Abstract
Metabolomic research tends to increase in popularity over the years, leading to the identification of new biomarkers related to specific health disorders. Saliva is one of the most newly introduced and systematically developed biofluids in the human body that can serve as an informative substance in the metabolomic profiling armamentarium. This review aims to analyze the current knowledge regarding the human salivary metabolome, its alterations due to physiological, environmental and external factors, as well as the limitations and drawbacks presented in the most recent research conducted, focusing on pre—analytical and analytical workflows. Furthermore, the use of the saliva metabolomic profile as a promising biomarker for several oral pathologies, such as oral cancer and periodontitis will be investigated.
1. Introduction
Saliva is the readily accessible biological fluid of the oral cavity [1]. It is produced by three major pairs of glands and hundreds of minor ones. The three major glands participating in the production of saliva are the parotid, submandibular and sublingual glands, which lead to the classification of parotid saliva, submandibular saliva and sublingual saliva respectively [2]. Additional classification of the secretion of saliva is based on the presence or absence of stimuli leading to its production. More precisely, at rest, unstimulated saliva is produced with its major portion originating from the submandibular glands (almost 70%), whereas stimulated saliva, induced by stimuli such as smell, taste ordrugs originates primarily from the parotid glands [1,3].
The term “whole mouth saliva” (WMS) is used in oral sciences to determine the transparent, clear, watery fluid composed by a mixture of the parotid saliva, the submandibular saliva and the sublingual saliva, combined with the secretions of minor salivary glands, the gingival crevicular fluid, eukaryotic cells (epithelial as well as leukocytic), food debris, microorganisms and their metabolites [4,5,6].
Due to its aqueous composition, saliva mainly consists of water (99% of its composition). Further components, such as mucus, digestive enzymes, growth factors, cytokines, immunoglobulins, antibacterial peptides, salts and low molecular weight metabolites are numbered among salivary products [7].
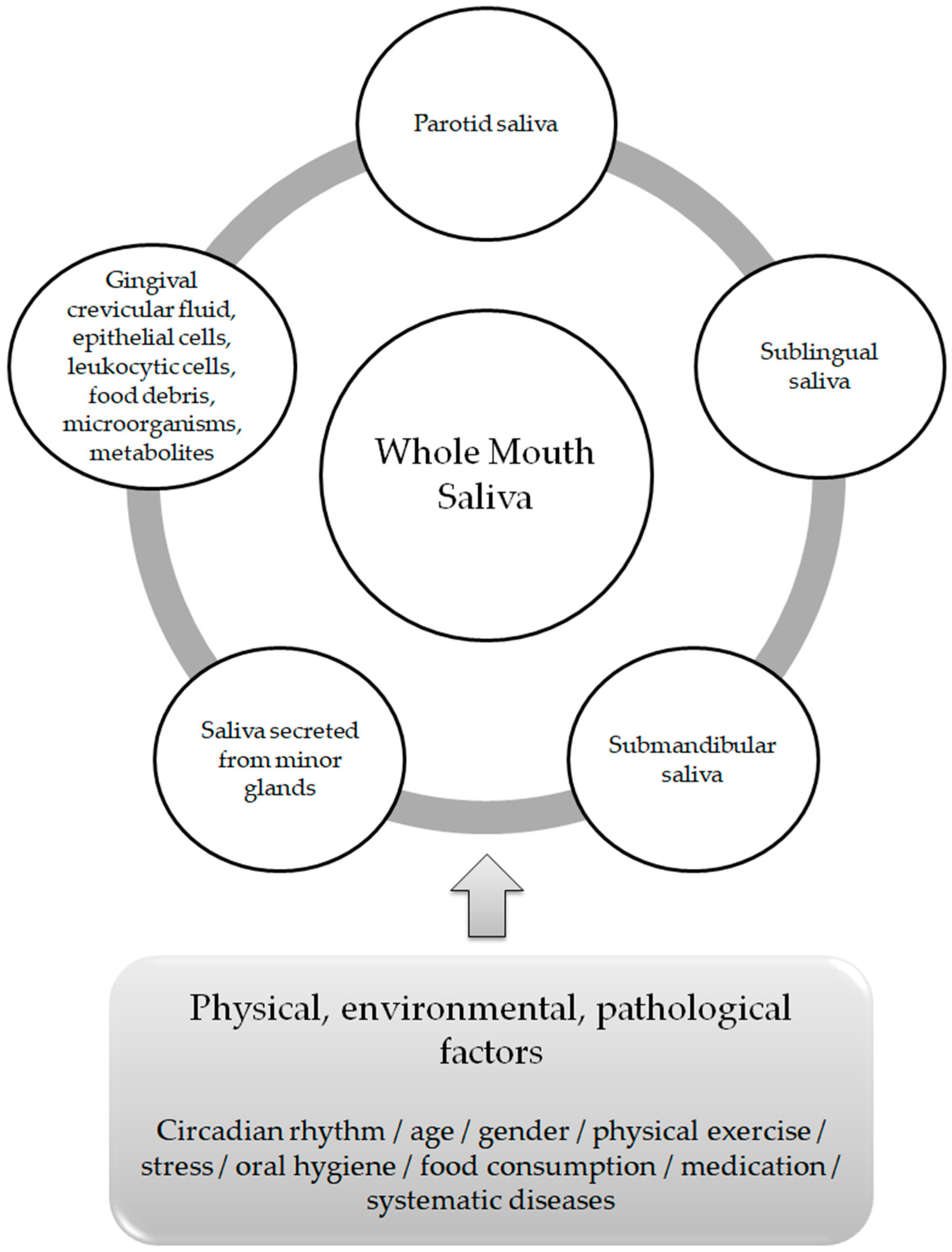
The average healthy person generates 0.75 to 1.5 L of saliva per day, with a greater volume secreted when the person is awake [8]. There is an evident differentiation in the composition, flow rate and volume of saliva both between individuals and within the same individual [9]. These variations depend on stimuli delivered by the sympathetic and parasympathetic systems of the autonomic nervous system (neural control) [10], as well as on physical, environmental and/or pathological factors, which include circadian rhythm, age, gender, physical exercise, oral hygiene, food consumption, medication andsystematic diseases [11] (Figure 1). Among its functions, lubrication and moisturization of the surfaces of the oral cavity, the pharynx and the esophagus, oral digestion, tissue and tooth integrity protection and antibacterial and antiviral defense, play a pivotal role in oral homeostasis and in the overall quality of life [12,13].
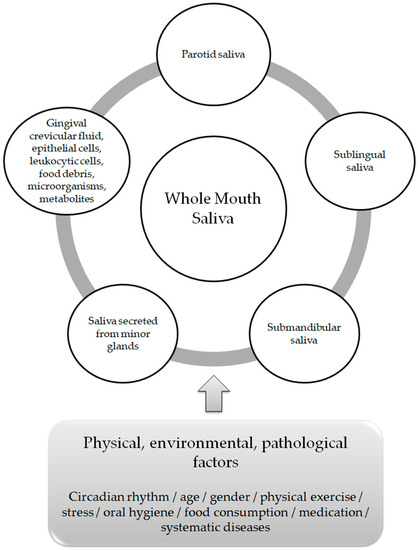
Figure 1.
