Salivary Metabolomic Analysis Reveals Amino Acid Metabolism Shift in SARS-CoV-2 Virus Activity and Post-Infection Condition
Abstract
:1. Introduction
2. Experimental Design
2.1. Subjects and Saliva Collection
2.2. Sample Preparation and NMR Measurements
2.3. Statistical Analysis
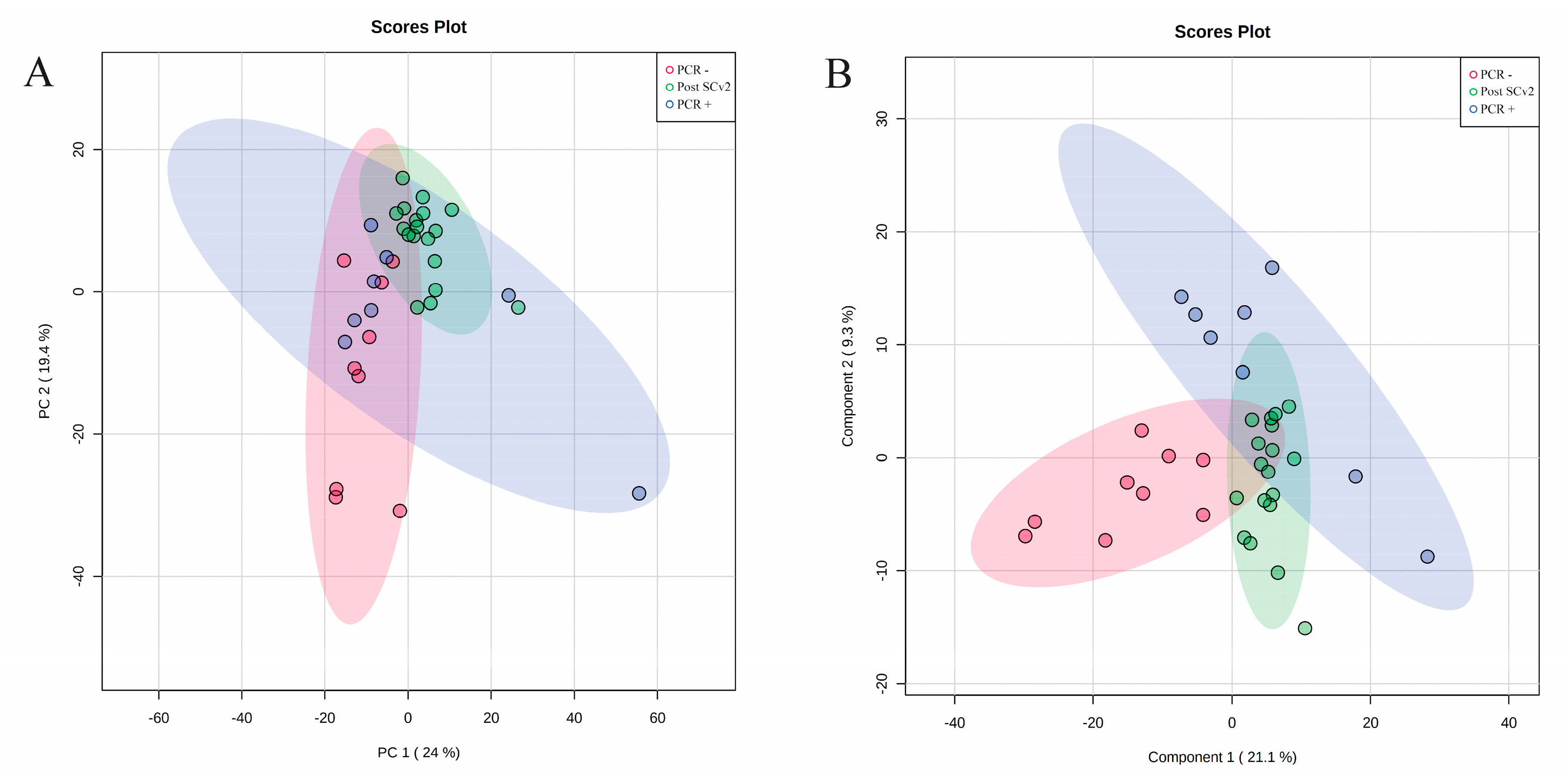
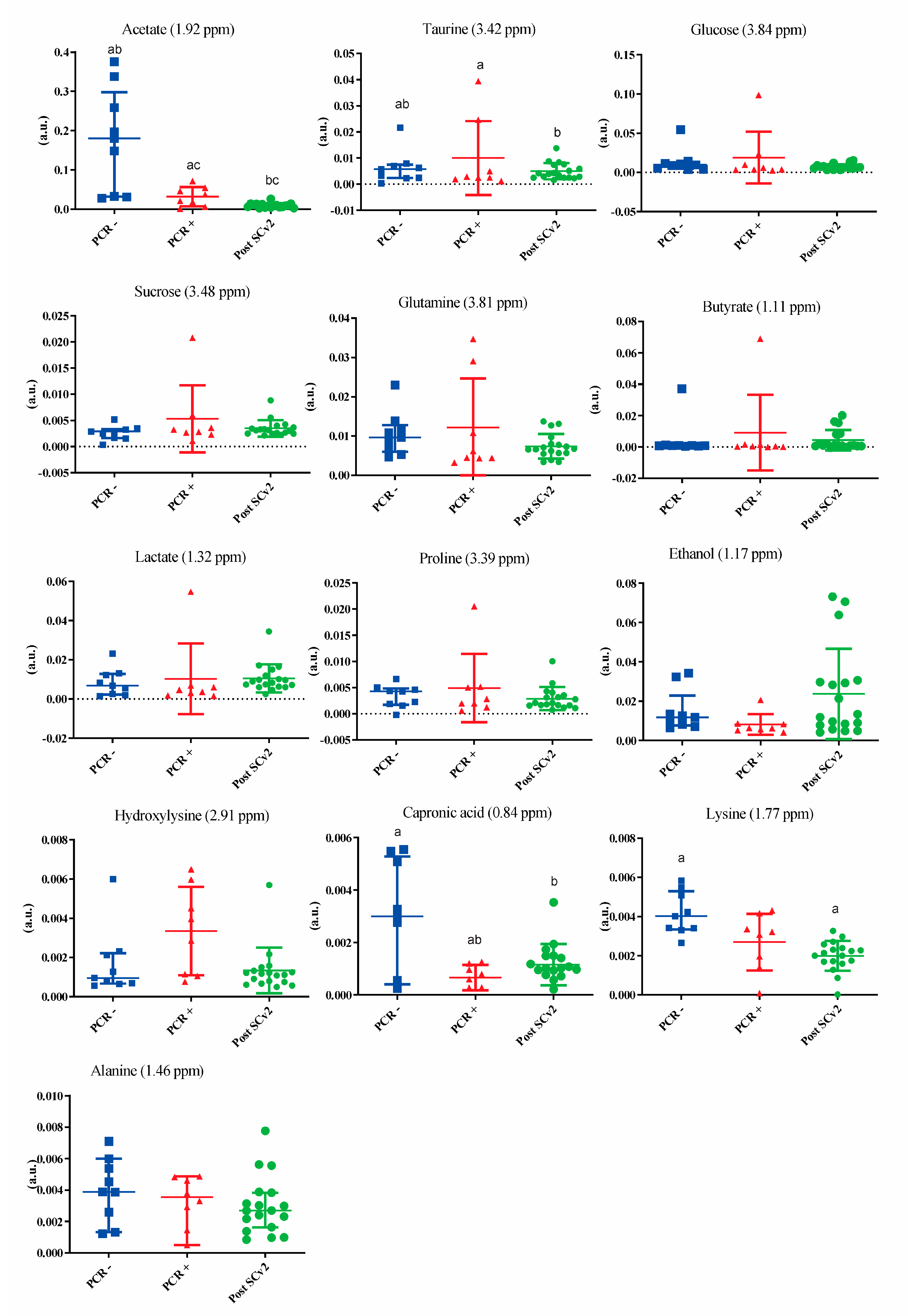
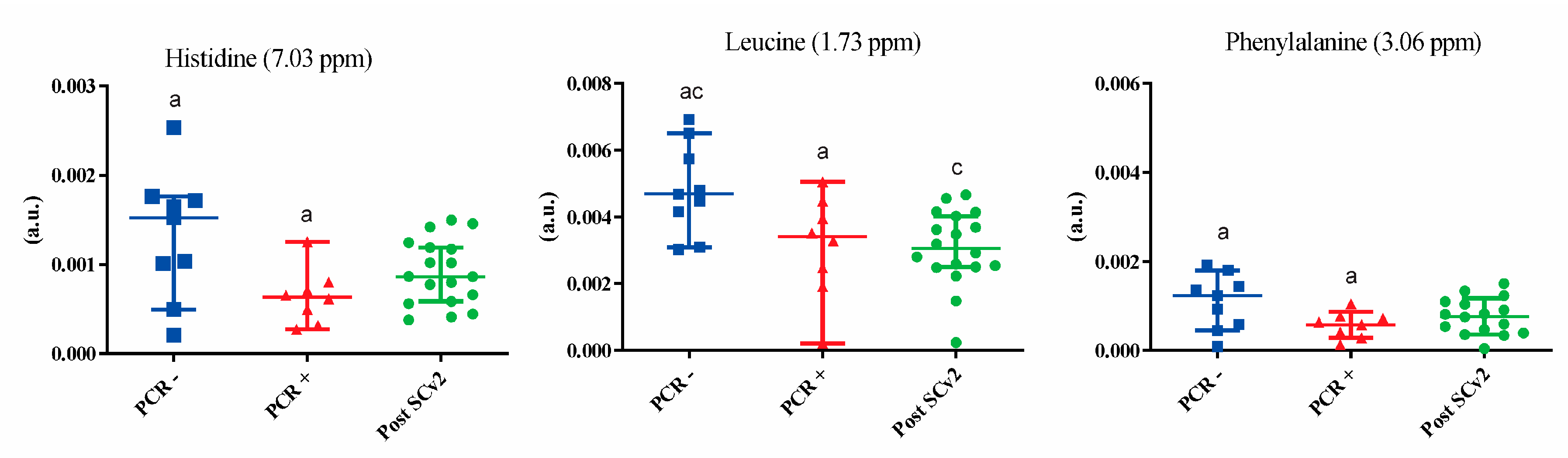
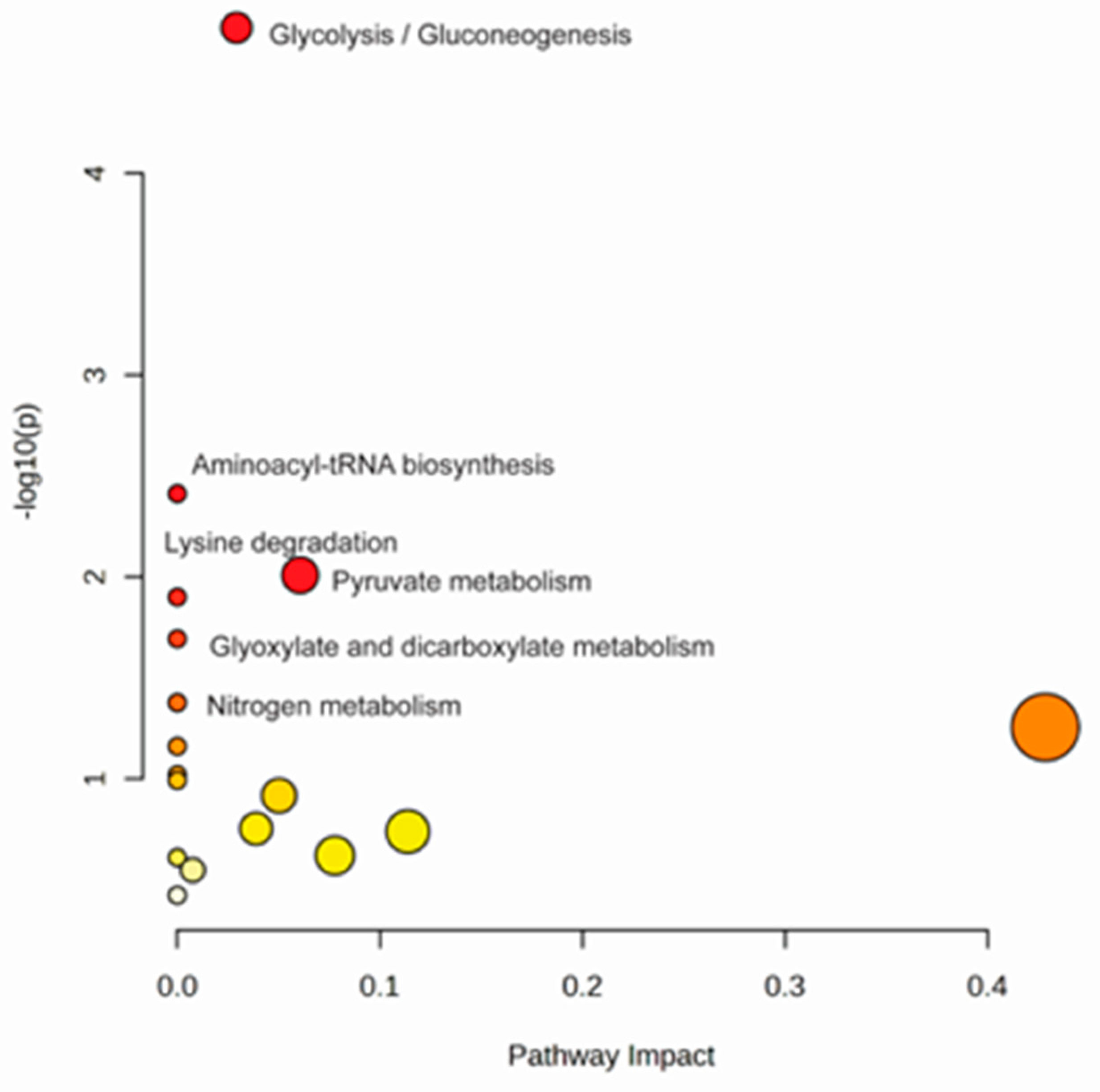
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hopkings, J. Mortality Analysis. Available online: https://coronavirus.jhu.edu/data/mortality (accessed on 29 September 2022).
- Worldometer. Coronavirus. Available online: https://www.worldometers.info/coronavirus/country/brazil/ (accessed on 29 September 2022).
- Globo, O. Ranking da Covid: Como o Brasil se compara a outros países em mortes, casos e vacinas aplicadas. São Paulo G 2021, 1, 2021. [Google Scholar]
- Globo, O. Governo Bolsonaro Pagou Influenciadores Para Defender Atendimento Precoce Contra COVID-19, diz Agência. 31 March 2021. 2022. Available online: https://apublica.org/2021/03/influenciadores-digitais-receberam-r-23-mil-do-governo-bolsonaro-para-propagandear-atendimento-precoce-contra-covid-19/ (accessed on 24 November 2022).
- Ferrante, L.; Duczmal, L.; Steinmetz, W.A.; Almeida, A.C.L.; Leão, J.; Vassão, R.C.; Tupinambás, U.; Fearnside, P.M. How Brazil’s President turned the country into a global epicenter of COVID-19. J. Public Health Policy 2021, 42, 439–451. [Google Scholar] [CrossRef]
- Lima, D.P.; Diniz, D.G.; Moimaz, S.A.; Sumida, D.H.; Okamoto, A.C. Saliva: Reflection of the body. Int. J. Infect. Dis. 2010, 14, e184–e188. [Google Scholar] [CrossRef] [PubMed]
- Mandel, I.D. The role of saliva in maintaining oral homeostasis. J. Am. Dent. Assoc. 1989, 119, 298–304. [Google Scholar] [CrossRef]
