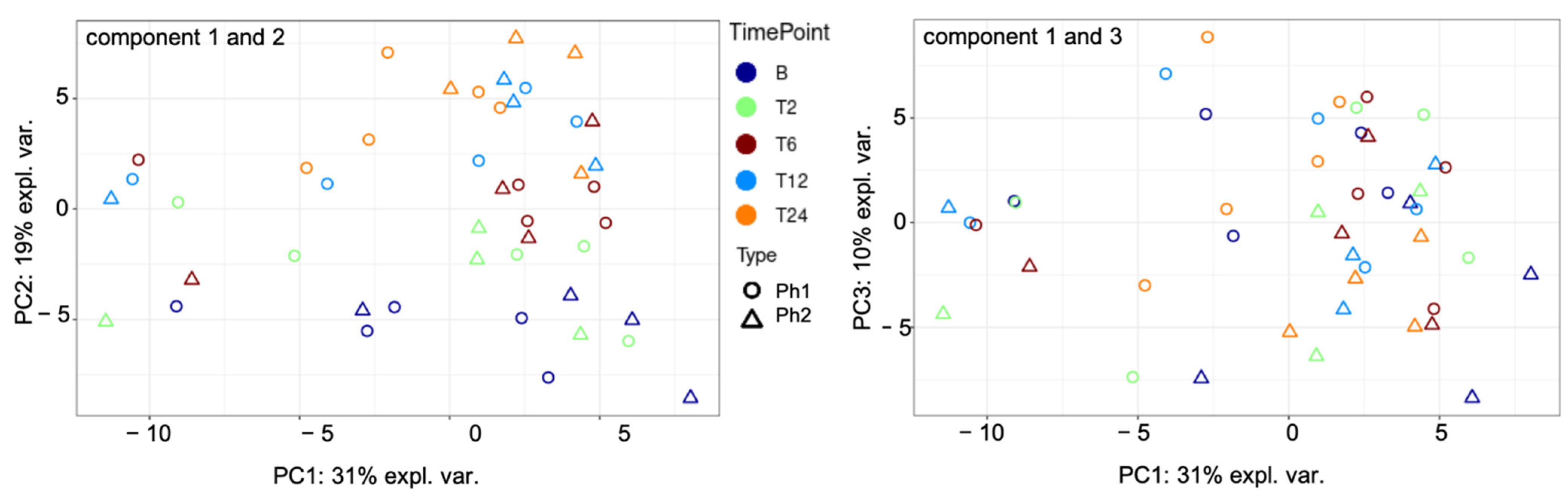
2.2. Unsupervised Cluster Analysis
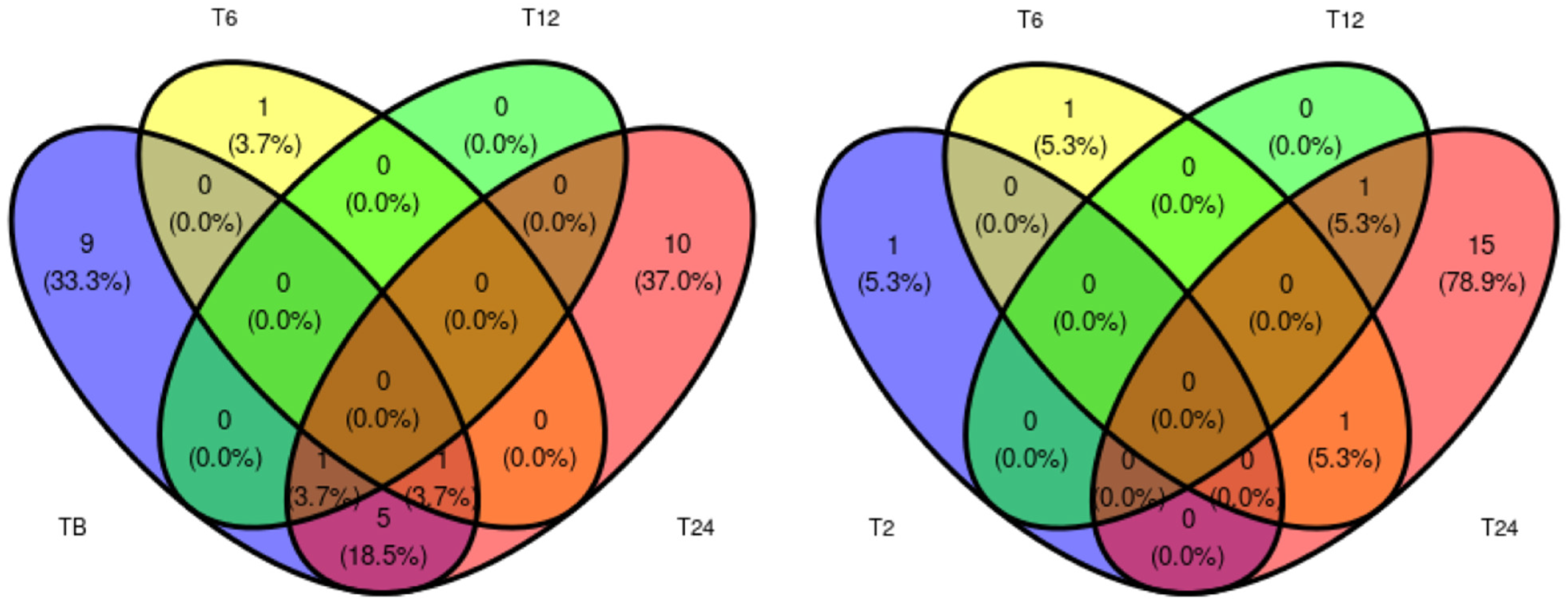
2.3. Proteins of Differential Abundance
2.4. Supervised Cluster Analysis
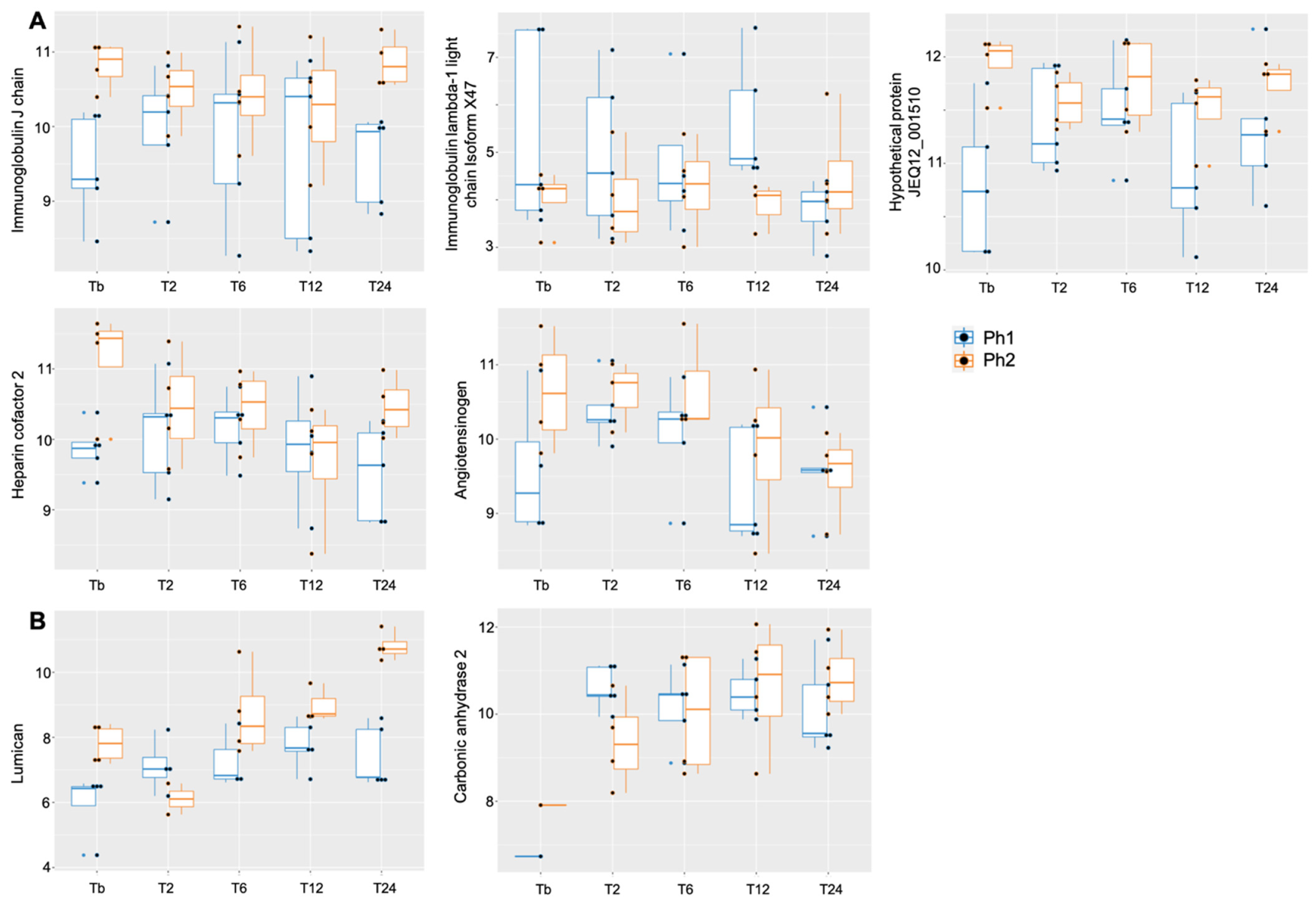
2.5. Analysis of Specific Proteins among Phenotypes over Time
2.6. Pathway Analysis
3. Discussion
4. Materials and Methods
4.1. Animal Model
4.2. Sample Collection and Processing
4.3. Protein Digestion Using Filter-Aided Sample Preparation (FASP) Method
4.4. Peptide Clean-Up
4.5. Mass Spectrometry
4.6. Data Processing and Quality Control
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACN | acetonitrile |
| AGT | angiotensinogen |
| APOA2 | apolipoprotein A-II |
| APOC3 | apolipoprotein C-III |
| ARDS | acute respiratory distress syndrome |
| ATS | American Thoracic Society |
| AUC | area under the curve |
| BCA | bicinchoninic acid |
| BLAST | Basic Local Alignment Search Tool |
| CA2 | carbonic anhydrase 2 |
| COVID-19 | coronavirus disease 2019 |
| CP | ceruloplasmin precursor |
| CVL | central venous line |
| DDA | data-dependent acquisition |
| DTT | dithiothreitol |
| ENSOARP00000000771 | uncharacterized protein |
| ENSOARP00000002890 | complement C4-like isoform X1 |
| ENSOARP00000011736 | hemoglobin subunit alpha |
| ENSOARP00000016410 | uncharacterized protein, belongs to serpin family |
| F2 | thrombin |
| FDR | false discovery rate |
| FiO2 | fraction of inspired oxygen |
| GPX3 | glutathione peroxidase 3 |
| HBB | hemoglobin subunit beta |
| HPLC | high-performance liquid chromatography |
| HPX | hemopexin |
| IAA | iodoacetamide |
| ITIH2 | inter-alpha-trypsin inhibitor heavy chain H2 |
| KNG1 | kininogen-1 isoform X2 |
| LC–MS/MS | liquid chromatography–mass spectrometry |
| LIS | Lung Injury Score |
| LMM | linear-mixed effects models |
| LOC101113086 | primary amine oxidase, lung isozyme |
| LPS | lipopolysaccharide |
| LUM | lumican |
| MERF | Medical Engineering Research Facility |
| MS TOF | mass spectrometry time-of-flight |
| MWCO | molecular weight cutoff |
| NHLBI | National Health Lung and Blood Institute |
| OA | oleic acid |
| P1 | human hypoinflammatory subphenotype |
| P2 | human hyperinflammatory subphenotype |
| PaO2 | partial pressure of oxygen in arterial blood |
| PC | principal component |
| PCA | principal component analysis |
| PF ratio | PaO2/FiO2 ratio |
| Ph1 | phenotype 1 |
| Ph2 | phenotype 2 |
| PHF21A | pHD finger protein 21A |
| PPI | protein–protein interaction |
| PLS-DA | partial least squares-discriminant analysis |
| SAA1 | serum amyloid A protein |
| SERPINC1 | antithrombin-III precursor |
| STRING | search tool for the retrieval of interacting genes/proteins |
| SWATH | sequential window acquisition of all theoretical mass spectra |
| TFA | trifluoroacetic acid |
| TOF | time of flight |
| TTR | transthyretin |
| QUT | Queensland University of Technology |
References
- Ashbaugh, D.G.; Bigelow, D.B.; Petty, T.L.; Levine, B.E. Acute Respiratory Distress in Adults. Lancet 1967, 2, 319–323. [Google Scholar] [CrossRef]
- Bellani, G.; Laffey, J.G.; Pham, T.; Fan, E.; Brochard, L.; Esteban, A.; Gattinoni, L.; van Haren, F.; Larsson, A.; McAuley, D.F.; et al. Epidemiology, Patterns of Care, and Mortality for Patients With Acute Respiratory Distress Syndrome in Intensive Care Units in 50 Countries. J. Am. Med. Assoc. 2016, 315, 788. [Google Scholar] [CrossRef]
