Influence of Symbiotic Fermentation Broth on Regulating Metabolism with Gut Microbiota and Metabolite Profiles Is Estimated Using a Third-Generation Sequencing Platform
Abstract
:1. Introduction
2. Materials and Methods
2.1. Production of SFBS
2.2. Ethics Statement
2.3. Animal Model
2.4. Analysis of Biochemical Variables
2.5. Isolation of Total Genomic DNA in the Fecal Samples
2.6. Profiling of Gut Microenvironment
2.7. Metabolites of SFBS Are Profiled Using an Untargeted LC-MS/MS Analysis
2.8. Statistical Analysis
3. Results
3.1. The Metabolite Profile of SFBS Is Assessed Using the LC-MS/MS Approach
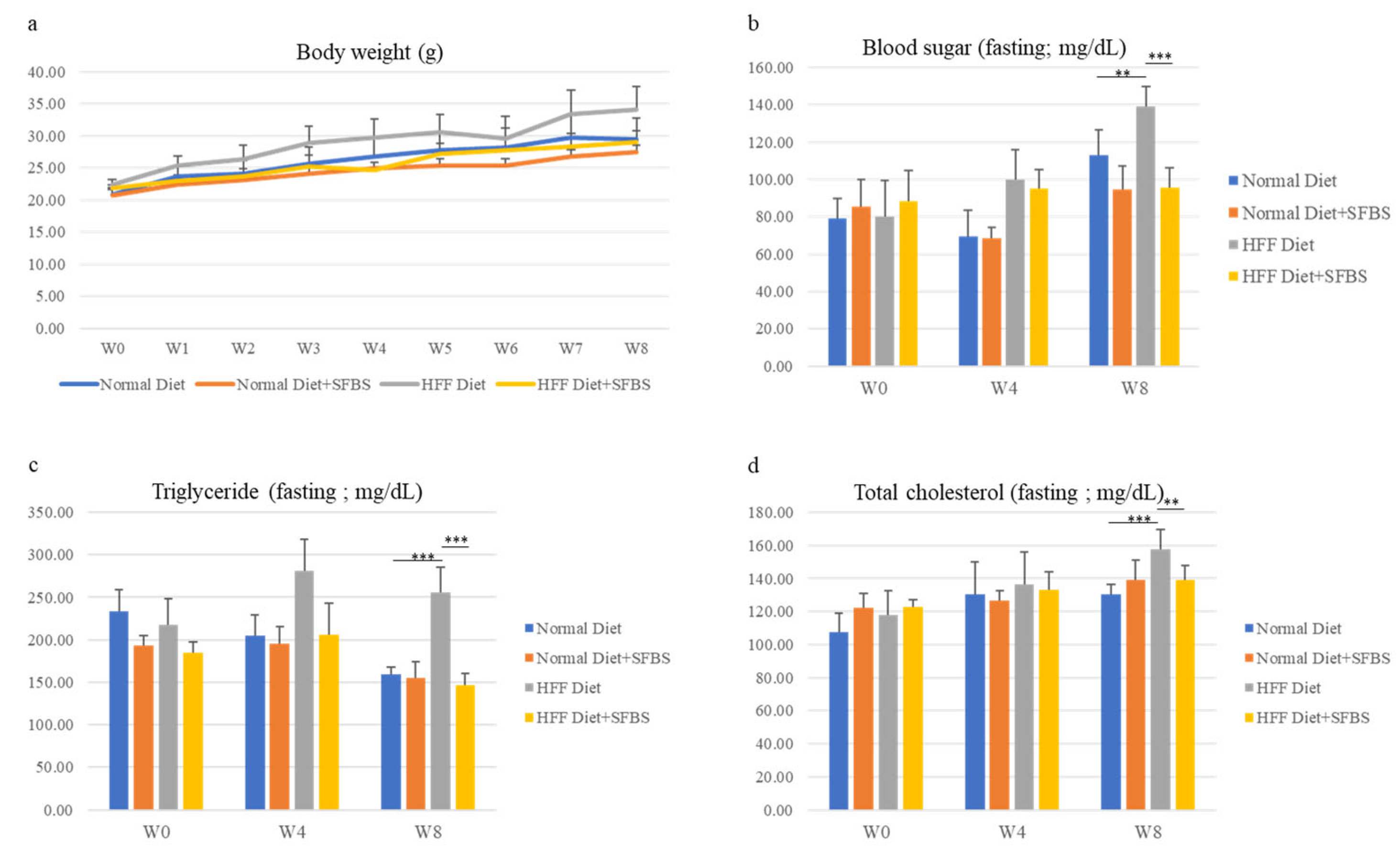
3.2. SFBS Supplementation Diminishes the Impact of a High-Fat-Fructose Diet on Biochemical Variables in Serum
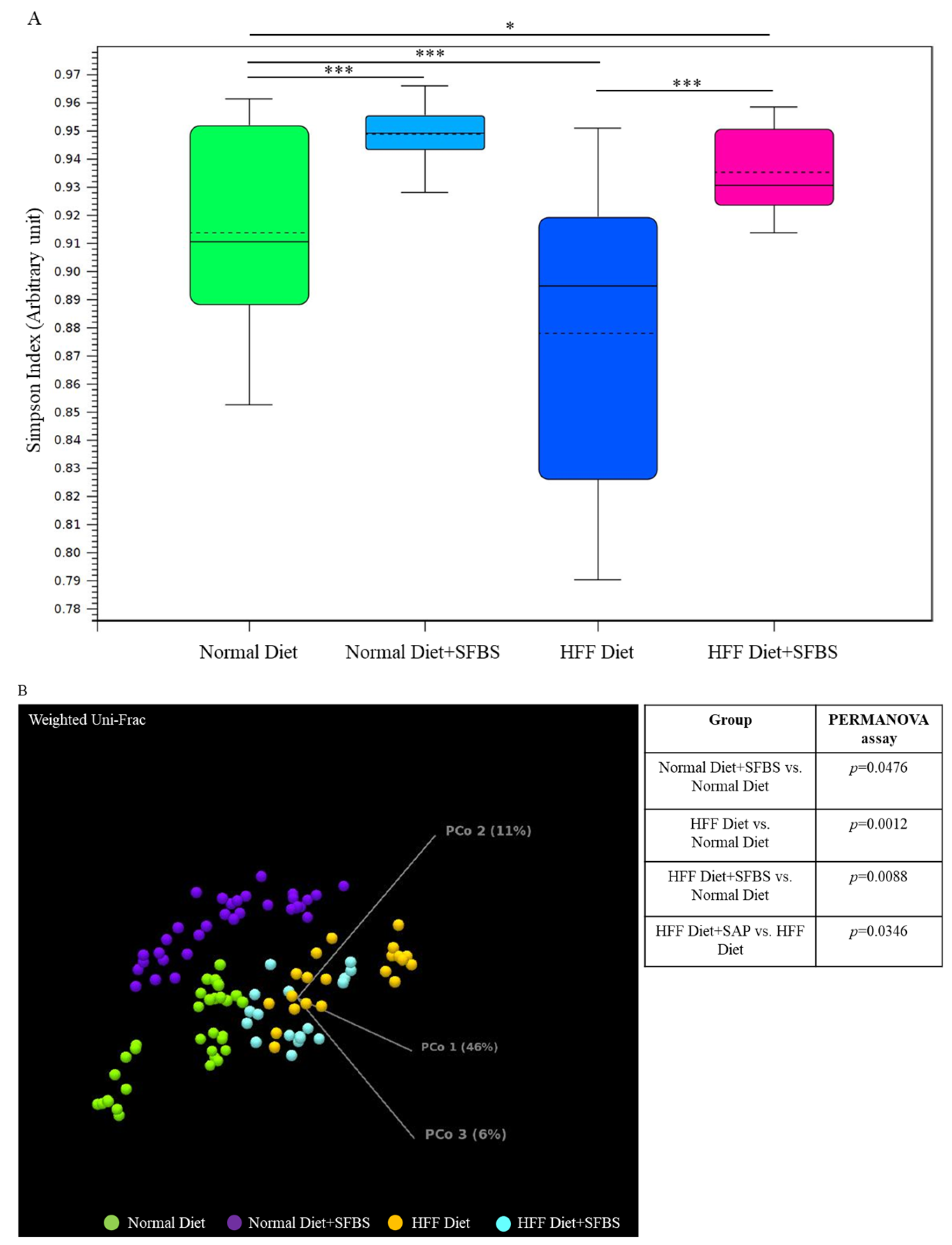
3.3. SFBS Supplementation Reprograms HFF Diet-Manipulated Gut Microenvironment
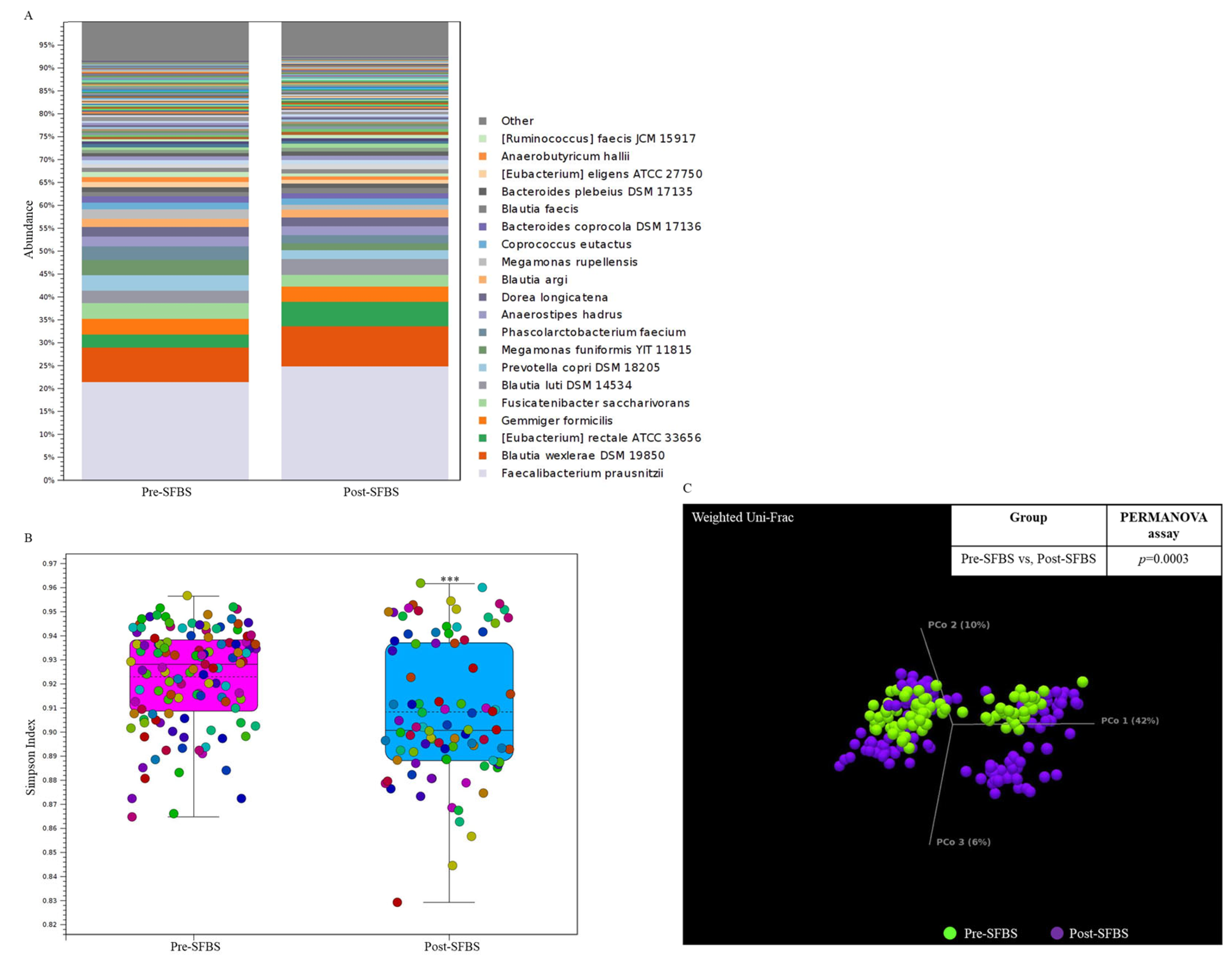