- Xu, H.; Zhong, L.; Deng, J.; Peng, J.; Dan, H.; Zeng, X.; Li, T.; Chen, Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral. Sci. 2020, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Salas Orozco, M.F.; Nino-Martinez, N.; Martinez-Castanon, G.A.; Patino Marin, N.; Samano Valencia, C.; Dipp Velazquez, F.A.; Sosa Munguia, P.D.C.; Casillas Santana, M.A. Presence of SARS-CoV-2 and Its Entry Factors in Oral Tissues and Cells: A Systematic Review. Medicina 2021, 57, 523. [Google Scholar] [CrossRef] [PubMed]
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wei, Q.; Alvarez, X.; Wang, H.; Du, Y.; Zhu, H.; Jiang, H.; Zhou, J.; Lam, P.; Zhang, L.; et al. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J. Virol. 2011, 85, 4025–4030. [Google Scholar] [CrossRef] [PubMed]
- Proctor, G.B.; Shaalan, A.M. Disease-Induced Changes in Salivary Gland Function and the Composition of Saliva. J. Dent. Res. 2021, 100, 1201–1209. [Google Scholar] [CrossRef]
- Costa Dos Santos Junior, G.; Pereira, C.M.; Kelly da Silva Fidalgo, T.; Valente, A.P. Saliva NMR-Based Metabolomics in the War Against COVID-19. Anal. Chem. 2020, 92, 15688–15692. [Google Scholar] [CrossRef] [PubMed]
- Danlos, F.X.; Grajeda-Iglesias, C.; Durand, S.; Sauvat, A.; Roumier, M.; Cantin, D.; Colomba, E.; Rohmer, J.; Pommeret, F.; Baciarello, G.; et al. Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death Dis. 2021, 12, 258. [Google Scholar] [CrossRef]
- Valdes, A.; Moreno, L.O.; Rello, S.R.; Orduna, A.; Bernardo, D.; Cifuentes, A. Metabolomics study of COVID-19 patients in four different clinical stages. Sci. Rep. 2022, 12, 1650. [Google Scholar] [CrossRef]
- Luporini, R.L.; Pott-Junior, H.; Di Medeiros Leal, M.C.B.; Castro, A.; Ferreira, A.G.; Cominetti, M.R.; de Freitas Anibal, F. Phenylalanine and COVID-19: Tracking disease severity markers. Int. Immunopharmacol. 2021, 101, 108313. [Google Scholar] [CrossRef] [PubMed]
