A CRR2-Dependent sRNA Sequence Supports Papillomavirus Vaccine Expression in Tobacco Chloroplasts
Abstract
1. Introduction
2. Experimental Design
2.1. Plant Growth
2.2. Vector Construction
2.3. Chloroplast Transformation
2.4. Immunoblot Analysis
2.5. RNA Extraction and RNA Gel Blot Analysis
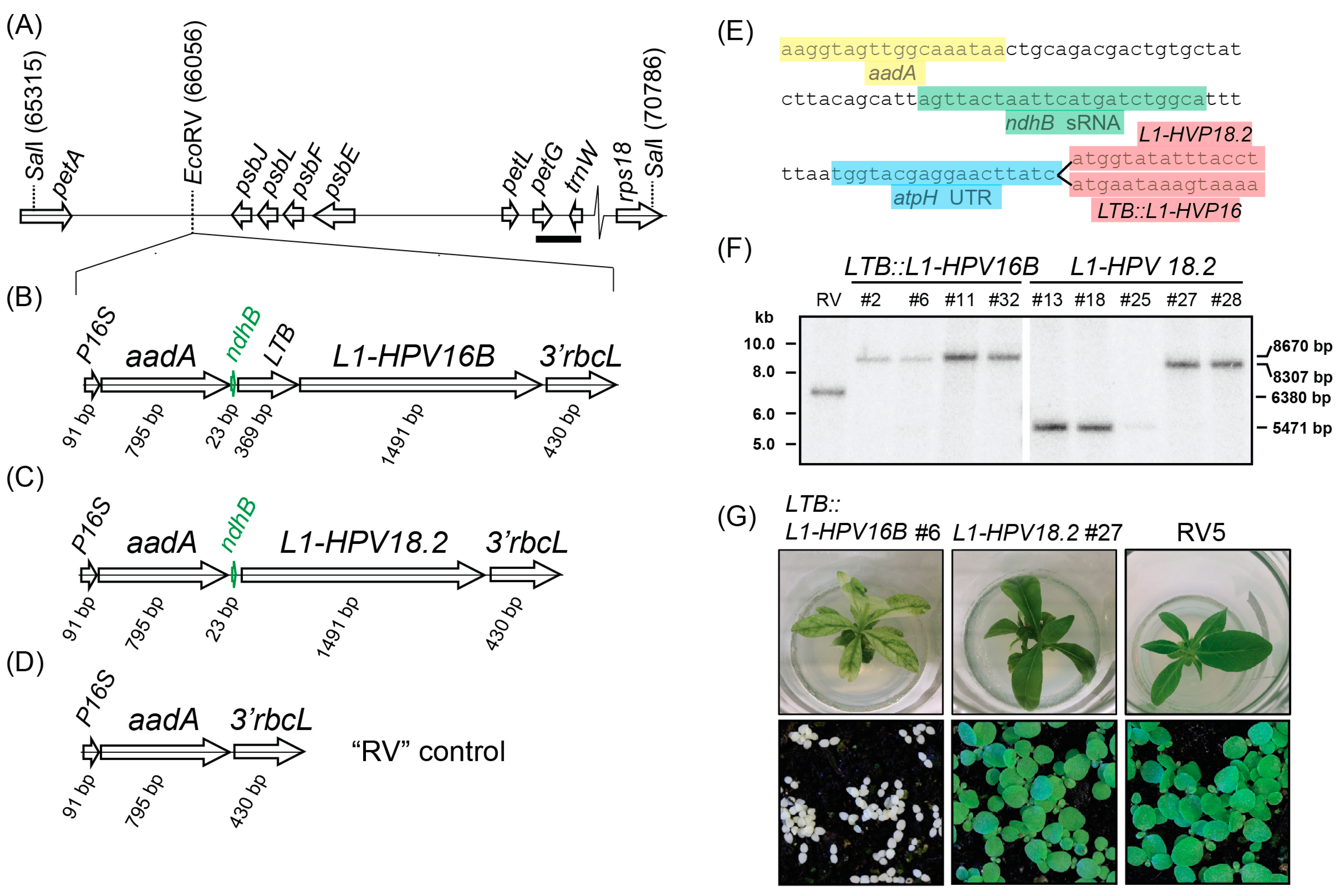
3. Results
3.1. Plant Lines Expressing Enterotoxin-L1 Fusion Proteins Show Impaired Chloroplast and Leaf Development
3.2. The L1 Proteins Accumulated in the Transplastomic Lines
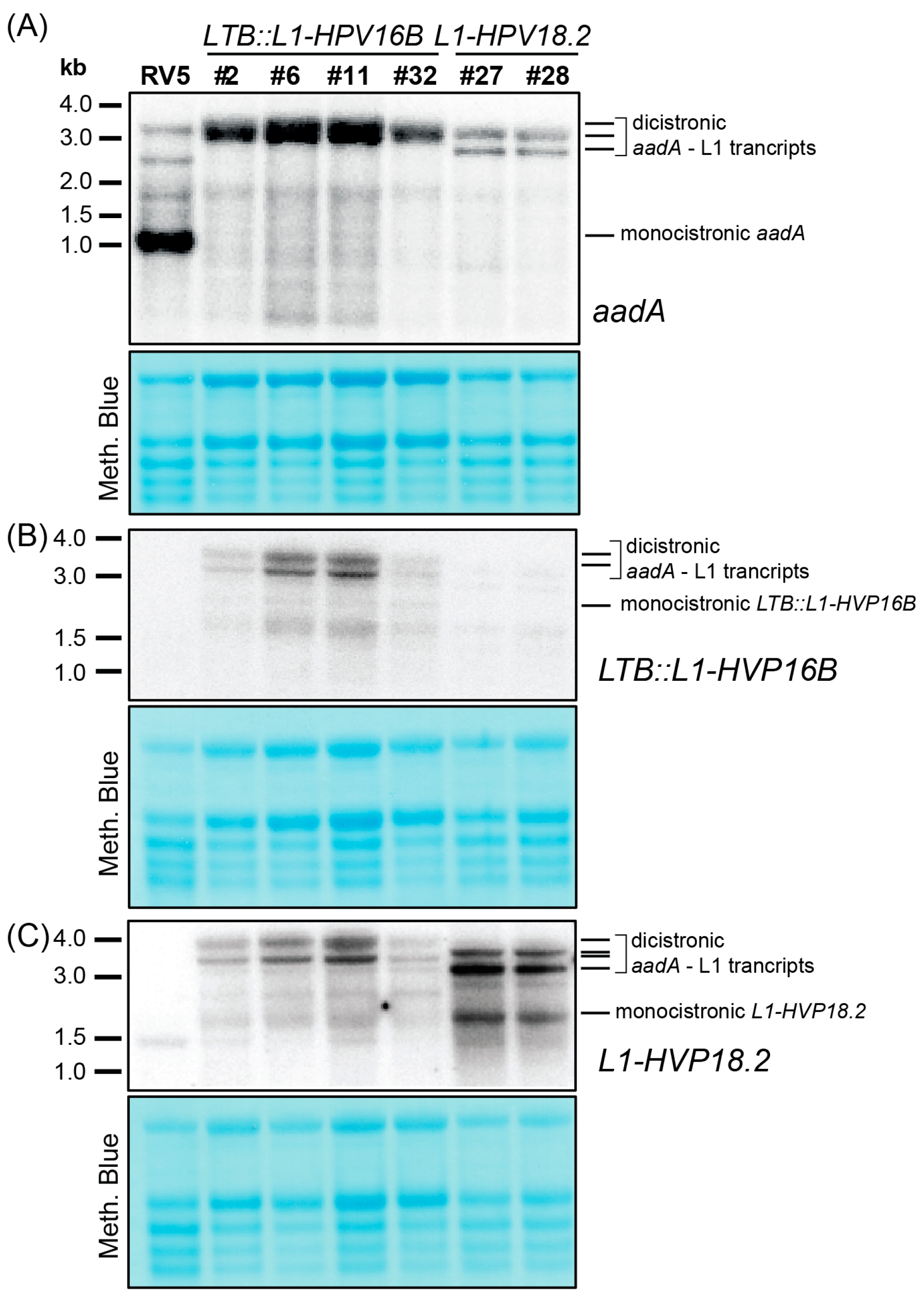
3.3. Mono- and Dicistronic L1 mRNAs Accumulated in Transplastomic Plants
3.4. Increased Expression of ndhB sRNA in Transplastomic Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Twyman, R.M.; Schillberg, S.; Fischer, R. Transgenic Plants in the Biopharmaceutical Market. Expert Opin. Emerg. Drugs 2005, 10, 185–218. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Weng, Y.; Dickey, A.; Wang, K.Y. Plants as Factories for Human Pharmaceuticals: Applications and Challenges. Int. J. Mol. Sci. 2015, 16, 28549–28565. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Michoux, F.; Lössl, A.G.; Nixon, P.J. Challenges and Perspectives in Commercializing Plastid Transformation Technology. J. Exp. Bot. 2016, 67, 5945–5960. [Google Scholar] [CrossRef] [PubMed]
- Maliga, P.; Tungsuchat-Huang, T.; Lutz, K.A. Transformation of the Plastid Genome in Tobacco: The Model System for Chloroplast Genome Engineering. Methods Mol. Biol. 2021, 2317, 135–153. [Google Scholar] [PubMed]
- Zhou, F.; Karcher, D.; Bock, R. Identification of a Plastid Intercistronic Expression Element (IEE) Facilitating the Expression of Stable Translatable Monocistronic mRNAs from Operons. Plant J. 2007, 52, 961–972. [Google Scholar] [CrossRef]
- Lu, Y.; Rijzaani, H.; Karcher, D.; Ruf, S.; Bock, R. Efficient Metabolic Pathway Engineering in Transgenic Tobacco and Tomato Plastids with Synthetic Multigene Operons. Proc. Natl. Acad. Sci. USA 2013, 110, E623–E632. [Google Scholar] [CrossRef]
- Bock, R. Engineering Plastid Genomes: Methods, Tools, and Applications in Basic Research and Biotechnology. Annu. Rev. Plant Biol. 2015, 66, 211–241. [Google Scholar] [CrossRef]
- McBride, K.E.; Svab, Z.; Schaaf, D.J.; Hogan, P.S.; Stalker, D.M.; Maliga, P. Amplification of a Chimeric Bacillus Gene in Chloroplasts Leads to an Extraordinary Level of an Insecticidal Protein in Tobacco. Biotechnology 1995, 13, 362–365. [Google Scholar] [CrossRef]