Whole Mouth Saliva components and factors affecting its composition.
Biotechnological advances and applications in the health sciences have led to the introduction of “omics” in medicine. Genomics, studying the structure, function, evolution and mapping of genes and transcriptomics, the field of biological study of mRNA molecules led to the formation of the appropriate conditions for circumstantial monitoring of smaller organ and/or cell compounds such as proteins (proteomics) and low molecular weight metabolites (metabolomics) [14,15].
A metabolite is a small molecule with a molecular weight typically less than 1500 Da [16]. The complete set of small molecular metabolites is called the “metabolome” [17]. “Metabolomics” is defined as the latest of the –omics technologies, investigating metabolites within biofluids, cells and tissues [18].
Τhe use of biofluids in the human body such as serum, plasma, urine and cerebrospinal fluid for metabolomic profiling on a variety of health disorders including cancer, infectious diseases, neurological diseases (Alzheimer’s disease, dementia) cardiovascular, rheumatological, renal, and respiratory diseases is scientifically established and acceptable [19]. On the contrary, scarce evidence is present when addressing salivary metabolites [20]. The salivary metabolome is considered as a critical asset in elucidating pathways identifying various local and systematic disorders, and it may be used as a key mediator in treatment design and modification as well as in treatment outcomes [21].
The aim of this narrative review is firstly, to shed light on the importance of the human salivary metabolome in health, secondarily to assess the analytical protocols and the limitations of salivary metabolomic studies and lastly to analyze the salivary metabolomic profile as a possible, sufficient and powerful biomarker of oral pathogenesis.
2. Human Salivary Metabolome Research and Its Limitations
A lot of studies focus on metabolite profiling in blood and urine, whereas scientific evidence for metabolic profiling in saliva is lacking. Takeda et al., concluded that the saliva metabolome proved to be comparable to the human serum and cerebrospinal fluid metabolomes in terms of chemical composition, strengthening the belief of homogeneity of compounds found in human saliva and human blood, independently of their different concentrations [20]. The well known positive correlation between salivary and plasma metabolite levels (e.g., glucose, lactate and pyruvate), as well as the fact that the proteomic and metabolomic alterations observed in saliva follow a similar pattern to the changes seen in blood, reinforce the use of saliva as an informative diagnostic biofluid [22,23].
Among the benefits of saliva are its ad libitum production, non-invasiveness, painlessness, relatively fast and cheap collection, minimal collector training, reduced anxiety when compared to blood collection and child friendlier approach when compared to blood collection, making it the perfect, informative, most readily available biofluid [5,24,25,26] The convenient analysis of saliva samples, the non-infectious collection process, the ease of its transportation and its disposable nature are further positive aspects of using saliva for metabolite profiling processes [27].
Most salivary metabolomic research on healthy subjects focuses on the identification of specific metabolites or metabolic species. This kind of research is characterized as targeted salivary metabolomic research [23]. The first important non—targeted metabolomic analysis of saliva was conducted by Silwood et al., in 2002, who identified more than 60 metabolites and quantified 11 salivary metabolites in healthy human saliva, along with an interesting intra and inter—subject variability in the concentrations of these molecules [28]. Likewise, Sugimoto et al. (2013) identified and quantified 148 salivary metabolites in healthy humans [29]. Dame et al. (2015) accomplished the identification and the quantification of a total of 308 salivary metabolites in healthy people [23]. Nowadays, more than 853 identified and quantified salivary metabolites or metabolite species are freely available at the Human Metabolome Database (HMDB) [30].
All salivary metabolomic research studies (either targeted or non-targeted) of healthy human samples, focus on multiple factors that tend to modify the concentration of the healthy saliva metabolome. The greatest factors affecting the healthy human saliva metabolome are: the collection method, where stimulated saliva presents a decrease in metabolite concentrations compared to unstimulated whole mouth saliva secretion samples [20]; the type of the gland that the saliva is secreted from, since submandibular gland saliva is more viscous than the serous parotid gland saliva [30]; the gender, where acetate, formate, glycine, lactate, methanol, propionate, propylene glycol, pyruvate and taurine were significantly higher in concentration in male rather than in female saliva samples [20,31], the smoking status, that leads to up- and/or down-regulations of metabolic concentrations [32], the diurnal cycle (circadian cycle), where specific salivary metabolites—mainly amino-acids- showed a clear diurnal variation in their concentration [33], the fasting conditions (diet),where longer time period between last diet and sample collection affected the salivary metabolomic profile [23,34] and the microflora of the oral cavity, but most precisely the host-microbiome interactions. The oral microbiome strongly affects the net metabolic composition of the WMS at rest and can lead to alterations in its composition upon exposure to exogenous substances. The updated literature indicates that certain WMS metabolites including short chain fatty Acids (SCFAs), are absent from the sterile parotid gland saliva, leading to the conclusion that some saliva metabolites present a strong correlation with the bacterial index of the WMS. Furthermore, the metabolic patterns of the WMS present greater inter—individual variations than those of plasma metabolites, possibly caused by the existence of diversity in the oral microbiota that modulates the WMS metabolites. Conversely, plasma metabolites are easily regulated due to host mechanisms [26,35,36]. A field of further investigation involves the reflection of the oral microbiome on the salivary metabolome, as well as the dynamic interactions of different biofluids.