- Bruzzone, C.; Bizkarguenaga, M.; Gil-Redondo, R.; Diercks, T.; Arana, E.; Garcia de Vicuna, A.; Seco, M.; Bosch, A.; Palazon, A.; San Juan, I.; et al. SARS-CoV-2 Infection Dysregulates the Metabolomic and Lipidomic Profiles of Serum. IScience 2020, 23, 101645. [Google Scholar] [CrossRef]
- Lawler, N.G.; Gray, N.; Kimhofer, T.; Boughton, B.; Gay, M.; Yang, R.; Morillon, A.C.; Chin, S.T.; Ryan, M.; Begum, S.; et al. Systemic Perturbations in Amine and Kynurenine Metabolism Associated with Acute SARS-CoV-2 Infection and Inflammatory Cytokine Responses. J. Proteome Res. 2021, 20, 2796–2811. [Google Scholar] [CrossRef]
- Silwood, C.J.; Lynch, E.; Claxson, A.W.; Grootveld, M.C. 1H and (13)C NMR spectroscopic analysis of human saliva. J. Dent. Res. 2002, 81, 422–427. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, L.R.; Martins, C.; Fidalgo, T.K.; Freitas-Fernandes, L.B.; de Oliveira Torres, R.; Soares, A.L.; Almeida, F.C.; Valente, A.P.; de Souza, I.P. Salivary Metabolite Fingerprint of Type 1 Diabetes in Young Children. J. Proteome Res. 2016, 15, 2491–2499. [Google Scholar] [CrossRef]
- Almeida, P.A.; Fidalgo, T.K.S.; Freitas-Fernandes, L.B.; Almeida, F.C.L.; Valente, A.P.; Souza, I.P.R. Salivary metabolic profile after hemodialysis among children and adolescents with chronic kidney disease. Metabolomics 2017, 13, 141–150. [Google Scholar] [CrossRef]
- Freitas-Fernandes, L.B.; Fidalgo, T.K.S.; de Almeida, P.A.; Souza, I.P.R.; Valente, A.P. Salivary metabolome of children and adolescents under peritoneal dialysis. Clin. Oral Investig. 2021, 25, 2345–2351. [Google Scholar] [CrossRef]
- Fidalgo, T.K.S.; Freitas-Fernandes, L.B.; Angeli, R.; Muniz, A.M.S.; Gonsalves, E.; Santos, R.; Nadal, J.; Almeida, J.C.L.; Valente, A.P.; Souza, I.P.R. Salivary metabolite signatures of children with and without dental caries lesions. Metabolomics 2013, in press. [Google Scholar] [CrossRef]