- Máca, J.; Jor, O.; Holub, M.; Sklienka, P.; Burša, F.; Burda, M.; Janout, V.; Ševčík, P. Past and Present ARDS Mortality Rates: A Systematic Review. Respir. Care 2017, 62, 113–122. [Google Scholar] [CrossRef]
- COVID-19 Excess Mortality Collaborators. Estimating Excess Mortality Due to the COVID-19 Pandemic: A Systematic Analysis of COVID-19-Related Mortality, 2020–2021. Lancet 2022, 399, 1513–1536. [Google Scholar] [CrossRef]
- Brower, R.G.; Matthay, M.A.; Morris, A.; Schoenfeld, D.; Thompson, B.T.; Wheeler, A. Ventilation with Lower Tidal Volumes as Compared with Traditional Tidal Volumes for Acute Lung Injury and the Acute Respiratory Distress Syndrome. N. Engl. J. Med. 2000, 342, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Brower, R.G.; Lanken, P.N.; MacIntyre, N.; Matthay, M.A.; Morris, A.; Ancukiewicz, M.; Schoenfeld, D.; Thompson, B.T. Higher versus Lower Positive End-Expiratory Pressures in Patients with the Acute Respiratory Distress Syndrome. N. Eng. J. Med. 2004, 351, 327–336. [Google Scholar] [CrossRef]
- Herridge, M.S.; Tansey, C.M.; Matté, A.; Tomlinson, G.; Diaz-Granados, N.; Cooper, A.; Guest, C.B.; Mazer, C.D.; Mehta, S.; Stewart, T.E.; et al. Functional Disability 5 Years after Acute Respiratory Distress Syndrome. N. Engl. J. Med. 2011, 364, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Calfee, C.S.; Delucchi, K.; Parsons, P.E.; Thompson, B.T.; Ware, L.B.; Matthay, M.A. Latent Class Analysis of ARDS Subphenotypes: Analysis of Data From Two Randomized Controlled Trials. Lancet Respir. Med. 2014, 2, 611–620. [Google Scholar] [CrossRef]
- Bos, L.D.; Schouten, L.R.; van Vught, L.A.; Wiewel, M.A.; Ong, D.S.Y.; Cremer, O.; Artigas, A.; Martin-Loeches, I.; Hoogendijk, A.J.; van der Poll, T.; et al. Identification and Validation of Distinct Biological Phenotypes in Patients with Acute Respiratory Distress Syndrome by Cluster Analysis. Thorax 2017, 72, 876–883. [Google Scholar] [CrossRef]
- Famous, K.R.; Delucchi, K.; Ware, L.B.; Kangelaris, K.N.; Liu, K.D.; Thompson, B.T.; Calfee, C.S. Acute Respiratory Distress Syndrome Subphenotypes Respond Differently to Randomized Fluid Management Strategy. Am. J. Respir. Crit. Care Med. 2017, 195, 331–338. [Google Scholar] [CrossRef]
- Calfee, C.S.; Delucchi, K.L.; Sinha, P.; Matthay, M.A.; Hackett, J.; Shankar-Hari, M.; McDowell, C.; Laffey, J.G.; O’Kane, C.M.; McAuley, D.F.; et al. Acute Respiratory Distress Syndrome Subphenotypes and Differential Response to Simvastatin: Secondary Analysis of a Randomised Controlled Trial. Lancet Respir. Med. 2018, 6, 691–698. [Google Scholar] [CrossRef]
- Sinha, P.; Delucchi, K.L.; Thompson, B.T.; McAuley, D.F.; Matthay, M.A.; Calfee, C.S. Latent Class Analysis of ARDS Subphenotypes: A Secondary Analysis of the Statins for Acutely Injured Lungs from Sepsis (SAILS) Study. Intensive Care Med. 2018, 44, 1859–1869. [Google Scholar] [CrossRef]
- Pratik, S.; Delucchi, K.; Chen, Y.; Zhuo, H.; Abbott, J.; Wang, C.; Wickersham, N.; McNeil, J.B.; Jauregui, A.; Ke, S.; et al. Latent Class Analysis derived Subphenotypes are generalisable to Observational Cohorts of Acute Respiratory Distress Syndrome: A Prospective Study. Thorax 2022, 77, 13–21. [Google Scholar] [CrossRef]
- Sinha, P.; Furfaro, D.; Cummings, M.J.; Abrams, D.; Delucchi, K.; Maddali, M.V.; He, J.; Thompson, A.; Murn, M.; Fountain, J.; et al. Latent Class Analysis Reveals COVID-19–Related Acute Respiratory Distress Syndrome Subgroups with Differential Responses to Corticosteroids. Am. J. Respir. Crit. Care Med. 2021, 204, 1274–1285. [Google Scholar] [CrossRef]
