Comparative Analysis of LC-ESI-IM-qToF-MS and FT-NIR Spectroscopy Approaches for the Authentication of Organic and Conventional Eggs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Chemicals
2.2. Samples
Sample Preparation
2.3. LC-ESI-IM-qToF-MS Data Acquisition
2.4. MS Data Processing
2.5. Fourier Transform near Infrared Spectroscopy
2.6. FT-NIR Spectra Preprocessing
2.7. Photometry
2.8. Multivariate Data Analysis
3. Results and Discussion
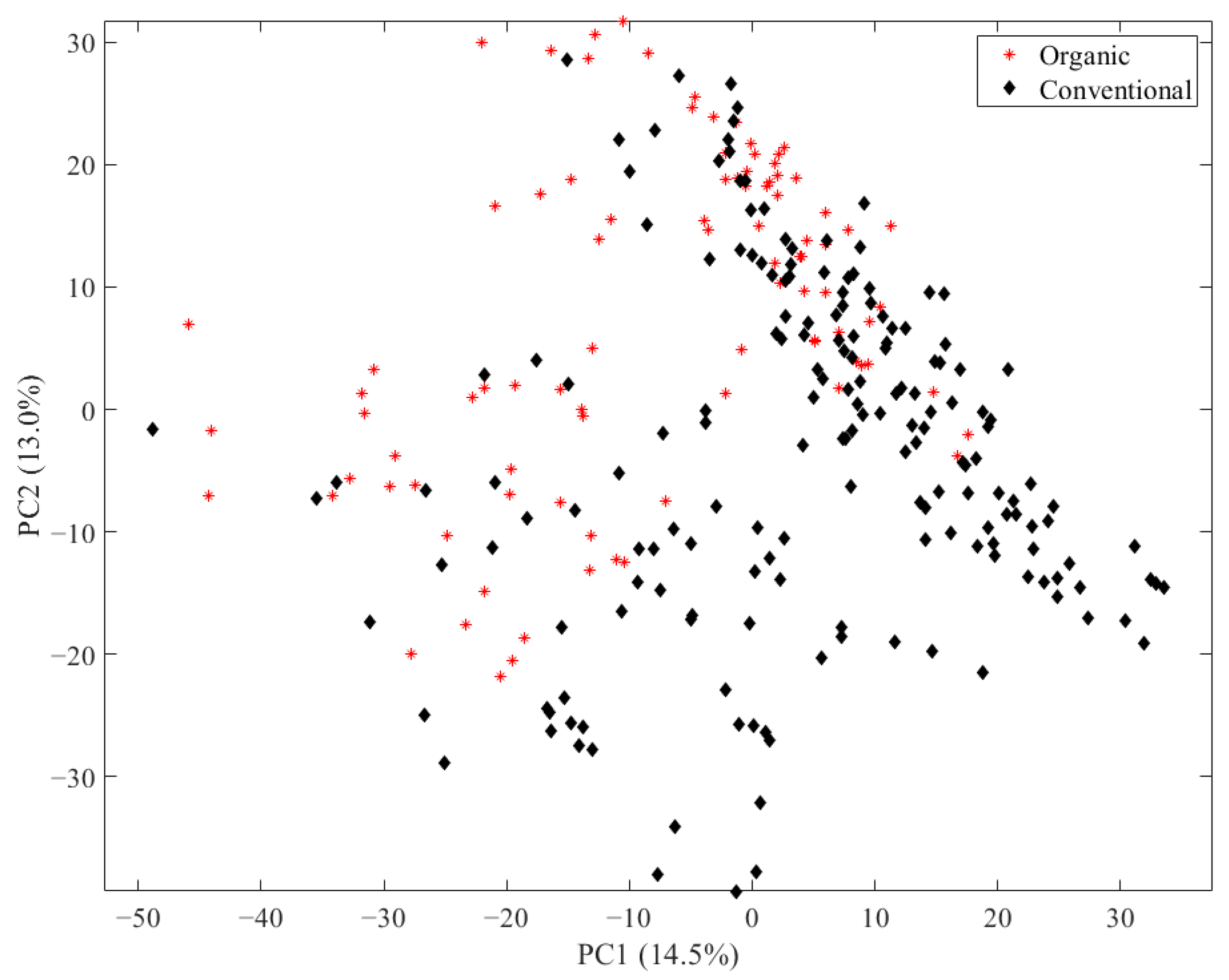
3.1. LC-ESI-IM-qToF-MS
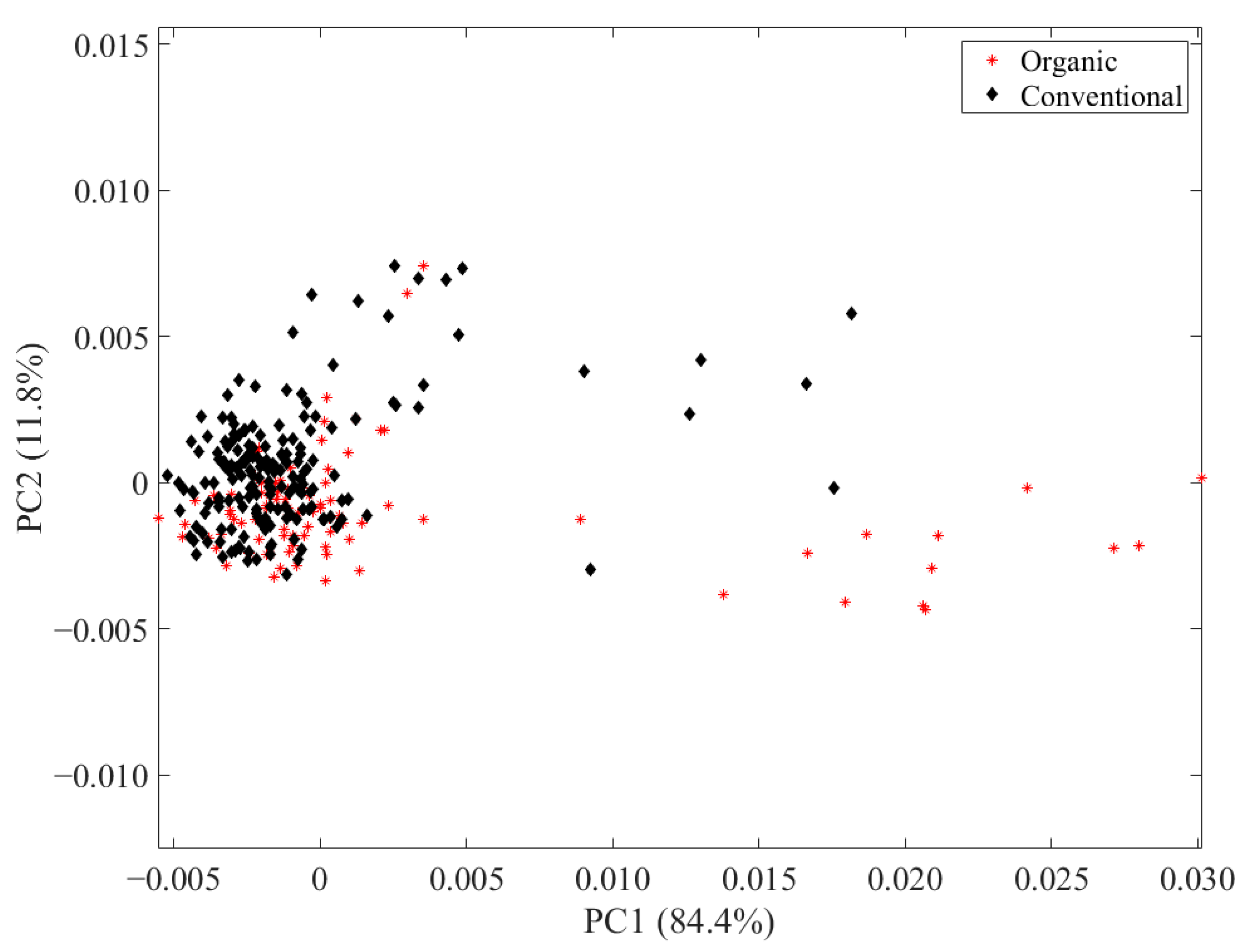