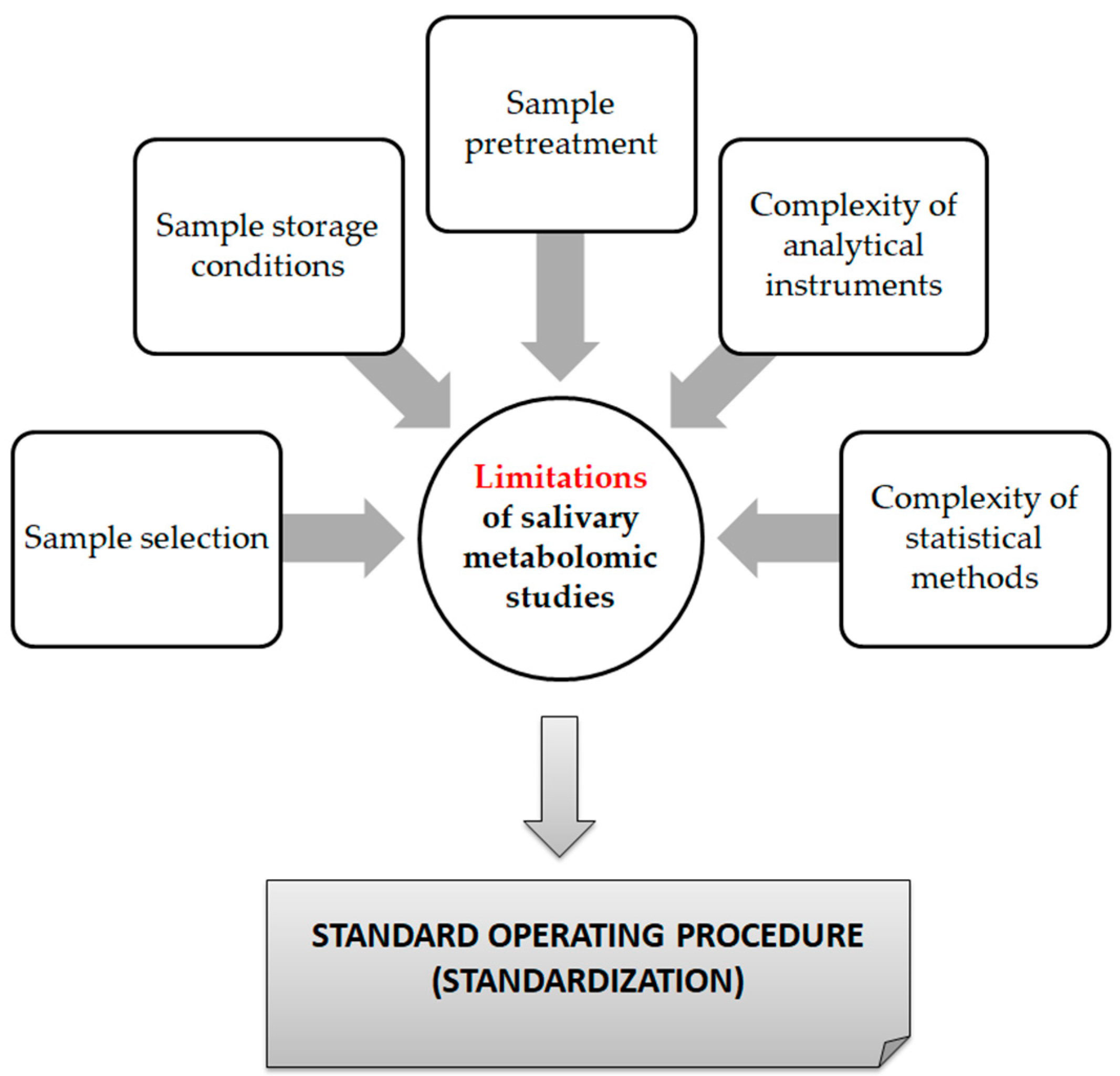
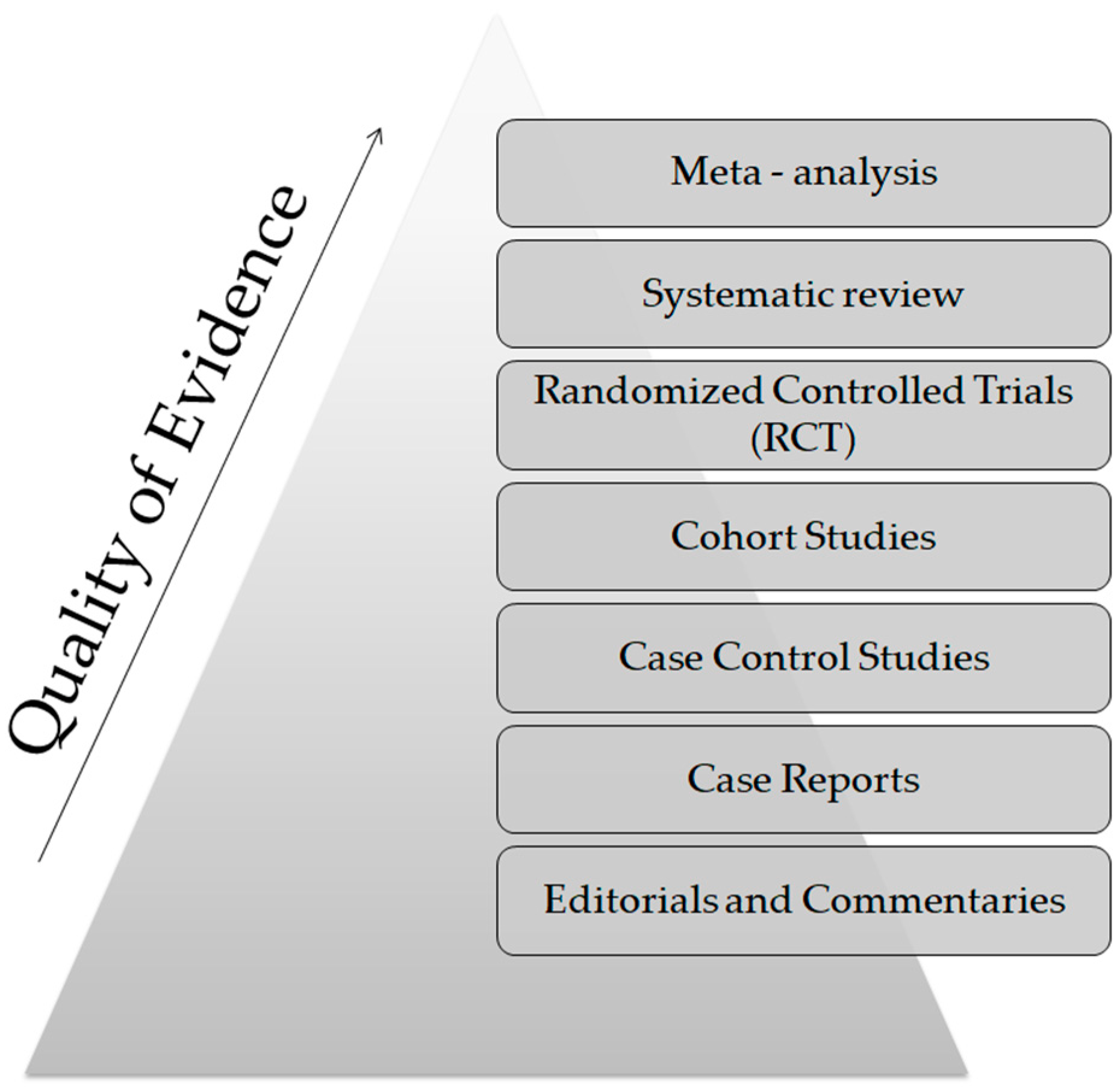
From a technical point of view, several analytical platforms are developed and integrated into the human saliva metabolomic profiling process. The two most renowned metabolite measurement technologies are nuclear magnetic resonance spectroscopy (HNMR) and mass spectrometry (MS) [20,23,24,37,38]. Subcategories of MS methods or additional combinatorial/conjunctive methods are mentioned below. Capillary electrophoresis time-of-flight mass spectrometry (CE-TOF-MS), gas-chromatography mass spectrometry (GC-MS), direct flow injection—liquid chromatography mass spectrometry (LC-MS), inductively coupled plasma mass spectrometry (ICP-MS) and high performance liquid chromatography (HPLC) [23,39,40]. Τhe description of each analytical platform is outside the scope of this mini-review. NMR is characterized as an untargeted metabolomics technique that leads to the identification and quantification of compounds, including short-chain organic acids, amino acids, alcohols, amines, sugars and pharmaceutical adjuvants. The advantage of this technique focuses on the minimal or no sample pre-treatment needed (deproteinization by centrifuging) and on its higher reproducibility compared to MS analytical platforms [41]. On the other hand, MS is an analytical method of high sensitivity, that identifies and/or quantifies a substance by measuring its mass and number of ions by the use of various ionization methods [42]. The combination of MS with other conjunctive methods has the advantage of greater metabolite identification even at lower concentrations [43,44,45]. The use of complex extraction methods and separation steps in order to detect and analyze both polar and non-polar organic acids is highlighted as a difficulty that complicates the identification procedure by mass spectrometry [41]. MS-based metabolomics require a well-designed pooled quality control sample (PQC) that is repeatedly analyzed throughout the sample batch and used for signal corrections compared to NMR-based metabolomic analysis [18,46]. Those characteristics are mentioned in order to understand that an additional limitation in salivary metabolomic studies exists due to the complexity of the analytical instruments used. Most studies only use one analytical platform and try to analyze one individual metabolome, which makes a potential comparison of salivary metabolic profiles between studies using different analytical technologies insignificant. At this point the allusion to the term “standard operating procedure” is of utmost importance (Figure 2). This term refers to the standardization and enactment of specific workflows as to the pre-analytical, analytical and post-analytical methods used [47,48]. The standardization of sample collection (for example, the use of unstimulated WMS in all studies), sample storage conditions (freezing temperatures), sample pretreatment (centrifugation for cell content removal and/or additional separation steps), sample analysis (by the use of more than one analytical platform) and statistical methods employed (principal component analysis—partial least-squares regression) would minimize the heterogeneous results in salivary metabolomic coverage [13,25,26], caused by separation difficulties, sensitivity differences, instrument detection differences, compound stability, solubility, and volatility [23,49,50], and give the opportunity to conduct comprehensive systematic reviews and meta-analyses, which are placed on top of the evidence-based science pyramid (Figure 3). Scientific studies are categorized based on the quality as well as the amount of evidence available, meaning that towards the base of the pyramid, the amount of evidence increases but simultaneously the quality of the evidence decreases.