- Oey, M.; Lohse, M.; Kreikemeyer, B.; Bock, R. Exhaustion of the Chloroplast Protein Synthesis Capacity by Massive Expression of a Highly Stable Protein Antibiotic. Plant J. 2009, 57, 436–445. [Google Scholar] [CrossRef]
- Svab, Z.; Maliga, P. High-Frequency Plastid Transformation in Tobacco by Selection for a Chimeric aadA Gene. Proc. Natl. Acad. Sci. USA 1993, 90, 913–917. [Google Scholar] [CrossRef]
- Ruf, S.; Karcher, D.; Bock, R. Determining the Transgene Containment Level Provided by Chloroplast Transformation. Proc. Natl. Acad. Sci. USA 2007, 104, 6998–7002. [Google Scholar] [CrossRef]
- Thyssen, G.; Svab, Z.; Maliga, P. Exceptional Inheritance of Plastids via Pollen in Nicotiana Sylvestris with No Detectable Paternal Mitochondrial DNA in the Progeny. Plant J. 2012, 72, 84–88. [Google Scholar] [CrossRef]
- Zoschke, R.; Bock, R. Chloroplast Translation: Structural and Functional Organization, Operational Control, and Regulation. Plant Cell 2018, 30, 745–770. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide Repeat Proteins in Plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef] [PubMed]
- Ruwe, H.; Schmitz-Linneweber, C. Short Non-Coding RNA Fragments Accumulating in Chloroplasts: Footprints of RNA Binding Proteins? Nucleic Acids Res. 2012, 40, 3106–3116. [Google Scholar] [CrossRef] [PubMed]
- Pfalz, J.; Bayraktar, O.A.; Prikryl, J.; Barkan, A. Site-Specific Binding of a PPR Protein Defines and Stabilizes 5′ and 3’ mRNA Termini in Chloroplasts. EMBO J. 2009, 28, 2042–2052. [Google Scholar] [CrossRef] [PubMed]
- Legen, J.; Ruf, S.; Kroop, X.; Wang, G.; Barkan, A.; Bock, R.; Schmitz-Linneweber, C. Stabilization and Translation of Synthetic Operon-Derived mRNAs in Chloroplasts by Sequences Representing PPR Protein-Binding Sites. Plant J. 2018, 94, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Endo, T.; Peltier, G.; Tasaka, M.; Shikanai, T. A Nucleus-Encoded Factor, CRR2, Is Essential for the Expression of Chloroplast ndhB in Arabidopsis. Plant J. 2003, 36, 541–549. [Google Scholar] [CrossRef]
- Horváth, E.M.; Peter, S.O.; Joët, T.; Rumeau, D.; Cournac, L.; Horváth, G.V.; Kavanagh, T.A.; Schäfer, C.; Peltier, G.; Medgyesy, P. Targeted Inactivation of the Plastid ndhB Gene in Tobacco Results in an Enhanced Sensitivity of Photosynthesis to Moderate Stomatal Closure. Plant Physiol. 2000, 123, 1337–1350. [Google Scholar] [CrossRef]