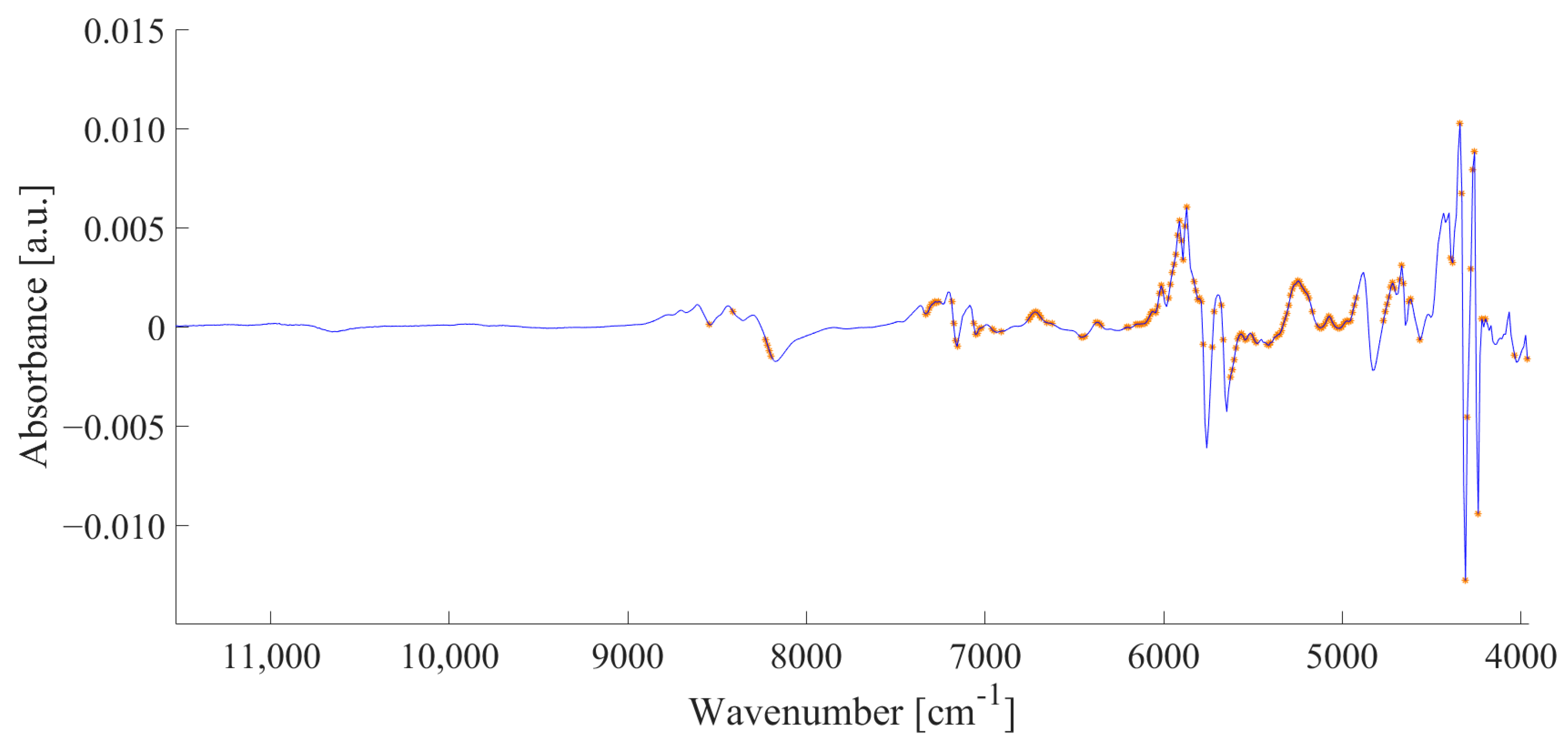
3.2. FT-NIR Spectroscopy
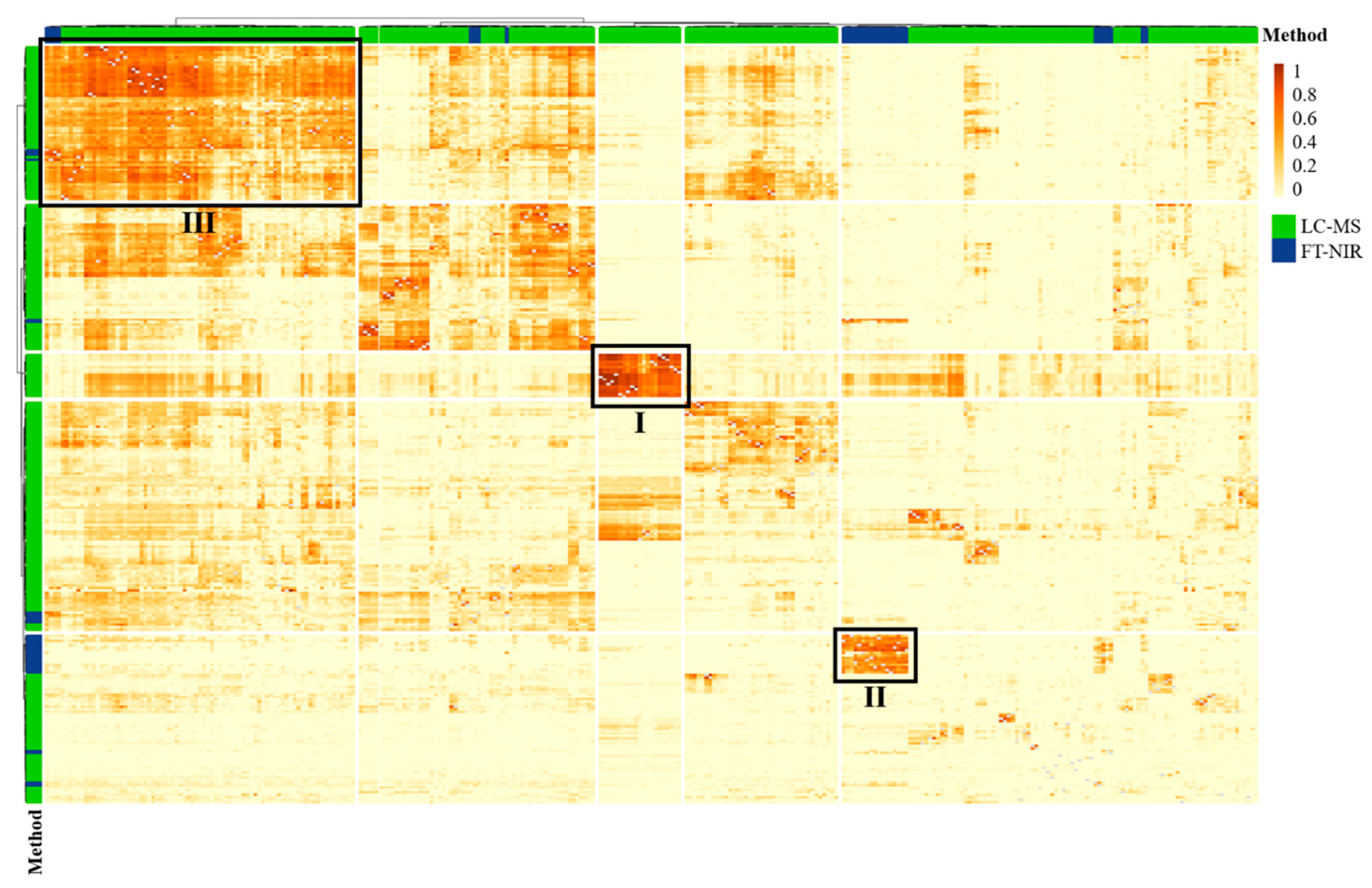
3.3. Data Fusion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capuano, E.; Boerrigter-Eenling, R.; van der Veer, G.; van Ruth, S.M. Analytical authentication of organic products: An overview of markers. J. Sci. Food Agric. 2013, 93, 12–28. [Google Scholar] [CrossRef]
- Küçükyılmaz, K.; Bozkurt, M.; Yamaner, Ç.; Çınar, M.; Çatlı, A.U.; Konak, R. Effect of an organic and conventional rearing system on the mineral content of hen eggs. Food Chem. 2012, 132, 989–992. [Google Scholar] [CrossRef]
- Giannenas, I.; Nisianakis, P.; Gavrill, A.; Kontopidis, G.; Kyriazakis, I. Trace mineral content of conventional, organic and courtyard eggs analysed by inductively coupled plasma mass spectrometry (ICP-MS). Food Chem. 2009, 114, 706–711. [Google Scholar] [CrossRef]
- Siderer, Y.; Maquet, A.; Anklam, E. Need for research to support consumer confidence in the growing organic food market. Trends Food Sci. Technol. 2005, 16, 332–343. [Google Scholar] [CrossRef]
- Council of Europe. Protocol of Amendment to the European Convention for the Protection of Animals Kept for Farming Purposes; European Treaty Series—No. 145; Council of Europe: Strasbourg, France, 1992. [Google Scholar]
- Commission Regulation (EC) No 589/2008 of 23 June 2008 Laying Down Detailed Rules for Implementing Council Regulation (EC) No 1234/2007 as Regards Marketing Standards for Eggs: VO (EG) No. 589/2008. 2008. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32008R0589 (accessed on 13 February 2023).