- Fidalgo, T.K.S.; Freitas-Fernandes, L.B.; Almeida, F.C.L.; Valente, A.P.; Souza, I.P.R. Longitudinal evaluation of salivary profile from children with dental caries before and after treatment. Metabolomics 2014, in press. [Google Scholar] [CrossRef]
- Ramadan, Z.; Jacobs, D.; Grigorov, M.; Kochhar, S. Metabolic profiling using principal component analysis, discriminant partial least squares, and genetic algorithms. Talanta 2006, 68, 1683–1691. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- van Velzen, E.J.; Westerhuis, J.A.; van Duynhoven, J.P.; van Dorsten, F.A.; Hoefsloot, H.C.; Jacobs, D.M.; Smit, S.; Draijer, R.; Kroner, C.I.; Smilde, A.K. Multilevel data analysis of a crossover designed human nutritional intervention study. J. Proteome Res. 2008, 7, 4483–4491. [Google Scholar] [CrossRef]
- Li, H.; Li, X.; Wu, Q.; Wang, X.; Qin, Z.; Wang, Y.; He, Y.; Wu, Q.; Li, L.; Chen, H. Plasma proteomic and metabolomic characterization of COVID-19 survivors 6 months after discharge. Cell Death Dis. 2022, 13, 235. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yin, Y.L.; Li, D.; Kim, S.W.; Wu, G. Amino acids and immune function. Br. J. Nutr. 2007, 98, 237–252. [Google Scholar] [CrossRef]
- Obled, C. Amino Acid Requirements in Inflammatory States; INRA, Theix, Unité de Nutrition et Métabolisme Protéique: Saint Genès Champanelle, France, 2003. [Google Scholar]
- Passlack, N.; Doherr, M.G.; Zentek, J. Effects of free amino acids on cytokine secretion and proliferative activity of feline T cells in an in vitro study using the cell line MYA-1. Cytotechnology 2016, 68, 1949–1961. [Google Scholar] [CrossRef] [PubMed]