- Ruwe, H.; Gutmann, B.; Schmitz-Linneweber, C.; Small, I.; Kindgren, P. The E Domain of CRR2 Participates in Sequence-Specific Recognition of RNA in Plastids. New Phytol. 2019, 222, 218–229. [Google Scholar] [CrossRef]
- Ruwe, H.; Wang, G.; Gusewski, S.; Schmitz-Linneweber, C. Systematic Analysis of Plant Mitochondrial and Chloroplast Small RNAs Suggests Organelle-Specific mRNA Stabilization Mechanisms. Nucleic Acids Res. 2016, 44, 7406–7417. [Google Scholar] [CrossRef] [PubMed]
- Parkin, D.M.; Bray, F. Chapter 2: The Burden of HPV-Related Cancers. Vaccine 2006, 24 (Suppl. S3), 11–25. [Google Scholar] [CrossRef] [PubMed]
- Ginsburg, O.; Bray, F.; Coleman, M.P.; Vanderpuye, V.; Eniu, A.; Kotha, S.R.; Sarker, M.; Huong, T.T.; Allemani, C.; Dvaladze, A.; et al. The Global Burden of Women’s Cancers: A Grand Challenge in Global Health. Lancet 2017, 389, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, N.D.; Parker, J.S.; Eberhard, D.A.; Patel, N.M.; Weck, K.E.; Sharpless, N.E.; Hu, Z.; Hayes, D.N.; Gulley, M.L. Identification of Human Papillomavirus Infection in Cancer Tissue by Targeted Next-Generation Sequencing. Appl. Immunohistochem. Mol. Morphol. 2016, 24, 490–495. [Google Scholar] [CrossRef]
- de Villiers, E.-M. Cross-Roads in the Classification of Papillomaviruses. Virology 2013, 445, 2–10. [Google Scholar] [CrossRef]
- de Sanjose, S.; Quint, W.G.; Alemany, L.; Geraets, D.T.; Klaustermeier, J.E.; Lloveras, B.; Tous, S.; Felix, A.; Bravo, L.E.; Shin, H.-R.; et al. Human Papillomavirus Genotype Attribution in Invasive Cervical Cancer: A Retrospective Cross-Sectional Worldwide Study. Lancet Oncol. 2010, 11, 1048–1056. [Google Scholar] [CrossRef]
- Wardak, S. Human Papillomavirus (HPV) and Cervical Cancer. Med. Dosw. Mikrobiol. 2016, 68, 73–84. [Google Scholar]
- Charde, S.H.; Warbhe, R.A. Human Papillomavirus Prevention by Vaccination: A Review Article. Cureus 2022, 14, e30037. [Google Scholar] [CrossRef]
- Marsian, J.; Lomonossoff, G.P. Molecular Pharming - VLPs Made in Plants. Curr. Opin. Biotechnol. 2016, 37, 201–206. [Google Scholar] [CrossRef]
- Müller, M.; Gissmann, L. A long way: History of the prophylactic papillomavirus vaccine. Dis. Markers. 2007, 23, 331–336. [Google Scholar] [CrossRef]
- Fernández-San Millán, A.; Ortigosa, S.M.; Hervás-Stubbs, S.; Corral-Martínez, P.; Seguí-Simarro, J.M.; Gaétan, J.; Coursaget, P.; Veramendi, J. Human Papillomavirus L1 Protein Expressed in Tobacco Chloroplasts Self-Assembles into Virus-like Particles That Are Highly Immunogenic. Plant Biotechnol. J. 2008, 6, 427–441. [Google Scholar] [CrossRef]