- Regulation (EU) No 1308/2013 of the European Parliament and of the Council of 17 December 2013 Establishing a Common Organisation of the Markets in Agricultural Products and Repealing Council Regulations (EEC) No 922/72, (EEC) No 234/79, (EC) No 1037/2001 and (EC) No 1234/2007: VO (EG) No. 1308/2013. 2013. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32013R1308 (accessed on 13 February 2023).
- Council Regulation (EU) 2018/848 of the European Parliament and of the Council of 30 May 2018 on Organic Production and Labelling of Organic Products and Repealing Council Regulation (EC) No 834/2007: VO (EG) No. 848/2018. 2018. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32018R0848 (accessed on 13 February 2023).
- Leenstra, F.; Maurer, V.; Galea, F.; Bestman, M.; Amsler-Kepalaite, Z.; Visscher, J.; Vermeij, I.; van Krimpen, M. Laying hen performance in different production systems; why do they differ and how to close the gap? Results of discussions with groups of farmers in The Netherlands, Switzerland and France, benchmarking and model calculations. Eur. Poult. Sci. 2014, 78, 1–10. [Google Scholar] [CrossRef]
- Willer, H.; Schaack, D.; Lernoud, J. Organic Farming and Market Development in Europe and the European Union. In FiBL & IFOAM—Organics International: The World of Organic Agriculture; Research Institute of Organic Agriculture FiBL and IFOAM-Organics International: Frick, Switzerland, 2018; pp. 218–255. [Google Scholar]
- European Union. Food Fraud Summary January 2018. Available online: https://knowledge4policy.ec.europa.eu/publication/food-fraud-summary-january-2018_en (accessed on 20 July 2023).
- European Union. Food Fraud Summary June 2018. Available online: https://knowledge4policy.ec.europa.eu/publication/food-fraud-summary-june-2018_en (accessed on 20 July 2023).
- European Union. Food Fraud Summary March 2018. Available online: https://knowledge4policy.ec.europa.eu/publication/food-fraud-summary-march-2018_en (accessed on 20 July 2023).
- Spink, J.; Moyer, D.C. Defining the public health threat of food fraud. J. Food Sci. 2011, 76, R157–R163. [Google Scholar] [CrossRef] [PubMed]
- Bannor, R.K.; Arthur, K.K.; Oppong, D.; Oppong-Kyeremeh, H. A comprehensive systematic review and bibliometric analysis of food fraud from a global perspective. J. Agric. Food Res. 2023, 14, 100686. [Google Scholar] [CrossRef]
- Mi, S.; Shang, K.; Zhang, C.-H.; Fan, Y.-Q. Characterization and discrimination of selected chicken eggs in China’s retail market based on multi-element and lipidomics analysis. Food Res. Int. 2019, 126, 108668. [Google Scholar] [CrossRef]
- Średnicka-Tober, D.; Barański, M.; Seal, C.; Sanderson, R.; Benbrook, C.; Steinshamn, H.; Gromadzka-Ostrowska, J.; Rembiałkowska, E.; Skwarło-Sońta, K.; Eyre, M.; et al. Composition differences between organic and conventional meat: A systematic literature review and meta-analysis. Br. J. Nutr. 2016, 115, 994–1011. [Google Scholar] [CrossRef]
- Tsiplakou, E.; Kotrotsios, V.; Hadjigeorgiou, I.; Zervas, G. Differences in sheep and goats milk fatty acid profile between conventional and organic farming systems. J. Dairy Res. 2010, 77, 343–349. [Google Scholar] [CrossRef]
- Borges, E.M.; Volmer, D.A.; Gallimberti, M.; de Souza, D.F.; de Souza, E.L.; Barbosa, F. Evaluation of macro- and microelement levels for verifying the authenticity of organic eggs by using chemometric techniques. Anal. Methods 2015, 7, 2577–2584. [Google Scholar] [CrossRef]
- Filipiak-Florkiewicz, A.; Deren, K.; Florkiewicz, A.; Topolska, K.; Juszczak, L.; Cieslik, E. The quality of eggs (organic and nutraceutical vs. conventional) and their technological properties. Poult. Sci. 2017, 96, 2480–2490. [Google Scholar] [CrossRef] [PubMed]
- Mugnai, C.; Sossidou, E.N.; Dal Bosco, A.; Ruggeri, S.; Mattioli, S.; Castellini, C. The effects of husbandry system on the grass intake and egg nutritive characteristics of laying hens. J. Sci. Food Agric. 2014, 94, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Rogers, K.M. Stable isotopes as a tool to differentiate eggs laid by caged, barn, free range, and organic hens. J. Agric. Food Chem. 2009, 57, 4236–4242. [Google Scholar] [CrossRef]