- Prescott, H.C.; Calfee, C.S.; Taylor Thompson, B.; Angus, D.C.; Liu, V.X. Toward Smarter Lumping and Smarter Splitting: Rethinking Strategies for Sepsis and Acute Respiratory Distress Syndrome Clinical Trial Design. Am. J. Respir. Crit. Care Med. 2016, 194, 147–155. [Google Scholar] [CrossRef]
- Ware, L.B.; Matthay, M.A.; Mebazaa, A. Designing an ARDS Trial for 2020 and beyond: Focus on Enrichment Strategies. Intensive Care Med. 2020, 46, 2153–2156. [Google Scholar] [CrossRef]
- Bos, L.D.J.; Laffey, J.G.; Ware, L.B.; Heijnen, N.F.L.; Sinha, P.; Patel, B.; Jabaudon, M.; Bastarache, J.A.; McAuley, D.F.; Summers, C.; et al. Towards a Biological Definition of ARDS: Are Treatable Traits the Solution? Intensive Care Med. Exp. 2022, 10, 8. [Google Scholar] [CrossRef]
- Chen, H.; Xie, J.; Su, N.; Wang, J.; Sun, Q.; Li, S.; Jin, J.; Zhou, J.; Mo, M.; Wei, Y.; et al. Corticosteroid Therapy Is Associated with Improved Outcome in Critically Ill Patients With COVID-19 With Hyperinflammatory Phenotype. Chest 2021, 159, 1793–1802. [Google Scholar] [CrossRef]
- Bime, C.; Casanova, N.; Oita, R.C.; Ndukum, J.; Lynn, H.; Camp, S.M.; Lussier, Y.; Abraham, I.; Carter, D.; Miller, E.J.; et al. Development of a Biomarker Mortality Risk Model in Acute Respiratory Distress Syndrome. Crit. Care 2019, 23, 410. [Google Scholar] [CrossRef]
- Calfee, C.S.; Janz, D.R.; Bernard, G.R.; May, A.K.; Kangelaris, K.N.; Matthay, M.A.; Ware, L.B. Distinct Molecular Phenotypes of Direct vs Indirect ARDS in Single-Center and Multicenter Studies. Chest 2015, 147, 1539–1548. [Google Scholar] [CrossRef]
- Calfee, C.S.; Ware, L.B.; Eisner, M.D.; Parsons, P.E.; Thompson, B.T.; Wickersham, N.; Matthay, M.A. Plasma Receptor for Advanced Glycation End Products and Clinical Outcomes in Acute Lung Injury. Thorax 2008, 63, 1083–1089. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.-P.; Zhang, Y.-Z.; Wan, Q.-Q. Non-Targeted Proteomics of Acute Respiratory Distress Syndrome: Clinical and Research Applications. Proteome Sci. 2021, 19, 5. [Google Scholar] [CrossRef] [PubMed]
- Sarma, A.; Christenson, S.A.; Zha, B.S.; Oliveira, A.; Neyton, L.P.A.; Mick, E.; Sinha, P.; Wilson, J.G.; Moazed, F.; Leligdowicz, A.; et al. Hyperinflammatory ARDS Is Characterized by Interferon-Stimulated Gene Expression, T-Cell Activation, and an Altered Metatranscriptome in Tracheal Aspirates. medRxiv 2022. [Google Scholar] [CrossRef]
- Shankar-Hari, M.; McAuley, D.F. Divide and Conquer: Identifying Acute Respiratory Distress Syndrome Subphenotypes. Thorax 2017, 72, 867–869. [Google Scholar] [CrossRef]
- Matthay, M.A.; Arabi, Y.M.; Siegel, E.R.; Ware, L.B.; Bos, L.D.J.; Sinha, P.; Beitler, J.R.; Wick, K.D.; Curley, M.A.Q.; Constantin, J.-M.; et al. Phenotypes and Personalized Medicine in the Acute Respiratory Distress Syndrome. Intensive Care Med. 2020, 46, 2136–2152. [Google Scholar] [CrossRef]
- Reddy, K.; Sinha, P.; Kane, C.M.O.; Gordon, A.C.; Calfee, C.S.; Mcauley, D.F. Subphenotypes in Critical Care: Translation into Clinical Practice. Lancet Respir. Med. 2020, 8, 631–643. [Google Scholar] [CrossRef]
- Millar, J.E.; Wildi, K.; Bartnikowski, N.; Bouquet, M.; Hyslop, K.; Passmore, M.R.; Ki, K.K.; See Hoe, L.E.; Obonyo, N.G.; Neyton, L.; et al. Characterizing Preclinical Sub-phenotypic Models of Acute Respiratory Distress Syndrome: An Experimental Ovine Study. Physiol. Rep. 2021, 9, e15048. [Google Scholar] [CrossRef]