- Waheed, M.T.; Thönes, N.; Müller, M.; Hassan, S.W.; Gottschamel, J.; Lössl, E.; Kaul, H.-P.; Lössl, A.G. Plastid expression of a double-pentameric vaccine candidate containing human papillomavirus-16 L1 antigen fused with LTB as adjuvant: Transplastomic plants show pleiotropic phenotypes. Plant Biotechnol. J. 2011, 9, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Regel, R.E.; Ivleva, N.B.; Zer, H.; Meurer, J.; Shestakov, S.V.; Herrmann, R.G.; Pakrasi, H.B.; Ohad, I. Deregulation of Electron Flow within Photosystem II in the Absence of the PsbJ Protein. J. Biol. Chem. 2001, 276, 41473–41478. [Google Scholar] [CrossRef] [PubMed]
- Koop, H.U.; Steinmüller, K.; Wagner, H.; Rössler, C.; Eibl, C.; Sacher, L. Integration of Foreign Sequences into the Tobacco Plastome via Polyethylene Glycol-Mediated Protoplast Transformation. Planta 1996, 199, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Kupsch, C.; Ruwe, H.; Gusewski, S.; Tillich, M.; Small, I.; Schmitz-Linneweber, C. Arabidopsis Chloroplast RNA Binding Proteins CP31A and CP29A Associate with Large Transcript Pools and Confer Cold Stress Tolerance by Influencing Multiple Chloroplast RNA Processing Steps. Plant Cell 2012, 24, 4266–4280. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W.; Rosenberg, A.H.; Dunn, J.J.; Dubendorff, J.W. Use of T7 RNA Polymerase to Direct Expression of Cloned Genes. Methods Enzymol. 1990, 185, 60–89. [Google Scholar]
- Rojas, M.; Yu, Q.; Williams-Carrier, R.; Maliga, P.; Barkan, A. Engineered PPR Proteins as Inducible Switches to Activate the Expression of Chloroplast Transgenes. Nat. Plants 2019, 5, 505–511. [Google Scholar] [CrossRef]
- Manavski, N.; Mathieu, S.; Rojas, M.; Méteignier, L.-V.; Brachmann, A.; Barkan, A.; Hammani, K. In Vivo Stabilization of Endogenous Chloroplast RNAs by Customized Artificial Pentatricopeptide Repeat Proteins. Nucleic Acids Res. 2021, 49, 5985–5997. [Google Scholar] [CrossRef]
- Waheed, M.T.; Gottschamel, J.; Hassan, S.W.; Lössl, A.G. Plant-Derived Vaccines: An Approach for Affordable Vaccines against Cervical Cancer. Hum. Vaccin. Immunother. 2012, 8, 403–406. [Google Scholar] [CrossRef]
- Yousefi, Z.; Aria, H.; Ghaedrahmati, F.; Bakhtiari, T.; Azizi, M.; Bastan, R.; Hosseini, R.; Eskandari, N. An Update on Human Papilloma Virus Vaccines: History, Types, Protection, and Efficacy. Front. Immunol. 2021, 12, 805695. [Google Scholar] [CrossRef]