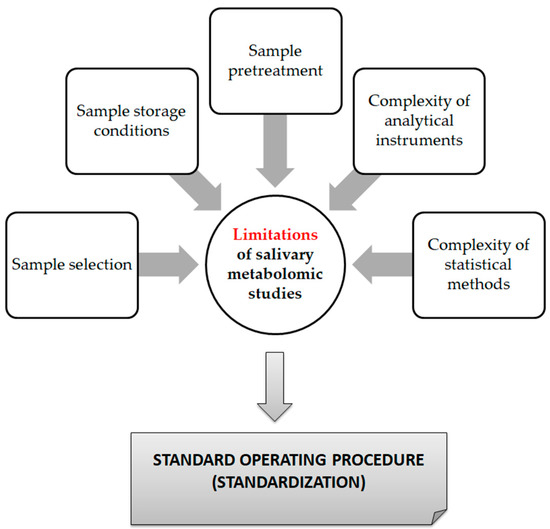
Figure 2.
Limitations of saliva metabolomic profile studies.
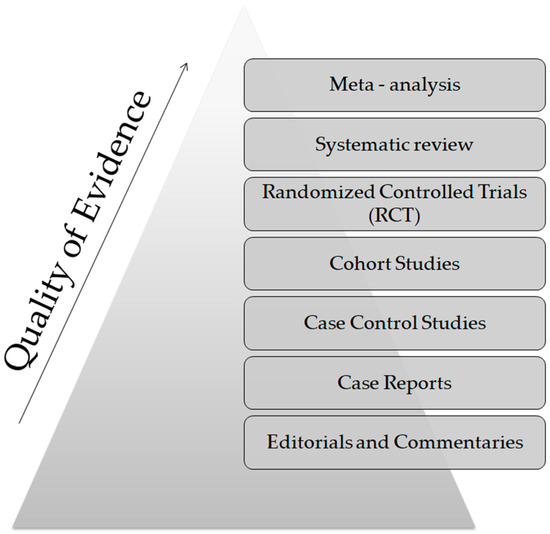
Figure 3.
The evidence—based pyramid.
3. The Human Salivary Metabolome and Oral Pathologies
According to the World Health Organization (WHO), a “biomarker” is defined as any substance, structure, or process that can be measured in the body or in its products and that can influence or predict the incidence of an outcome or disease [51]. Over the years, clinical studies have focused on detecting specific salivary metabolites associated with specific oral diseases, thus characterizing these metabolites as diagnostic biomarkers. This remains questionable, as one salivary metabolite’s quantitative alteration may simply indicate a non-specific pathological shift, incidental to insufficient diagnostic specificity. On the other hand, multivariate analysis may offer greater accuracy for putative biomarker findings [26]. The early “fingerprints” of changes in a wide range of diseases could be discovered by the study of the metabolic profile of saliva. Focusing on oral pathology and its association with salivary metabolomics, efforts have been made in the fields of oral cancer and periodontal diseases.
As mentioned by Khurshid et al., in 2018, oral cancer is the 6th most common cancer worldwide, and its late detection is highly associated with its high mortality and morbidity rates. Approximately 60–80% of patients with oral cancer are diagnosed at a late-stage. Hundreds of salivary biomarkers (using genomics, transcriptomics and proteomics) have already been identified, including cytokines (IL-8, IL-1b, TNF-α), defensin-1, P53, tissue polypeptide-specific antigen, dual specificity phosphatase, profilin, cofilin-1, transferrin etc. [52]. Human saliva metabolomics and its contribution to oral cancer diagnosis is a field of continuous investigation, since saliva is in direct contact with the mucosa and cancerous cells of the oral cavity. The validation of specific saliva metabolomic biomarkers for oral cancer may lead to early stage detection and a more appropriate treatment modality for the patient [13,21,25,26]. Several studies, with different protocols, conducted over the last few years identified different saliva metabolite concentrations either between healthy controls and oral squamous cell carcinoma subjects or between OSCC subjects and premalignant lesion subjects (oral lichen planus, oral leukoplakia, precancerous dysplasia, keratosis). Their samples, analytical and discrimination methods and outcomes are summarized in Table 1 [53,54,55,56,57,58,59,60,61,62,63,64,65,66,67]. A notable diversity of candidate biomarkers is presented in these studies. More precisely, Sugimoto et al. (2010) [55] and Wang et al. (2014) [57,58] both mentioned increased choline and betaine and decreased L-carnitine in patients with OSCC compared to healthy controls. Choline is also found among other metabolites as a potential biomarker in the research of Ohshima et al., in 2017 [60]. Glycine, proline and ornithine were found in three independent studies by Lohavanichbutr et al. (2018), Ishakawa et al. (2019) and Tantray et al. (2022) as potential oral cancer biomarkers [61,62,67]. However, different studies identified different groups of metabolites that were either upregulated or downregulated in OSCC and precancerous samples compared to healthy controls. These deviations are associated with limitations concerning sample size, population tested, and analytical methods used—factors that lead to heterogeneous results despite the similar study designs of the studies. The limitations of this kind of study lie in two factors: firstly, the evaluation of the specificity of the salivary metabolite biomarker against other inflammatory diseases, for instance, periodontitis, because a possible overlap in the metabolite biomarkers between periodontitis and oral cancer could lead to serious misdiagnosis, and secondarily, the study design, the analytical instruments used, the discrimination methods, and the absence of cross-validation of the analytical equipment between different laboratories [13].