- Enkhbaatar, P.; Nelson, C.; Salsbury, J.R.; Carmical, J.R.; Torres, K.E.O.; Herndon, D.; Prough, D.S.; Luan, L.; Sherwood, E.R. Comparison of Gene Expression by Sheep and Human Blood Stimulated with the TLR4 Agonists Lipopolysaccharide and Monophosphoryl Lipid A. PLoS ONE 2015, 10, e0144345. [Google Scholar] [CrossRef]
- Livingstone, S.A.; Wildi, K.S.; Dalton, H.J.; Usman, A.; Ki, K.K.; Passmore, M.R.; Li Bassi, G.; Suen, J.Y.; Fraser, J.F. Coagulation Dysfunction in Acute Respiratory Distress Syndrome and Its Potential Impact in Inflammatory Subphenotypes. Front. Med. 2021, 8, 723217. [Google Scholar] [CrossRef]
- Ranieri, V.M.; Rubenfeld, G.D.; Thompson, B.T.; Ferguson, N.D.; Caldwell, E.; Fan, E.; Camporota, L.; Slutsky, A.S. Acute Respiratory Distress Syndrome: The Berlin Definition. J. Am. Med. Assoc. 2012, 307, 2526–2533. [Google Scholar] [CrossRef]
- Chen, X.; Shan, Q.; Jiang, L.; Zhu, B.; Xi, X. Quantitative Proteomic Analysis by ITRAQ for Identification of Candidate Biomarkers in Plasma from Acute Respiratory Distress Syndrome Patients. Biochem. Biophys. Res. Commun. 2013, 441, 1–6. [Google Scholar] [CrossRef]
- Wu, A.; Hinds, C.J.; Thiemermann, C. High-Density Lipoproteins in Sepsis and Septic Shock: Metabolism, Actions, and Therapeutic Applications. Shock 2004, 21, 210–221. [Google Scholar] [CrossRef]
- Gonçalves-de-Albuquerque, C.F.; Silva, A.R.; Burth, P.; Castro-Faria, M.V.; Castro-Faria-Neto, H.C. Acute Respiratory Distress Syndrome: Role of Oleic Acid-Triggered Lung Injury and Inflammation. Mediat. Inflamm. 2015, 2015, 260465. [Google Scholar] [CrossRef]
- Baba, H.; Ishiwata, T.; Takashi, E.; Xu, G.; Asano, G. Expression and Localization of Lumican in the Ischemic and Reperfused Rat Heart. Jpn. Circ. J. 2001, 65, 445–450. [Google Scholar] [CrossRef]
- Dong, H.; Li, J.; Lv, Y.; Zhou, Y.; Wang, G.; Hu, S.; He, X.; Yang, P.; Zhou, Z.; Xiang, X.; et al. Comparative Analysis of the Alveolar Macrophage Proteome in ALI/ARDS Patients between the Exudative Phase and Recovery Phase. BMC Immunol. 2013, 14, 25. [Google Scholar] [CrossRef][Green Version]
- Forhead, A.J.; Pipkin, F.B.; Sutherland, M.F.; Fowden, A.L. Changes in the Maternal and Fetal Renin-Angiotensin Systems in Response to Angiotensin II Type 1 Receptor Blockade and Angiotensin-Converting Enzyme Inhibition in Pregnant Sheep during Late Gestation. Exp. Physiol. 1997, 82, 761–776. [Google Scholar] [CrossRef]
- Wei, H.; Wang, J.Y. Role of Polymeric Immunoglobulin Receptor in Iga and Igm Transcytosis. Int. J. Mol. Sci. 2021, 22, 2284. [Google Scholar] [CrossRef]
- Chavakis, T.; Boeckel, N.; Santoso, S.; Voss, R.; Isordia-Salas, I.; Pixley, R.A.; Morgenstern, E.; Colman, R.W.; Preissner, K.T. Inhibition of Platelet Adhesion and Aggregation by a Defined Region (Gly-486-Lys-502) of High Molecular Weight Kininogen. J. Biol. Chem. 2002, 277, 23157–23164. [Google Scholar] [CrossRef]
- Fujikura, T. Histochemical Studies of Pulmonary Hyaline Membranes. Keio J. Med. 1955, 4, 175–190. [Google Scholar] [CrossRef]
- Bergenstam, R.; Edlund, T.; Zettergren, L. Hyaline Membrane Disease: The Influence of High Oxygen Concentration on Ciliary Activity in the Respiratory Tract: An Experimental Study on Rabbits. Acta Paediatr. 1958, 47, 527–533. [Google Scholar] [CrossRef]
- Parker, J.C.; Hernandez, L.A.; Longenecker, G.L.; Peevy, K.; Johnson, W. Lung Edema Caused by High Peak Inspiratory Pressures in Dogs: Role of Increased Microvascular Filtration Pressure and Permeability. Am. Rev. Respir. Dis. 1990, 142, 321–328. [Google Scholar] [CrossRef]