- Masoodi, M.; Peschka, M.; Schmiedel, S.; Haddad, M.; Frye, M.; Maas, C.; Lohse, A.; Huber, S.; Kirchhof, P.; Nofer, J.R.; et al. Disturbed lipid and amino acid metabolisms in COVID-19 patients. J. Mol. Med. 2022, 100, 555–568. [Google Scholar] [CrossRef]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5, e140327. [Google Scholar] [CrossRef]
- Wu, P.; Chen, D.; Ding, W.; Wu, P.; Hou, H.; Bai, Y.; Zhou, Y.; Li, K.; Xiang, S.; Liu, P.; et al. The trans-omics landscape of COVID-19. Nat. Commun. 2021, 12, 4543. [Google Scholar] [CrossRef]
- Miyajima, M. Amino acids: Key sources for immunometabolites and immunotransmitters. Int. Immunol. 2020, 32, 435–446. [Google Scholar] [CrossRef]
- Ozturk, A.; Bayraktar, N.; Bayraktar, M.; Ibrahim, B.; Bozok, T.; Resat, C.M. Evaluation of amino acid profile in serum of patients with Covid-19 for providing a new treatment strategy. J. Med. Biochem. 2022, 41, 526–533. [Google Scholar] [CrossRef]
- Flaherty, G.T.; Hession, P.; Liew, C.H.; Lim, B.C.W.; Leong, T.K.; Lim, V.; Sulaiman, L.H. COVID-19 in adult patients with pre-existing chronic cardiac, respiratory and metabolic disease: A critical literature review with clinical recommendations. Trop. Dis. Travel Med. Vaccines 2020, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Paez-Franco, J.C.; Torres-Ruiz, J.; Sosa-Hernandez, V.A.; Cervantes-Diaz, R.; Romero-Ramirez, S.; Perez-Fragoso, A.; Meza-Sanchez, D.E.; German-Acacio, J.M.; Maravillas-Montero, J.L.; Mejia-Dominguez, N.R.; et al. Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID-19 patients. Sci. Rep. 2021, 11, 6350. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Wang, T. Metabolic Reprogramming in COVID-19. Int. J. Mol. Sci. 2021, 22, 11475. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Perlman, S. Immune responses in influenza A virus and human coronavirus infections: An ongoing battle between the virus and host. Curr. Opin. Virol. 