Table 1.
Research focused on salivary metabolomics serving as biomarkers in oral cancer.
The same opportunities, but simultaneously the same concerns, are detected in the use of salivary metabolites in periodontitis diagnosis and treatment. Periodontal diseases are characterized as one of the two main causes of tooth loss and are inextricably linked to connective tissue loss, periodontal pocket formation, and progressive bone degradation [21,68]. The development of new saliva metabolite biomarkers may eliminate tooth loss due to early diagnosis of the severity of the periodontal condition. In several studies that are listed in Table 2, an upregulation or downregulation of specific salivary metabolite species is detected (e.g., increased levels of fatty acids, phenylphenol, dipeptides leucylisoleucine, serylisoleucine, arachidonate, and dihomo-linolate) [55,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83]. Besides saliva, the gingival crevicular fluid (GCF) is a great source of possible biomarkers for periodontal diseases. It includes a variety of host and microbial enzymes, endotoxins, nucleic acids, carbohydrates, lipids, degradation products of several metabolic pathways, cytokines and immunoglobulins. It is revealed that GCF is a more ideal biofluid for the diagnosis of periodontal diseases (and the differentiation of healthy patients, gingivitis patients, and periodontitis subjects) compared to serum and saliva, since it contains biomarkers that reflect inflammation, immune response and tissue destruction at the sight of periodontal lesions [84]. Metabolites associated with periodontal variables are clearly linked to tissue destruction, host defense mechanisms and bacterial metabolism [76]. When interpreting the presence of “salivary metabolites” in periodontitis patients, caution is needed, because they may be products of the developed microflora (oral microbiome metabolites). The bacterial metabolite phenylacetate is strongly correlated with periodontal disease. Salivary metabolomics may also be used as prognostic biomarkers of non-surgically and/or surgically treated periodontal disease. A change in the salivary metabolomic profile of a diseased patient after treatment entails the modification of a previously “diseased” specimen into a newly “healthy” one.

Table 2.
Research focused on salivary metabolomics serving as biomarkers in periodontal disease.
4. Conclusions
Conclusively, salivary metabolomics form a promising newly established research field. The drawbacks of using saliva metabolites as putative biological indicators of oral or systematic health disorders focus on the small sample sizes of the studies conducted and on the great challenges of the implementation of these technologies into clinical diagnostics. The validation of salivary biomarkers may be accomplished by elevating metabolomic research from a “case-control-study” design to a “large-scale validation study” design. The simultaneous observation of salivary metabolomics and microbiomics would enlighten the pathogenetic mechanisms of oral diseases. The primary goal of salivary metabolomic profiling is to distinguish the type of inflammation itself and not to simply compare an inflammation status to that of a healthy control. Salivary metabolomics may open new horizons in clarifying pathogenesis, as well as in disease monitoring and treatment outcome assessment. Mapping the human saliva metabolome of individuals at many standpoints is equivalent to the establishment of more personalized treatment and follow-up protocols. All in all, the oral cavity is a complex organ with numerous factors affecting its salivary metabolic profile, a fact that may ramify accurate research findings.
Author Contributions
Conceptualization, K.T. and E.P.; validation, K.T. and E.P.; formal analysis, K.T.; investigation, K.T.; data curation, K.T.; writing—original draft preparation, K.T.; writing—review and editing, K.T. and E.P.; visualization, K.T.; supervision, E.P.; project administration, K.T. and E.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- de Almeida, P.D.V.; Gregio, A.M.T.; Machado, M.A.N.; de Lima, A.A.S.; Azevedo, L.R. Saliva composition and functions: A comprehensive review. J. Contemp. Dent. Pract. 2008, 9, 72–80. [Google Scholar]
- Humphrey, S.P.; Williamson, R.T. A review of saliva: Normal composition, flow, and function. J. Prosthet Dent. 2001, 85, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Dodds, M.W.; Johnson, D.A.; Yeh, C.K. Health benefits of saliva: A review. J. Dent. 2005, 33, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Amerongen, A.V.; Veerman, E.C. Saliva—The defender of the oral cavity. Oral Dis. 2002, 8, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Pappa, E.; Kousvelari, E.; Vastardis, H. Saliva in the “Omics” era: A promising tool in paediatrics. Oral Dis. 2019, 25, 16–25. [Google Scholar] [CrossRef]
- Proctor, G.B. The physiology of salivary secretion. Periodontology 2016, 70, 11–25. [Google Scholar] [CrossRef]
- Soini, H.A.; Klouckova, I.; Wiesler, D.; Oberzaucher, E.; Grammer, K.; Dixon, S.J.; Xu, Y.; Brereton, R.G.; Penn, D.G.; Novotny, M.V. Analysis of volatile organic compounds in human saliva by a static sorptive extraction method and gas chromatography-mass spectrometry. J. Chem. Ecol. 2010, 36, 1035–1042. [Google Scholar] [CrossRef]
- Sanchez-Pablo, M.A.; Gonzalez-Garcia, V.; del Castillo-Rueda, A. Study of total stimulated saliva flow and hyperpigmentation in the oral mucosa of patients diagnosed with hereditary hemochromatosis. Series of 25 cases. Med. Oral Patol. Oral Cir. Bucal. 2012, 17, e45–e49. [Google Scholar] [CrossRef]
- Hsiao, Y.C.; Chu, L.J.; Chen, Y.T.; Chi, L.M.; Chien, K.Y.; Chiang, W.F.; Chang, Y.T.; Chen, S.F.; Wang, W.S.; Chuang, Y.N. Variability assessment of 90 salivary proteins in intraday and interday samples from healthy donors by multiple reaction monitoring-mass spectrometry. Proteom.–Clin. Appl. 2018, 12, 1700039. [Google Scholar] [CrossRef]