- Tsuno, K.; Prato, P.; Kolobow, T. Acute Lung Injury from Mechanical Ventilation at Moderately High Airway Pressures. J. Appl. Physiol. 1990, 69, 956–961. [Google Scholar] [CrossRef]
- Albert, R.K.; Leasa, D.; Sanderson, M.; Robertson, H.T.; Hlastala, M.P. The Prone Position Improves Arterial Oxygenation and Reduces Shunt in Oleic-Acid-Induced Acute Lung Injury. Am. Rev. Respir. Dis. 1987, 135, 628–633. [Google Scholar] [CrossRef]
- Lamm, W.J.; Graham, M.M.; Albert, R.K. Mechanism by Which the Prone Position Improves Oxygenation in Acute Lung Injury. Am. J. Respir. Crit. Care Med. 1994, 150, 184–193. [Google Scholar] [CrossRef]
- Bowler, R.P.; Duda, B.; Chan, E.D.; Enghild, J.J.; Ware, L.B.; Matthay, M.A.; Duncan, M.W. Proteomic Analysis of Pulmonary Edema Fluid and Plasma in Patients with Acute Lung Injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004, 286, L1095–L1104. [Google Scholar] [CrossRef]
- Schnapp, L.M.; Donohoe, S.; Chen, J.; Sunde, D.A.; Kelly, P.M.; Ruzinski, J.; Martin, T.; Goodlett, D.R. Mining the Acute Respiratory Distress Syndrome Proteome: Identification of the Insulin-like Growth Factor (IGF)/IGF-Binding Protein-3 Pathway in Acute Lung Injury. Am. J. Pathol. 2006, 169, 86–95. [Google Scholar] [CrossRef]
- Chemonges, S.; Gupta, R.; Mills, P.C.; Kopp, S.R.; Sadowski, P. Characterisation of the Circulating Acellular Proteome of Healthy Sheep Using LC-MS/MS-Based Proteomics Analysis of Serum. Proteome Sci. 2017, 15, 11. [Google Scholar] [CrossRef]
- Matute-Bello, G.; Downey, G.; Moore, B.B.; Groshong, S.D.; Matthay, M.A.; Slutsky, A.S.; Kuebler, W.M. An Official American Thoracic Society Workshop Report: Features and Measurements of Experimental Acute Lung Injury in Animals. Am. J. Respir. Cell. Mol. Biol. 2011, 44, 725–738. [Google Scholar] [CrossRef]
- Kulkarni, H.S.; Lee, J.S.; Bastarache, J.A.; Kuebler, W.M.; Downey, G.P.; Albaiceta, G.M.; Altemeier, W.A.; Artigas, A.; Bates, J.H.T.; Calfee, C.S.; et al. Update on the Features and Measurements of Experimental Acute Lung Injury in Animals: An Official American Thoracic Society Workshop Report. Am. J. Respir. Cell Mol. Biol. 2022, 66, e1–e14. [Google Scholar] [CrossRef]
- Mercat, A.; Richard, J.-C.M.; Vielle, B.; Jaber, S.; Osman, D.; Diehl, J.-L.; Lefrant, J.-Y.; Prat, G.; Richecoeur, J.; Nieszkowska, A.; et al. Positive End-Expiratory Pressure Setting in Adults with Acute Lung Injury and Acute Respiratory Distress Syndrome: A Randomized Controlled Trial. J. Am. Med. Assoc. 2008, 299, 646–655. [Google Scholar] [CrossRef]
- Bouquet, M.; Passmore, M.R.; See Hoe, L.E.; Tung, J.-P.; Simonova, G.; Boon, A.-C.; Fraser, J.F. Development and Validation of ELISAs for the Quantitation of Interleukin (IL)-1β, IL-6, IL-8 and IL-10 in Ovine Plasma. J. Immunol. Methods 2020, 486, 112835. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.L.; Phung, T.K.; Bruschi, M.; Janusz, A.; Stewart, J.; Meehan, J.; Healy, P.; Nouwens, A.S.; Fox, G.P.; Vickers, C.E. Process Proteomics of Beer Reveals a Dynamic Proteome with Extensive Modi Fi Cations. J. Proteome Res. 2018, 17, 1647–1653. [Google Scholar] [CrossRef] [PubMed]
- Kerr, E.D.; Phung, T.K.; Caboche, C.H.; Fox, G.P.; Platz, G.J.; Schulz, B.L. The Intrinsic and Regulated Proteomes of Barley Seeds in Response to Fungal Infection. Anal. Biochem. 2019, 580, 30–35. [Google Scholar] [CrossRef]