2018, 28, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Kieninger, M.; Sinning, A.; Vadasz, T.; Gruber, M.; Gronwald, W.; Zeman, F.; Lunz, D.; Dienemann, T.; Schmid, S.; Graf, B.; et al. Lower blood pH as a strong prognostic factor for fatal outcomes in critically ill COVID-19 patients at an intensive care unit: A multivariable analysis. PLoS ONE 2021, 16, e0258018. [Google Scholar] [CrossRef]
- Wannemacher, R.W., Jr.; Klainer, A.S.; Dinterman, R.E.; Beisel, W.R. The significance and mechanism of an increased serum phenylalanine-tyrosine ratio during infection. Am. J. Clin. Nutr. 1976, 29, 997–1006. [Google Scholar] [CrossRef]
- Barberis, E.; Timo, S.; Amede, E.; Vanella, V.V.; Puricelli, C.; Cappellano, G.; Raineri, D.; Cittone, M.G.; Rizzi, E.; Pedrinelli, A.R.; et al. Large-Scale Plasma Analysis Revealed New Mechanisms and Molecules Associated with the Host Response to SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 8623. [Google Scholar] [CrossRef]
- Kimura-Ohba, S.; Asaka, M.N.; Utsumi, D.; Takabatake, Y.; Takahashi, A.; Yasutomi, Y.; Isaka, Y.; Kimura, T. d-Alanine as a biomarker and a therapeutic option for severe influenza virus infection and COVID-19. Biochim. Biophys. Acta Mol. Basis Dis. 2023, 1869, 166584. [Google Scholar] [CrossRef] [PubMed]
- Letieri, A.S.; Freitas-Fernandes, L.B.; Souza, I.P.R.; Valente, A.P.; Fidalgo, T.K.S. Metabolomic Signatures of In Vitro Biofilm Maturation of Streptococcus mutans. Curr. Microbiol. 2022, 79, 86. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Liu, C.; Li, D.; Huang, Q.; Liu, D.; Zhang, Y.; Ye, C.; Zhou, D.; Wang, Y.; Tan, Y.; et al. Metabolomic analyses reveal new stage-specific features of COVID-19. Eur. Respir. J. 2022, 59, 2100284. [Google Scholar] [CrossRef] [PubMed]
| Signals/Symptoms | PCR− | PCR+ | Post-SARS-CoV-2 Infection |
| Smokers Ex-smokers | 0% (n = 0) 0% (n = 0) | 0% (n = 0) 0% (n = 0) | 0% (n = 0) 44.44 (n = 8) |