- Samman, S.; Kung, F.P.; Carter, L.M.; Foster, M.J.; Ahmad, Z.I.; Phuyal, J.L.; Petocz, P. Fatty acid composition of certified organic, conventional and omega-3 eggs. Food Chem. 2009, 116, 911–914. [Google Scholar] [CrossRef]
- Tres, A.; van Ruth, S.M. Verification of organic feed identity by fatty acid fingerprinting. J. Agric. Food Chem. 2011, 59, 8816–8821. [Google Scholar] [CrossRef]
- Vincevica-Gaile, Z.; Gaga, K.; Klavins, M. Food and Environment: Trace Element Content of Hen Eggs from Different Housing Types. APCBEE Procedia 2013, 5, 221–226. [Google Scholar] [CrossRef] [Green Version]
- Puertas, G.; Vázquez, M. Fraud detection in hen housing system declared on the eggs’ label: An accuracy method based on UV-VIS-NIR spectroscopy and chemometrics. Food Chem. 2019, 288, 8–14. [Google Scholar] [CrossRef]
- Puertas, G.; Cazón, P.; Vázquez, M. A quick method for fraud detection in egg labels based on egg centrifugation plasma. Food Chem. 2023, 402, 134507. [Google Scholar] [CrossRef]
- Breiman, L. Out-of-Bag Estimation; Technical Report; University of Californi: Berkeley, CA, USA, 1996. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Biau, G.; Scornet, E. A random forest guided tour. TEST 2016, 25, 197–227. [Google Scholar] [CrossRef] [Green Version]
- Seifert, S.; Gundlach, S.; Szymczak, S. Surrogate minimal depth as an importance measure for variables in random forests. Bioinformatics 2019, 35, 3663–3671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishwaran, H.; Kogalur, U.B.; Gorodeski, E.Z.; Minn, A.J.; Lauer, M.S. High-Dimensional Variable Selection for Survival Data. J. Am. Stat. Assoc. 2010, 105, 205–217. [Google Scholar] [CrossRef]
- Wenck, S.; Creydt, M.; Hansen, J.; Gärber, F.; Fischer, M.; Seifert, S. Opening the Random Forest Black Box of the Metabolome by the Application of Surrogate Minimal Depth. Metabolites 2021, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Shakiba, N.; Gerdes, A.; Holz, N.; Wenck, S.; Bachmann, R.; Schneider, T.; Seifert, S.; Fischer, M.; Hackl, T. Determination of the geographical origin of hazelnuts (Corylus avellana L.) by Near-Infrared spectroscopy (NIR) and a Low-Level Fusion with nuclear magnetic resonance (NMR). Microchem. J. 2022, 174, 107066. [Google Scholar] [CrossRef]
- Ackermann, S.M.; Lachenmeier, D.W.; Kuballa, T.; Schütz, B.; Spraul, M.; Bunzel, M. NMR-based differentiation of conventionally from organically produced chicken eggs in Germany. Magn. Reson. Chem. 2019, 57, 579–588. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, D527–D532. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Tu, J.; Xiong, X.; Shen, X.; Zhu, Z.-J. LipidCCS: Prediction of Collision Cross-Section Values for Lipids with High Precision To Support Ion Mobility-Mass Spectrometry-Based Lipidomics. Anal. Chem. 2017, 89, 9559–9566. [Google Scholar] [CrossRef]
- Geladi, P.; MacDougall, D.; Martens, H. Linearization and Scatter-Correction for Near-Infrared Reflectance Spectra of Meat. Appl. Spectrosc. 1985, 39, 491–500. [Google Scholar] [CrossRef]
- Rinnan, Å.; van den Berg, F.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Arndt, M.; Drees, A.; Ahlers, C.; Fischer, M. Determination of the Geographical Origin of Walnuts (Juglans regia L.) Using Near-Infrared Spectroscopy and Chemometrics. Foods 2020, 9, 1860. [Google Scholar] [CrossRef] [PubMed]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Arndt, M.; Rurik, M.; Drees, A.; Ahlers, C.; Feldmann, S.; Kohlbacher, O.; Fischer, M. Food authentication: Determination of the geographical origin of almonds (Prunus dulcis Mill.) via near-infrared spectroscopy. Microchem. J. 2021, 160, 105702. [Google Scholar] [CrossRef]
- Engel, J.; Gerretzen, J.; Szymańska, E.; Jansen, J.J.; Downey, G.; Blanchet, L.; Buydens, L.M. Breaking with trends in pre-processing? TrAC Trends Anal. Chem. 2013, 50, 96–106. [Google Scholar] [CrossRef]
- Wright, M.N.; Ziegler, A. ranger: A fast implementation of random forests for high dimensional data in C++ and R. arXiv 2015, arXiv:1508.04409. [Google Scholar] [CrossRef] [Green Version]
- Kolde, R. Pheatmap: Pretty Heatmaps, R Package Version 1.0. 12; R Package: Madison, WI, USA, 2019.