- Casals, M.; Girabent-Farrés, M.; Carrasco, J.L. Methodological Quality and Reporting of Generalized Linear Mixed Models in Clinical Medicine (2000–2012): A Systematic Review. PLoS ONE 2014, 9, e112653. [Google Scholar] [CrossRef]
- Barr, D.J.; Levy, R.; Scheepers, C.; Tily, H.J. Random Effects Structure for Confirmatory Hypothesis Testing: Keep It Maximal. J. Mem. Lang. 2013, 68, 255–278. [Google Scholar] [CrossRef]


| (A) Component 1 | Ph1 | Ph2 | GroupContrib | Importance |
|---|---|---|---|---|
| hypothetical protein JEQ12_008126 | −0.50842 | 0.635523 | Ph2 | −0.49715 |
| hypothetical protein JEQ12_002713 | −0.46116 | 0.576452 | Ph2 | −0.4048 |
| immunoglobulin J chain | −0.43948 | 0.549347 | Ph2 | −0.36243 |
| hypothetical protein JEQ12_001510 | −0.40883 | 0.511038 | Ph2 | −0.30254 |
| lumican | −0.46124 | 0.569764 | Ph2 | −0.26074 |
| alpha-1-macroglobulin-like isoform X1 | −0.4179 | 0.459686 | Ph2 | −0.22226 |
| hypothetical protein JEQ12_014972 | −0.36751 | 0.459382 | Ph2 | −0.22178 |
| clusterin | −0.36404 | 0.455049 | Ph2 | −0.21501 |
| hypothetical protein JEQ12_010483 | −0.35937 | 0.449214 | Ph2 | −0.20589 |
| inter-alpha-trypsin inhibitor heavy chain H2 isoform X2 | −0.34194 | 0.427429 | Ph2 | −0.17183 |
| hypothetical protein JEQ12_008129, partial | −0.35521 | 0.448681 | Ph2 | −0.16998 |
| hypothetical protein JEQ12_008015 | −0.33536 | 0.419202 | Ph2 | −0.15897 |
| heparin cofactor 2 | −0.32167 | 0.386004 | Ph2 | −0.10707 |
| adiponectin isoform X1 | −0.36314 | 0.453928 | Ph2 | −0.07133 |
| hemoglobin subunit beta | 0.279596 | −0.34949 | Ph1 | 0.049993 |
| hypothetical protein JEQ12_003887 | −0.27908 | 0.348852 | Ph2 | −0.04899 |
| sex hormone-binding globulin isoform X3 | −0.34802 | 0.386691 | Ph2 | −0.04769 |
| fibronectin isoform X8 | −0.2735 | 0.34187 | Ph2 | −0.03807 |
| complement component C8 gamma chain | −0.39878 | 0.451948 | Ph2 | −0.03353 |
| short palate, lung and nasal epithelium carcinoma-associated protein 2B-like | −0.2696 | 0.37445 | Ph2 | −0.03047 |
| (B) Component 2 | Ph1 | Ph2 | GroupContrib | Importance |
| fibrinogen gamma chain isoform X1 | 0.244913 | −0.30614 | Ph1 | 0.995488 |
| hemopexin | 0.093814 | −0.11727 | Ph1 | 0.089809 |
| hypothetical protein JEQ12_017492 | 0.083199 | −0.104 | Ph1 | 0.030618 |
| (C) Component 3 | Ph1 | Ph2 | GroupContrib | Importance |
| serum paraoxonase/arylesterase 1 isoform X1 | −0.04943 | 0.061784 | Ph2 | 0.562009 |
| retinol-binding protein 4 | 0.056336 | −0.07116 | Ph1 | 0.49437 |
| serpin A3–8 | −0.21267 | 0.265832 | Ph2 | −0.39709 |
| alpha-2-macroglobulin isoform X3 | −0.22798 | 0.28498 | Ph2 | 0.350947 |
| thyroxine-binding globulin precursor | 0.11401 | −0.10801 | Ph1 | 0.255706 |
| hypothetical protein JEQ12_010483 | −0.35937 | 0.449214 | Ph2 | −0.18727 |
| carboxypeptidase N subunit 2 | 0.119992 | −0.10736 | Ph1 | 0.141224 |
| PHD finger protein 21A isoform X13 | 0.07898 | −0.09478 | Ph1 | 0.133177 |
| immunoglobulin lambda variable 1–40 isoform X18 | 0.047427 | −0.06324 | Ph1 | 0.130715 |
| hypothetical protein JEQ12_012143 | 0.229434 | −0.28679 | Ph1 | 0.060661 |
| (A) p < 0.1 among Ph1 and Ph2 | Ph1/Ph2 | Time | Interaction | Constant |
|---|---|---|---|---|
| immunoglobulin J chain | 0.9 (−0.09, 1.88), p < 0.1 | 0.002 (−0.13, 0.14) | −0.02 (−0.22, 0.18) | 9.72 (9.06, 10.37), p < 0.01 |
| heparin cofactor 2 | 0.9 (−0.0004, 1.80), p < 0.1 | −0.09 (−0.27, 0.09) | −0.12 (−0.39, 0.15) | 10.17 (9.57, 10.77), p < 0.01 |