| Lost smell or taste | 0% (n = 0) | 25.0% (n = 2) | 55.55% (n = 10) |
| Fever | 0% (n = 0) | 37.5% (n= 3) | 44.44 (n = 8) |
| Caught | 22.22 (n = 2) | 37.5% (n = 3) | 100% (n = 18) |
| Malaise symptom | 33.33% (n = 3) | 62.5% (n = 5) | 83.33 % (n = 15) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Silva Fidalgo, T.K.; Freitas-Fernandes, L.B.; Marques, B.B.F.; de Araújo, C.S.; da Silva, B.J.; Guimarães, T.C.; Fischer, R.G.; Tinoco, E.M.B.; Valente, A.P. Salivary Metabolomic Analysis Reveals Amino Acid Metabolism Shift in SARS-CoV-2 Virus Activity and Post-Infection Condition. Metabolites 2023, 13, 263. https://doi.org/10.3390/metabo13020263
da Silva Fidalgo TK, Freitas-Fernandes LB, Marques BBF, de Araújo CS, da Silva BJ, Guimarães TC, Fischer RG, Tinoco EMB, Valente AP. Salivary Metabolomic Analysis Reveals Amino Acid Metabolism Shift in SARS-CoV-2 Virus Activity and Post-Infection Condition. Metabolites. 2023; 13(2):263. https://doi.org/10.3390/metabo13020263
Chicago/Turabian Styleda Silva Fidalgo, Tatiana Kelly, Liana Bastos Freitas-Fernandes, Barbara Bruno Fagundes Marques, Caroline Souza de Araújo, Bruno Jefferson da Silva, Taísa Coelho Guimarães, Ricardo Guimarães Fischer, Eduardo Muniz Barretto Tinoco, and Ana Paula Valente. 2023. "Salivary Metabolomic Analysis Reveals Amino Acid Metabolism Shift in SARS-CoV-2 Virus Activity and Post-Infection Condition" Metabolites 13, no. 2: 263. https://doi.org/10.3390/metabo13020263
APA Styleda Silva Fidalgo, T. K., Freitas-Fernandes, L. B., Marques, B. B. F., de Araújo, C. S., da Silva, B. J., Guimarães, T. C., Fischer, R. G., Tinoco, E. M. B., & Valente, A. P. (2023). Salivary Metabolomic Analysis Reveals Amino Acid Metabolism Shift in SARS-CoV-2 Virus Activity and Post-Infection Condition. Metabolites, 13(2), 263. https://doi.org/10.3390/metabo13020263