- Schütz, D.; Achten, E.; Creydt, M.; Riedl, J.; Fischer, M. Non-Targeted LC-MS Metabolomics Approach towards an Authentication of the Geographical Origin of Grain Maize (Zea mays L.) Samples. Foods 2021, 10, 2160. [Google Scholar] [CrossRef]
- Klockmann, S.; Reiner, E.; Bachmann, R.; Hackl, T.; Fischer, M. Food Fingerprinting: Metabolomic Approaches for Geographical Origin Discrimination of Hazelnuts (Corylus avellana) by UPLC-QTOF-MS. J. Agric. Food Chem. 2016, 64, 9253–9262. [Google Scholar] [CrossRef]
- Creydt, M.; Flügge, F.; Dammann, R.; Schütze, B.; Günther, U.L.; Fischer, M. Food Fingerprinting: LC-ESI-IM-QTOF-Based Identification of Blumeatin as a New Marker Metabolite for the Detection of Origanum majorana Admixtures to O. onites/vulgare. Metabolites 2023, 13, 673. [Google Scholar] [CrossRef]
- Esatbeyoglu, T.; Rimbach, G. Canthaxanthin: From molecule to function. Mol. Nutr. Food Res. 2017, 61, 1600469. [Google Scholar] [CrossRef]
- van Ruth, S.; Alewijn, M.; Rogers, K.; Newton-Smith, E.; Tena, N.; Bollen, M.; Koot, A. Authentication of organic and conventional eggs by carotenoid profiling. Food Chem. 2011, 126, 1299–1305. [Google Scholar] [CrossRef]
- Huck, C.W.; Popp, M.; Scherz, H.; Bonn, G.K. Development and evaluation of a new method for the determination of the carotenoid content in selected vegetables by HPLC and HPLC-MS-MS. J. Chromatogr. Sci. 2000, 38, 441–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bovskova, H.; Mikova, K.; Panovska, Z. Evaluation of Egg Yolk Colour. Czech J. Food Sci. 2014, 32, 213–217. [Google Scholar] [CrossRef] [Green Version]
- Saleh, A.A.; Gawish, E.; Mahmoud, S.F.; Amber, K.; Awad, W.; Alzawqari, M.H.; Shukry, M.; Abdel-Moneim, A.-M.E. Effect of Natural and Chemical Colorant Supplementation on Performance, Egg-Quality Characteristics, Yolk Fatty-Acid Profile, and Blood Constituents in Laying Hens. Sustainability 2021, 13, 4503. [Google Scholar] [CrossRef]
- Spada, F.P.; Selani, M.M.; Coelho, A.A.D.; Savino, V.J.M.; Rodella, A.A.; Souza, M.C.; Fischer, F.S.; Lemes, D.E.A.; Canniatti-Brazaca, S.G. Influence of natural and synthetic carotenoids on the color of egg yolk. Sci. Agric. 2016, 73, 234–242. [Google Scholar] [CrossRef] [Green Version]
- Frick, A.A.; Weyermann, C. An untargeted lipidomic approach for qualitative determination of latent fingermark glycerides using UPLC-IMS-QToF-MSE. Analyst 2019, 144, 3590–3600. [Google Scholar] [CrossRef]
- Byrdwell, W.C.; Neff, W.E. Dual parallel electrospray ionization and atmospheric pressure chemical ionization mass spectrometry (MS), MS/MS and MS/MS/MS for the analysis of triacylglycerols and triacylglycerol oxidation products. Rapid Commun. Mass Spectrom. 2002, 16, 300–319. [Google Scholar] [CrossRef] [PubMed]
- Milinsk, M.C.; Murakami, A.E.; Gomes, S.; Matsushita, M.; de Souza, N.E. Fatty acid profile of egg yolk lipids from hens fed diets rich in n-3 fatty acids. Food Chem. 2003, 83, 287–292. [Google Scholar] [CrossRef]