- Proctor, G.B.; Carpenter, G.H. Regulation of salivary gland function by autonomic nerves. Auton. Neurosci. Basic Clin. 2007, 133, 3–18. [Google Scholar] [CrossRef]
- Helmerhorst, E.J.; Dawes, C.; Oppenheim, F.G. The complexity of oral physiology and its impact on salivary diagnostics. Oral Dis. 2018, 24, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Hanning, C.; Hanning, M.; Kensche, A.; Carpenter, G. The mucosal pellicle- An underestimated factor in oral physiology. Arch. Oral Biol. 2017, 80, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Panneerselvam, K.; Ishikawa, S.; Krishnan, R.; Sugimoto, M. Salivary Metabolomics for Oral Cancer Detection: A Narrative Review. Metabolites 2022, 12, 436. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Córdoba, B.; Santiago-García, J. Saliva: A fluid of study for OMICS. OMICS 2014, 18, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Kaczor-Urbanowicz, K.E.; Martin Carreras-Presas, C.; Aro, K.; Tu, M.; Garcia-Godoy, F.; Wong, D.T. Saliva diagnostics—Current views and directions. Exp. Biol. Med. 2017, 242, 459–472. [Google Scholar] [CrossRef]
- Chaleckis, R.; Meister, I.; Zhang, P.; Wheelock, C.E. Challenges, progress and promises of metabolite annotation for LC-MS-based metabolomics. Curr. Opin. Biotechnol. 2019, 55, 44–50. [Google Scholar] [CrossRef]
- Wong, D.T. Salivaomics. J. Am. Dent. Assoc. 2012, 143, 19–24. [Google Scholar] [CrossRef]
- Hynne, H.; Sandås, E.M.; Elgstøen, K.B.P.; Rootwelt, H.; Utheim, T.P.; Galtung, H.K.; Jensen, J.L. Saliva Metabolomics in Dry Mouth Patients with Head and Neck Cancer or Sjögren’s Syndrome. Cells 2022, 11, 323. [Google Scholar] [CrossRef]
- Evans, E.D.; Duvallet, C.; Chu, N.D.; Oberst, M.K.; Murphy, M.A.; Rockafellow, I.; Sontag, D.; Alm, E.J. Predicting human health from biofluid-based metabolomics using machine learning. Sci. Rep. 2020, 10, 17635. [Google Scholar] [CrossRef]
- Takeda, I.; Stretch, C.; Barnaby, P.; Bhatnager, K.; Rankin, K.; Fu, H.; Weljie, A.; Jha, N.; Slupsky, C. Understanding the human salivary metabolome. NMR Biomed. 2009, 22, 577–584. [Google Scholar] [CrossRef]
- Mikkonen, J.J.; Singh, S.P.; Herrala, M.; Lappalainen, R.; Myllymaa, S.; Kullaa, A.M. Salivary metabolomics in the diagnosis of oral cancer and periodontal diseases. J. Periodontal Res. 2016, 51, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Chiappin, S.; Antonelli, G.; Gatti, R.; De Palo, E.F. Saliva specimen: A new laboratory tool for diagnostic and basic investigation. Clin. Chim. Acta 2007, 383, 30–40. [Google Scholar] [CrossRef]
- Dame, Z.T.; Aziat, F.; Mandal, R.; Krishnamurthy, R.; Bouatra, S.; Borzouie, S.; Guo, A.C.; Sajed, T.; Deng, L.; Lin, H.; et al. The human saliva metabolome. Metabolomics 2015, 11, 1864–1883. [Google Scholar] [CrossRef]
- Pfaffe, T.; Cooper-White, J.; Beyerlein, P.; Kostner, K.; Punyadeera, C. Diagnostic potential of saliva: Current state and future applications. Clin. Chem. 2011, 57, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Hyvärinen, E.; Savolainen, M.; Mikkonen, J.J.W.; Kullaa, A.M. Salivary Metabolomics for Diagnosis and Monitoring Diseases: Challenges and Possibilities. Metabolites 2021, 11, 587. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.; Carpenter, G.; So, P.W. Salivary Metabolomics: From Diagnostic Biomarker Discovery to Investigating Biological Function. Metabolites 2020, 10, 47. [Google Scholar] [CrossRef] [PubMed]
- Khurshid, Z.; Zohaib, S.; Najeeb, S.; Zafar, M.S.; Slowey, P.D.; Almas, K. Human Saliva Collection Devices for Proteomics: An Update. Int. J. Mol. Sci. 2016, 17, 846. [Google Scholar] [CrossRef]
- Silwood, C.; Lynch, E.; Claxson, A.; Grootveld, M. 1h and 13c nmr spectroscopic analysis of human saliva. J. Dent. Res. 2002, 81, 422–427. [Google Scholar] [CrossRef]
- Sugimoto, M.; Saruta, J.; Matsuki, C.; To, M.; Onuma, H.; Kaneko, M.; Soga, T.; Tomita, M.; Tsukinoki, K. Physiological and environmental parameters associated with mass spectrometry-based salivary metabolomic profiles. Metabolomics. 2013, 9, 454–463. [Google Scholar] [CrossRef]
- Human Metabolome Database. Available online: https://hmdb.ca (accessed on 29 January 2023).
- Lukacs, J.; Largaespada, L. Explaining sex differences in dental caries prevalence: Saliva, hormones, and ‘‘life-history’’ etiologies. Am. J. Hum. Biol. 2006, 18, 540–555. [Google Scholar] [CrossRef]
- Mueller, D.C.; Piller, M.; Niessner, R.; Scherer, M.; Scherer, G. Untargeted metabolomic profiling in saliva of smokers and nonsmokers by a validated GC-TOF-MS method. J. Proteome Res. 2014, 13, 1602–1613. [Google Scholar] [CrossRef] [PubMed]
- Dallmann, R.; Viola, A.U.; Tarokh, L.; Cajochen, C.; Brown, S.A. The human circadian metabolome. Proc. Nati. Acad. Sci. USA 2012, 109, 2625–2629. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Sugimoto, M.; Kitabatake, K.; Tu, M.; Sugano, A.; Yamamori, I.; Iba, A.; Yusa, K.; Kaneko, M.; Ota, S.; et al. Effect of Timing of Collection of Salivary Metabolomic Biomarkers on Oral Cancer Detection. Amino Acids 2017, 49, 761–770. [Google Scholar] [CrossRef]
- Gardner, A.; Parkes, H.G.; So, P.-W.; Carpenter, G.H. Determining bacterial and host contributions to the human salivary metabolome. J. Oral Microbiol. 2019, 11, 1617014. [Google Scholar] [CrossRef]
- Belstrøm, D. The salivary microbiota in health and disease. J. Oral Microbiol. 2020, 12, 1723975. [Google Scholar] [CrossRef]
- Wishart, D.S.; Cheng, L.L.; Copié, V.; Edison, A.S.; Eghbalnia, H.R.; Hoch, J.C.; Gouveia, G.J.; Pathmasiri, W.; Powers, R.; Schock, T.B.; et al. NMR and Metabolomics—A Roadmap for the Future. Metabolites 2022, 12, 678. [Google Scholar] [CrossRef] [PubMed]