- Kombe Kombe, A.J.; Li, B.; Zahid, A.; Mengist, H.M.; Bounda, G.-A.; Zhou, Y.; Jin, T. Epidemiology and Burden of Human Papillomavirus and Related Diseases, Molecular Pathogenesis, and Vaccine Evaluation. Front Public Health 2020, 8, 552028. [Google Scholar] [CrossRef] [PubMed]
- Karcher, D.; Bock, R. The Amino Acid Sequence of a Plastid Protein Is Developmentally Regulated by RNA Editing. J. Biol. Chem. 2002, 277, 5570–5574. [Google Scholar] [CrossRef] [PubMed]
- Waheed, M.T.; Thönes, N.; Müller, M.; Hassan, S.W.; Razavi, N.M.; Lössl, E.; Kaul, H.-P.; Lössl, A.G. Transplastomic Expression of a Modified Human Papillomavirus L1 Protein Leading to the Assembly of Capsomeres in Tobacco: A Step towards Cost-Effective Second-Generation Vaccines. Transgenic Res. 2011, 20, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Lenzi, P.; Scotti, N.; Alagna, F.; Tornesello, M.L.; Pompa, A.; Vitale, A.; De Stradis, A.; Monti, L.; Grillo, S.; Buonaguro, F.M.; et al. Translational Fusion of Chloroplast-Expressed Human Papillomavirus Type 16 L1 Capsid Protein Enhances Antigen Accumulation in Transplastomic Tobacco. Transgenic Res. 2008, 17, 1091–1102. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Tillich, M.; Herrmann, R.G.; Maier, R.M. Heterologous, Splicing-Dependent RNA Editing in Chloroplasts: Allotetraploidy Provides Trans-Factors. EMBO J. 2001, 20, 4874–4883. [Google Scholar] [CrossRef]
- Millar, D.G.; Hirst, T.R.; Snider, D.P. Escherichia Coli Heat-Labile Enterotoxin B Subunit Is a More Potent Mucosal Adjuvant than Its Vlosely Related Homologue, the B Subunit of Cholera Toxin. Infect. Immun. 2001, 69, 3476–3482. [Google Scholar] [CrossRef]
- Sapp, M.; Fligge, C.; Petzak, I.; Harris, J.R.; Streeck, R.E. Papillomavirus Assembly Requires Trimerization of the Major Capsid Protein by Disulfides between Two Highly Conserved Cysteines. J. Virol. 1998, 72, 6186–6189. [Google Scholar] [CrossRef]
- Svab, Z.; Maliga, P. Mutation Proximal to the tRNA Binding Region of the Nicotiana Plastid 16S rRNA Confers Resistance to Spectinomycin. Mol. Gen. Genet. 1991, 228, 316–319. [Google Scholar] [CrossRef]
- Fromm, H.; Edelman, M.; Aviv, D.; Galun, E. The Molecular Basis for rRNA-Dependent Spectinomycin Resistance in Nicotiana Chloroplasts. EMBO J. 1987, 6, 3233–3237. [Google Scholar] [CrossRef]