| immunoglobulin lambda-1 light chain isoform X47 | −1.35 (−2.85, 0.16), p < 0.1 | −0.43 (−1.38, 0.53) | 0.41 (−1.02, 1.84) | 5.37 (4.37, 6.37), p < 0.01 |
| angiotensinogen | 0.88 (−0.14, 1.89), p < 0.1 | −0.12 (−0.30, 0.06) | −0.16 (−0.43, 0.11) | 10.16 (9.48, 10.84), p < 0.01 |
| hypothetical protein JEQ12_001510 | 0.84 (0.18, 1.50), p < 0.05 | 0.06 (−0.06, 0.17) | −0.11 (−0.28, 0.06) | 11.02 (10.57, 11.46), p < 0.01 |
| (B) p < 0.1 over time | Ph1/Ph2 | Time | Interaction | Constant |
| apolipoprotein C-III | −0.14 (−1.73, 1.46) | −0.48 (−0.79, −0.16), p < 0.01 | 0.17 (−0.31, 0.64) | 9.51 (8.45, 10.58), p < 0.01 |
| ceruloplasmin isoform X2 | 0.09 (−0.78, 0.96) | 0.1 (−0.02, 0.23), p < 0.1 | 0.02 (−0.17, 0.20) | 12.34 (11.76, 12.91), p < 0.01 |
| complement C4-like isoform X1 | 0.01 (−1.06, 1.09) | −0.25 (−0.43, −0.06), p < 0.05 | −0.03 (−0.31, 0.26) | 11.02 (10.31, 11.74), p < 0.01 |
| inter-alpha-trypsin inhibitor heavy chain H2 isoform X2 | 0.61 (−0.14, 1.36) | −0.22 (−0.33, −0.12), p < 0.01 | −0.02 (−0.18, 0.14) | 11.57 (11.07, 12.07), p < 0.01 |
| prothrombin precursor | 0.34 (−1.10, 1.79) | −0.27 (−0.48, −0.05), p < 0.05 | 0.08 (−0.24, 0.40) | 10.98 (10.02, 11.95), p < 0.01 |
| serpin A3–7 isoform X2 | 0.16 (−0.63, 0.95) | 0.19 (0.05, 0.34), p < 0.01 | −0.1 (−0.32, 0.12) | 10.36 (9.83, 10.88), p < 0.01 |
| serpin A3–8 | 0..35 (−0.84, 1.53) | 0.47 (0.28, 0.65), p < 0.01 | 0.07 (−0.20, 0.35) | 8.86 (8.07, 9.65), p < 0.01 |
| serpin A3–6-like | −1.55 (−4.04, 0.93) | −0.42 (−0.89, 0.05), p < 0.1 | 0.28 (−0.40, 0.95) | 9.63 (7.92, 11.35), p < 0.01 |
| serum amyloid A protein | 0.29 (−1.64, 2.21) | 1.42 (1.06, 1.79), p < 0.01 | −0.03 (−0.55, 0.49) | 4.09 (2.73, 5.46), p < 0.01 |
| glutathione peroxidase 3 | −0.39 (−1.82, 1.04) | 0.27 (−0.01, 0.55), p < 0.1 | 0.31 (−0.07, 0.70) | 5.69 (4.64, 6.74), p < 0.01 |
| synaptotagmin-like protein 4 isoform X3 | −0.21 (−1.10, 0.69) | 0.18 (0.01, 0.34), p < 0.05 | −0.01 (−0.26, 0.24) | 16.43 (15.84, 17.03), p < 0.01 |
| serum paraoxonase/arylesterase 1 isoform X1 | 0.35 (−0.38, 1.08) | −0.11 (−0.22, 0.01), p < 0.1 | −0.1 (−0.27, 0.08) | 11.6 (11.11, 12.08), p < 0.01 |
| transthyretin precursor | −1.35 (−4.77, 2.06) | −0.48 (−0.97, 0.01), p < 0.1 | 0.1 (−0.67, 0.87) | 11.47 (9.21, 13.73), p < 0.01 |
| lumican | 0.2 (−1.22, 1.62) | 0.35 (0.07, 0.62), p < 0.05 | 0.47 (0.06, 0.87), p < 0.05 | 6.57 (5.08, 7.02), p < 0.01 |
| zinc finger protein 264-like isoform X1 | 0.28 (−0.58, 1.14) | 0.17 (−0.01, 0.36), p < 0.1 | −0.2 (−0.46, 0.06) | 12.15 (11.54, 12.75), p < 0.01 |
| Hemopexin | −0.8 (−2.22, 0.61) | −0.38 (−0.63, −0.14), p < 0.01 | 0.19 (−0.18, 0.55) | 12.93 (11.98, 13.87), p < 0.01 |
| complement C3 | 0.08 (−0.66, 0.82) | −0.12 (−0.25, 0.01), p < 0.1 | −0.001 (−0.20, 0.20) | 9.53 (9.03, 10.02), p < 0.01 |
| hemoglobin subunit beta | −1.74 (−4.02, 0.53) | 0.44 (0.02, 0.85), p < 0.05 | 0.17 (−0.45, 0.80) | 13.25 (11.74, 14.77), p < 0.01 |
| apolipoprotein A-II | 0.58 (−0.45, 1.61) | −0.18 (−0.38, 0.03), p < 0.1 | −0.08 (−0.39, 0.23) | 12.09 (11.40, 12.78), p < 0.01 |
| PHD finger protein 21A isoform X13 | −0.05 (−1.18, 1.08) | 0.22 (0.001, 0.44), p < 0.05 | −0.04 (−0.37, 0.29) | 14.89 (14.13, 15.64), p < 0.01 |
| hypothetical protein JEQ12_002713 | 0.54 (−0.15, 1.22) | −0.11 (−0.24, 0.01), p < 0.1 | 0.03 (−0.16, 0.22) | 12.24 (11.78, 12.69), p < 0.01 |