- Gowda, G.A.N.; Djukovic, D. Overview of Mass Spectrometry-Based Metabolomics: Opportunities and Challenges. In Mass Spectrometry in Metabolomics. Methods in Molecular Biology, 1st ed.; Raftery, D., Ed.; Humana Press: New York, NY, USA, 2014; Volume 1198, pp. 3–12. [Google Scholar] [CrossRef]
- Naz, S.; Moreira dos Santos, D.C.; García, A.; Barbas, C. Analytical protocols based on LC-MS, GC-MS and CE-MS for nontargeted metabolomics of biological tissues. Bioanalysis 2014, 6, 1657–1677. [Google Scholar] [CrossRef]
- Drouin, N.; Ramautar, R. Capillary Electrophoresis-Mass Spectrometry for Metabolomics: Possibilities and Perspectives. Adv. Exp. Med. Biol. 2021, 1336, 159–178. [Google Scholar] [CrossRef] [PubMed]
- Vignoli, A.; Ghini, V.; Meoni, G.; Licari, C.; Takis, P.G.; Tenori, L.; Turano, P.; Luchinat, C. High-Throughput Metabolomics by 1D NMR. Angew. Chem. Int. Ed. Engl. 2019, 58, 968–994. [Google Scholar] [CrossRef]
- Zeki, Ö.C.; Eylem, C.C.; Reçber, T.; Kır, S.; Nemutlu, E. Integration of GC-MS and LC-MS for untargeted metabolomics profiling. J. Pharm. Biomed. Anal. 2020, 190, 113509. [Google Scholar] [CrossRef]
- Emwas, A.H. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol. Biol. 2015, 1277, 161–193. [Google Scholar] [CrossRef]
- Marshall, D.D.; Powers, R. Beyond the paradigm: Combining mass spectrometry and nuclear magnetic resonance for metabolomics. Prog. Nucl. Magn. Reson. Spectrosc. 2017, 100, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Delvaux, A.; Rathahao-Paris, E.; Alves, S. Different ion mobility-mass spectrometry coupling techniques to promote metabolomics. Mass. Spectrom. Rev. 2022, 41, 695–721. [Google Scholar] [CrossRef] [PubMed]
- Aristizabal-Henao, J.J.; Lemas, D.J.; Griffin, E.K.; Costa, K.A.; Camacho, C.; Bowden, J.A. Metabolomic Profiling of Biological Reference Materials using a Multiplatform High-Resolution Mass Spectrometric Approach. J. Am. Soc. Mass. Spectrom. 2021, 32, 2481–2489. [Google Scholar] [CrossRef] [PubMed]
- Pezzatti, J.; Boccard, J.; Codesido, S.; Gagnebin, Y.; Joshi, A.; Picard, D.; González-Ruiz, V.; Rudaz, S. Implementation of liquid chromatography-high resolution mass spectrometry methods for untargeted metabolomic analyses of biological samples: A tutorial. Anal. Chim. Acta 2020, 1105, 28–44. [Google Scholar] [CrossRef]
- Martias, C.; Baroukh, N.; Mavel, S.; Blasco, H.; Lefèvre, A.; Roch, L.; Montigny, F.; Gatien, J.; Schibler, L.; Dufour-Rainfray, D.; et al. Optimization of Sample Preparation for Metabolomics Exploration of Urine, Feces, Blood and Saliva in Humans Using Combined NMR and UHPLC-HRMS Platforms. Molecules 2021, 26, 4111. [Google Scholar] [CrossRef] [PubMed]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The human serum metabolome. PLoS ONE 2011, 6, 16957. [Google Scholar] [CrossRef] [PubMed]
- Bouatra, S.; Aziat, F.; Mandal, R.; Guo, A.C.; Wilson, M.R.; Knox, C.; Bjorndahl, T.C.; Krishnamurthy, R.; Saleem, F.; Liu, P.; et al. The human urine metabolome. PLoS ONE 2013, 8, e73076. [Google Scholar] [CrossRef]
- WHO. International Programme on Chemical Safety Biomarkers in Risk Assessment: Validity and Validation. Environmental Health Criteria 222. Available online: http://www.inchem.org/documents/ehc/ehc/ehc222.htm (accessed on 16 April 2021).
- Khurshid, Z.; Zafar, M.S.; Khan, R.S.; Najeeb, S.; Slowey, P.D.; Rehman, I.U. Role of Salivary Biomarkers in Oral Cancer Detection. Adv. Clin. Chem. 2018, 86, 23–70. [Google Scholar] [CrossRef]
- Yan, S.-K.; Wei, B.; Lin, Z.; Yang, Y.; Zhou, Z.; Zhang, W. A metabonomic approach to the diagnosis of oral squamous cell carcinoma, oral lichen planus and oral leukoplakia. Oral. Oncol. 2008, 44, 477–483. [Google Scholar] [CrossRef]
- Jou, Y.J.; Lin, C.; Lai, C.; Chen, C.H.; Kao, J.Y.; Chen, S.Y.; Tsai, M.-H.; Huang, S.-H.; Lin, C.-W. Proteomic identification of salivary transferrin as a biomarker for early detection of oral cancer. Anal. Chim. Acta 2010, 681, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef]
- Wei, J.; Xie, G.; Zhou, Z.; Shi, P.; Qiu, Y.; Zheng, X.; Chen, T.; Su, M.; Zhao, A.; Jia, W. Salivary metabolite signatures of oral cancer and leukoplakia. Int. J. Cancer 2011, 129, 2207–2217. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, P.; Wang, X.; Duan, Y. Investigation and identification of potential biomarkers in human saliva for the early diagnosis of oral squamous cell carcinoma. Clin. Chim. Acta 2014, 427, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, P.; Wang, X.; Duan, Y. The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. Sci. Rep. 2014, 4, 6802. [Google Scholar] [CrossRef]
- Ishikawa, S.; Sugimoto, M.; Kitabatake, K.; Sugano, A.; Nakamura, M.; Kaneko, M.; Ota, S.; Hiwatari, K.; Enomoto, A.; Soga, T. Identification of salivary metabolomic biomarkers for oral cancer screening. Sci. Rep. 2016, 6, 31520. [Google Scholar] [CrossRef]
- Ohshima, M.; Sugahara, K.; Kasahara, K.; Katakura, A. Metabolomic analysis of the saliva of japanese patients with oral squamous cell carcinoma. Oncol. Rep. 2017, 37, 2727–2734. [Google Scholar] [CrossRef]
- Lohavanichbutr, P.; Zhang, Y.; Wang, P.; Gu, H.; Gowda, G.A.N.; Djukovic, D.; Buas, M.F.; Raftery, D.; Chen, C. Salivary Metabolite Profiling Distinguishes Patients with Oral Cavity Squamous Cell Carcinoma from Normal Controls. PLoS ONE 2018, 13, e0204249. [Google Scholar] [CrossRef]
- Ishikawa, S.; Wong, D.T.W.; Sugimoto, M.; Gleber-Netto, F.O.; Li, F.; Tu, M.; Zhang, Y.; Akin, D.; Iino, M. Identification of salivary metabolites for oral squamous cell carcinoma and oral epithelial dysplasia screening from persistent suspicious oral mucosal lesions. Clin. Oral Investig. 2019, 23, 3557–3563. [Google Scholar] [CrossRef]
- Sridharan, G.; Ramani, P.; Patankar, S.; Vijayaraghavan, R. Evaluation of Salivary Metabolomics in Oral Leukoplakia and Oral Squamous Cell Carcinoma. J. Oral Pathol. Med. 2019, 48, 299–306. [Google Scholar] [CrossRef]