- Kim, B.-M.; Kang, T.-J. Expression of B Subunit of E. Coli Heat-Labile Enterotoxin in the Progenies of Transgenic Tobacco Bred by Crossing Nuclear- and Chloroplast-Transgenic Lines. Protein Expr. Purif. 2019, 155, 54–58. [Google Scholar] [CrossRef]
- Kang, T.-J.; Han, S.-C.; Kim, M.-Y.; Kim, Y.-S.; Yang, M.-S. Expression of Non-Toxic Mutant of Escherichia Coli Heat-Labile Enterotoxin in Tobacco Chloroplasts. Protein Exp. Purif. 2004, 38, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.-J.; Loc, N.-H.; Jang, M.-O.; Jang, Y.-S.; Kim, Y.-S.; Seo, J.-E.; Yang, M.-S. Expression of the B Subunit of E. Coli Heat-Labile Enterotoxin in the Chloroplasts of Plants and Its Characterization. Transgenic Res. 2003, 12, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.W.; Waheed, M.T.; Müller, M.; Clarke, J.L.; Shinwari, Z.K.; Lössl, A.G. Expression of HPV-16 L1 Capsomeres with Glutathione-S-Transferase as a Fusion Protein in Tobacco Plastids: An Approach for a Capsomere-Based HPV Vaccine. Hum. Vaccin. Immunother. 2014, 10, 2975–2982. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Chan, H.-T.; Pasoreck, E.K. Vaccination via Chloroplast Genetics: Affordable Protein Drugs for the Prevention and Treatment of Inherited or Infectious Human Diseases. Annu. Rev. Genet. 2016, 50, 595–618. [Google Scholar] [CrossRef] [PubMed]
- Emadpour, M.; Karcher, D.; Bock, R. Boosting Riboswitch Efficiency by RNA Amplification. Nucleic Acids Res. 2015, 43, e66. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Legen, J.; Dühnen, S.; Gauert, A.; Götz, M.; Schmitz-Linneweber, C. A CRR2-Dependent sRNA Sequence Supports Papillomavirus Vaccine Expression in Tobacco Chloroplasts. Metabolites 2023, 13, 315. https://doi.org/10.3390/metabo13030315
Legen J, Dühnen S, Gauert A, Götz M, Schmitz-Linneweber C. A CRR2-Dependent sRNA Sequence Supports Papillomavirus Vaccine Expression in Tobacco Chloroplasts. Metabolites. 2023; 13(3):315. https://doi.org/10.3390/metabo13030315
Chicago/Turabian StyleLegen, Julia, Sara Dühnen, Anton Gauert, Michael Götz, and Christian Schmitz-Linneweber. 2023. "A CRR2-Dependent sRNA Sequence Supports Papillomavirus Vaccine Expression in Tobacco Chloroplasts" Metabolites 13, no. 3: 315. https://doi.org/10.3390/metabo13030315
APA StyleLegen, J., Dühnen, S., Gauert, A., Götz, M., & Schmitz-Linneweber, C. (2023). A CRR2-Dependent sRNA Sequence Supports Papillomavirus Vaccine Expression in Tobacco Chloroplasts. Metabolites, 13(3), 315. https://doi.org/10.3390/metabo13030315