- Workman, J., Jr.; Weyer, L. Practical Guide and Spectral Atlas for Interpretive Near-Infrared Spectroscopy; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Brenna, O.V.; Berardo, N. Application of near-infrared reflectance spectroscopy (NIRS) to the evaluation of carotenoids content in maize. J. Agric. Food Chem. 2004, 52, 5577–5582. [Google Scholar] [CrossRef]
- Nokkaew, R. Determination of Carotenoids and DOBI content in crude Palm Oil by Spectroscopy Techniques: Comparison of Raman and FT-NIR Spectroscopy. GEOMATE 2019, 16, 92–98. [Google Scholar] [CrossRef]
- de Oliveira, G.A.; de Castilhos, F.; Renard, C.M.-G.C.; Bureau, S. Comparison of NIR and MIR spectroscopic methods for determination of individual sugars, organic acids and carotenoids in passion fruit. Food Res. Int. 2014, 60, 154–162. [Google Scholar] [CrossRef]
- Ruiz, D.; Reich, M.; Bureau, S.; Renard, C.M.G.C.; Audergon, J.-M. Application of reflectance colorimeter measurements and infrared spectroscopy methods to rapid and nondestructive evaluation of carotenoids content in apricot (Prunus armeniaca L.). J. Agric. Food Chem. 2008, 56, 4916–4922. [Google Scholar] [CrossRef] [PubMed]
- Creydt, M.; Arndt, M.; Hudzik, D.; Fischer, M. Plant Metabolomics: Evaluation of Different Extraction Parameters for Nontargeted UPLC-ESI-QTOF-Mass Spectrometry at the Example of White Asparagus officinalis. J. Agric. Food Chem. 2018, 66, 12876–12887. [Google Scholar] [CrossRef] [PubMed]

| Predicted | ||||
|---|---|---|---|---|
| Organic (%) | Conventional (%) | Sensitivity (%) | ||
| True | Organic (%) | 97.8 | 2.2 | 97.8 |
| Conventional (%) | 4.5 | 95.5 | 95.5 | |
| Specificity (%) | 91.8 | 98.8 | 96.3 | |
| Predicted | ||||
|---|---|---|---|---|
| Organic (%) | Conventional (%) | Sensitivity (%) | ||
| True | Organic (%) | 71.7 | 28.3 | 71.7 |
| Conventional (%) | 15.7 | 84.3 | 84.3 | |
| Specificity (%) | 70.2 | 85.2 | 80.0 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lösel, H.; Brockelt, J.; Gärber, F.; Teipel, J.; Kuballa, T.; Seifert, S.; Fischer, M. Comparative Analysis of LC-ESI-IM-qToF-MS and FT-NIR Spectroscopy Approaches for the Authentication of Organic and Conventional Eggs. Metabolites 2023, 13, 882. https://doi.org/10.3390/metabo13080882
Lösel H, Brockelt J, Gärber F, Teipel J, Kuballa T, Seifert S, Fischer M. Comparative Analysis of LC-ESI-IM-qToF-MS and FT-NIR Spectroscopy Approaches for the Authentication of Organic and Conventional Eggs. Metabolites. 2023; 13(8):882. https://doi.org/10.3390/metabo13080882
Chicago/Turabian StyleLösel, Henri, Johannes Brockelt, Florian Gärber, Jan Teipel, Thomas Kuballa, Stephan Seifert, and Markus Fischer. 2023. "Comparative Analysis of LC-ESI-IM-qToF-MS and FT-NIR Spectroscopy Approaches for the Authentication of Organic and Conventional Eggs" Metabolites 13, no. 8: 882. https://doi.org/10.3390/metabo13080882
APA StyleLösel, H., Brockelt, J., Gärber, F., Teipel, J., Kuballa, T., Seifert, S., & Fischer, M. (2023). Comparative Analysis of LC-ESI-IM-qToF-MS and FT-NIR Spectroscopy Approaches for the Authentication of Organic and Conventional Eggs. Metabolites, 13(8), 882. https://doi.org/10.3390/metabo13080882