| hypothetical protein JEQ12_008125 | −0.42 (−1.05, 0.22) | −0.15 (−0.26, −0.04), p < 0.01 | 0.14 (−0.03, 0.31) | 17.7 (17.28, 18.12), p < 0.01 |
| hypothetical protein JEQ12_008126 | 0.4 (−0.19, 1.00) | −0.12 (−0.21, −0.02), p < 0.05 | 0.08 (−0.06, 0.22) | 15.48 (15.09, 15.88), p < 0.01 |
| hypothetical protein JEQ12_008387 | 0.34 (−0.33, 1.01) | 0.53 (0.39, 0.66), p < 0.01 | −0.05 (−0.25, 0.15) | 10.18 (9.73, 10.62), p < 0.01 |
| hypothetical protein JEQ12_005133 | −0.17 (−1.93, 1.58) | 1.72 (1.39, 2.04), p < 0.01 | 0.02 (−0.49, 0.53) | 3.73 (2.63, 4.83), p < 0.01 |
| hypothetical protein JEQ12_003887 | 0.59 (−0.72, 1.91) | −0.22 (−0.46, 0.02), p < 0.1 | 0.03 (−0.33, 0.38) | 13.07 (12.20, 13.95), p < 0.01 |
| hypothetical protein JEQ12_012143 | −0.51 (−2.55, 1.53) | 0.58 (0.17, 0.99), p < 0.01 | −0.13 (−0.74, 0.49) | 13.01 (11.65, 14.37), p < 0.01 |
| (C) p < 0.1 for group:time interaction | Ph1/Ph2 | Time | Interaction | Constant |
| lumican | 0.2 (−1.22, 1.62) | 0.35 (0.07, 0.62), p < 0.05 | 0.47 (0.06, 0.87), p < 0.05 | 6.57 (5.08, 7.02), p < 0.01 |
| carbonic anhydrase 2 | −1.36 (−3.16, 0.43) | 0.17 (−0.13, 0.47) | 0.39 (−0.06, 0.83), p < 0.1 | 9.62 (8.41, 10.83), p < 0.01 |
| Input in BLAST | Identified Protein | Accession Number | Perc. Identity | Query Cover |
|---|---|---|---|---|
| hypothetical protein JEQ12_001510 | kininogen-1 isoform X2 | XP_004003107.2 | 99.77% | 100% |
| hypothetical protein JEQ12_002713 | primary amine oxidase, liver isozyme | XP_027830273.2 | 99.86% | 100% |
| hypothetical protein JEQ12_008126 | immunoglobulin mu chain | AAA51379.1 | 99.79% | 73% |
| hypothetical protein JEQ12_008387 | inter-alpha-trypsin inhibitor heavy chain H4 isoform X2 | XP_004018440.3 | 99.56% | 100% |
| hypothetical protein JEQ12_005133 | haptoglobin isoform X2 | XP_004015160.1 | 99.75% | 100% |
| hypothetical protein JEQ12_003887 | antithrombin-III precursor | NP_001009393.1 | 99.57% | 100% |
| hypothetical protein JEQ12_012143 | hemoglobin subunit alpha | EGW10374.1 | 100% | 97% |
| hypothetical protein JEQ12_008125 | Ig gamma 1 chain | CAA49451.1 | 99.70% | 81% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wildi, K.; Bouquet, M.; Ainola, C.; Livingstone, S.; Colombo, S.M.; Heinsar, S.; Sato, N.; Sato, K.; Wilson, E.; Abbate, G.; et al. Differential Protein Expression among Two Different Ovine ARDS Phenotypes—A Preclinical Randomized Study. Metabolites 2022, 12, 655. https://doi.org/10.3390/metabo12070655
Wildi K, Bouquet M, Ainola C, Livingstone S, Colombo SM, Heinsar S, Sato N, Sato K, Wilson E, Abbate G, et al. Differential Protein Expression among Two Different Ovine ARDS Phenotypes—A Preclinical Randomized Study. Metabolites. 2022; 12(7):655. https://doi.org/10.3390/metabo12070655
Chicago/Turabian StyleWildi, Karin, Mahe Bouquet, Carmen Ainola, Samantha Livingstone, Sebastiano Maria Colombo, Silver Heinsar, Noriko Sato, Kei Sato, Emily Wilson, Gabriella Abbate, and et al. 2022. "Differential Protein Expression among Two Different Ovine ARDS Phenotypes—A Preclinical Randomized Study" Metabolites 12, no. 7: 655. https://doi.org/10.3390/metabo12070655
APA StyleWildi, K., Bouquet, M., Ainola, C., Livingstone, S., Colombo, S. M., Heinsar, S., Sato, N., Sato, K., Wilson, E., Abbate, G., Passmore, M. R., Hyslop, K., Liu, K., Li Bassi, G., Suen, J. Y., & Fraser, J. F. (2022). Differential Protein Expression among Two Different Ovine ARDS Phenotypes—A Preclinical Randomized Study. Metabolites, 12(7), 655. https://doi.org/10.3390/metabo12070655