- Ishikawa, S.; Hiraka, T.; Kirii, K.; Sugimoto, M.; Shimamoto, H.; Sugano, A.; Kitabatake, K.; Toyoguchi, Y.; Kanoto, M.; Nemoto, K.; et al. Relationship between Standard Uptake Values of Positron Emission Tomography/Computed Tomography and Salivary Metabolites in Oral Cancer: A Pilot Study. J. Clin. Med. 2020, 9, 3958. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Yang, X.; Narayanan, R.; Shankar, V.; Ethiraj, S.; Wang, X.; Duan, N.; Ni, Y.H.; Hu, Q.; Zare, R.N. Oral squamous cell carcinoma diagnosed from saliva metabolic profiling. Proc. Natl. Acad. Sci. USA 2020, 117, 16167–16173. [Google Scholar] [CrossRef] [PubMed]
- de Sá Alves, M.; de Sá Rodrigues, N.; Bandeira, C.M.; Chagas, J.F.S.; Pascoal, M.B.N.; Nepomuceno, G.L.J.T.; da Silva Martinho, H.; Alves, M.G.O.; Mendes, M.A.; Dias, M.; et al. Identification of Possible Salivary Metabolic Biomarkers and Altered Metabolic Pathways in South American Patients Diagnosed with Oral Squamous Cell Carcinoma. Metabolites 2021, 11, 650. [Google Scholar] [CrossRef] [PubMed]
- Tantray, S.; Sharma, S.; Prabhat, K.; Nasrullah, N.; Gupta, M. Salivary metabolite signatures of oral cancer and leukoplakia through gas chromatography-mass spectrometry. J. Oral Maxillofac. Pathol. 2022, 26, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Ozmeric, N. Advances in periodontal disease markers. Clin. Chim. Acta 2004, 343, 1–16. [Google Scholar] [CrossRef]
- Barnes, V.M.; Ciancio, S.G.; Shibly, O.; Xu, T.; Devizio, W.; Trivedi, H.M.; Guo, L.; Jönsson, T.J. Metabolomics reveals elevated macromolecular degradation in periodontal disease. J. Dent. Res. 2011, 90, 1293–1297. [Google Scholar] [CrossRef]
- Aimetti, M.; Cacciatore, S.; Graziano, A.; Tenori, L. Metabonomic analysis of saliva reveals generalized chronic periodontitis signature. Metabolomics 2012, 8, 465–474. [Google Scholar] [CrossRef]
- Huang, Y.; Zhu, M.; Li, Z.; Sa, R.; Chu, Q.; Zhang, Q.; Zhang, H.; Tang, W.; Zhang, M.; Yin, H. Mass spectrometry-based metabolomic profiling identifies alterations in salivary redox status and fatty acid metabolism in response to inflammation and oxidative stress in periodontal disease. Free Radic. Biol. Med. 2014, 70, 223–232. [Google Scholar] [CrossRef]
- Barnes, V.M.; Kennedy, A.D.; Panagakos, F.; Devizio, W.; Trivedi, H.M.; Jönsson, T.; Guo, L.; Cervi, S.; Scannapieco, F.A. Global Metabolomic Analysis of Human Saliva and Plasma from Healthy and Diabetic Subjects, with and without Periodontal Disease. PLoS ONE 2014, 9, e105181. [Google Scholar] [CrossRef]
- Kuboniwa, M.; Sakanaka, A.; Hashino, E.; Bamba, T.; Fukusaki, E.; Amano, A. Prediction of Periodontal Inflammation via Metabolic Profiling of Saliva. J. Dent. Res. 2016, 95, 1381–1386. [Google Scholar] [CrossRef]
- Ozeki, M.; Nozaki, T.; Aoki, J.; Bamba, T.; Jensen, K.R.; Murakami, S.; Toyoda, M. Metabolomic Analysis of Gingival Crevicular Fluid Using Gas Chromatography/Mass Spectrometry. Mass. Spectrom. 2016, 5, A0047. [Google Scholar] [CrossRef]
- Rzeznik, M.; Triba, M.N.; Levy, P.; Jungo, S.; Botosoa, E.; Duchemann, B.; Le Moyec, L.; Bernaudin, J.F.; Savarin, P.; Guez, D. Identification of a discriminative metabolomic fingerprint of potential clinical relevance in saliva of patients with periodontitis using 1H nuclear magnetic resonance (NMR) spectroscopy. PLoS ONE 2017, 12, e0182767. [Google Scholar] [CrossRef]
- Liebsch, C.; Pitchika, V.; Pink, C.; Samietz, S.; Kastenmüller, G.; Artati, A.; Suhre, K.; Adamski, J.; Nauck, M.; Völzke, H.; et al. The Saliva Metabolome in Association to Oral Health Status. J. Dent. Res. 2019, 98, 642–651. [Google Scholar] [CrossRef]
- Singh, M.P.; Saxena, M.; Saimbi, C.S.; Siddiqui, M.H.; Roy, R. Post-periodontal surgery propounds early repair salivary biomarkers by 1H NMR based metabolomics. Metabolomics 2019, 15, 141. [Google Scholar] [CrossRef] [PubMed]
- Romano, F.; Meoni, G.; Manavella, V.; Baima, G.; Mariani, G.M.; Cacciatore, S.; Tenori, L.; Aimetti, M. Effect of non-surgical periodontal therapy on salivary metabolic fingerprint of generalized chronic periodontitis using nuclear magnetic resonance spectroscopy. Arch. Oral Biol. 2019, 97, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Gawron, K.; Wojtowicz, W.; Łazarz-Bartyzel, K.; Łamasz, A.; Qasem, B.; Mydel, P.; Chomyszyn-Gajewska, M.; Potempa, J.; Mlynarz, P. Metabolomic Status of The Oral Cavity in Chronic Periodontitis. In Vivo 2019, 33, 1165–1174. [Google Scholar] [CrossRef]
- Schulte, F.; King, O.D.; Paster, B.J.; Moscicki, A.B.; Yao, T.J.; Van Dyke, R.B.; Shiboski, C.; Ryder, M.; Seage, G.; Hardt, M. Pediatric HIV/AIDS Cohort Study. Salivary metabolite levels in perinatally HIV-infected youth with periodontal disease. Metabolomics 2020, 16, 98. [Google Scholar] [CrossRef] [PubMed]
- Citterio, F.; Romano, F.; Meoni, G.; Iaderosa, G.; Grossi, S.; Sobrero, A.; Dego, F.; Corana, M.; Berta, G.N.; Tenori, L.; et al. Changes in the Salivary Metabolic Profile of Generalized Periodontitis Patients after Non-surgical Periodontal Therapy: A Metabolomic Analysis Using Nuclear Magnetic Resonance Spectroscopy. J. Clin. Med. 2020, 9, 3977. [Google Scholar] [CrossRef]
- Rodrigues, W.F.; Miguel, C.B.; Agostinho, F.; da Silva, G.V.; Lazo-Chica, J.E.; Scapin, S.M.N.; Napimoga, M.H.; Trindade-da-Silva, C.A.; Krieger, J.E.; Pereira, A.D.C.; et al. Metabolomic Evaluation of Chronic Periodontal Disease in Older Adults. Mediat. Inflamm. 2021, 2021, 1796204. [Google Scholar] [CrossRef]
- Overmyer, K.A.; Rhoads, T.W.; Merrill, A.E.; Ye, Z.; Westphall, M.S.; Acharya, A.; Shukla, S.K.; Coon, J.J. Proteomics, Lipidomics, Metabolomics, and 16S DNA Sequencing of Dental Plaque From Patients With Diabetes and Periodontal Disease. Mol. Cell Proteom. 2021, 20, 100126. [Google Scholar] [CrossRef]
- Barros, S.P.; Williams, R.; Offenbacher, S.; Morelli, T. Gingival crevicular fluid as a source of biomarkers for periodontitis. Periodontol 2000 2016, 70, 53–64. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).