3.4. SFBS Supplementation Reprograms the Gut Microenvironment of Enrolled Participants
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, J.; Wei, H.; Zhou, Y.; Szeto, C.H.; Li, C.; Lin, Y.; Coker, O.O.; Lau, H.C.H.; Chan, A.W.H.; Sung, J.J.Y.; et al. High-fat diet promotes colorectal tumorigenesis through modulating gut microbiota and metabolites. Gastroenterol. 2022, 162, 135–149. [Google Scholar] [CrossRef]
- Zepeda-Hernández, A.; Garcia-Amezquita, L.E.; Requena, T.; García-Cayuela, T. Probiotics, prebiotics, and synbiotics added to dairy products: Uses and applications to manage type 2 diabetes. Food Res. Int. 2021, 142, 110208. [Google Scholar] [CrossRef] [PubMed]
- Kusumo, P.D.; Maulahela, H.; Utari, A.P.; Surono, I.S.; Soebandrio, A.; Abdullah, M. Probiotic Lactobacillus plantarum IS 10506 supplementation increase SCFA of women with functional constipation. Iran. J. Microbiol. 2019, 11, 389–396. [Google Scholar] [PubMed]
- Rezende, E.S.V.; Lima, G.C.; Naves, M.M.V. Dietary fibers as beneficial microbiota modulators: A proposal classification by prebiotic categories. Nutrition 2021, 89, 111217. [Google Scholar] [CrossRef]
- Menne, E.; Guggenbuhl, N.; Roberfroid, M. Fn-type chicory inulin hydrolysate has a prebiotic effect in humans. J. Nutr. 2000, 130, 1197–1199. [Google Scholar] [CrossRef] [PubMed]
- Guaragni, A.; Boiago, M.M.; Bottari, N.B.; Morsch, V.M.; Lopes, T.F.; Schafer da Silva, A. Feed supplementation with inulin on broiler performance and meat quality challenged with Clostridium perfringens: Infection and prebiotic impacts. Microb. Pathog. 2020, 139, 103889. [Google Scholar] [CrossRef]
- Miao, M.; Wang, Q.; Wang, X.; Fan, C.; Luan, T.; Yan, L.; Zhang, Y.; Zeng, X.; Dai, Y.; Li, P. The protective effects of inulin-type fructans against high-fat/sucrose diet-induced gestational diabetes mice in association with gut microbiota regulation. Front. Microbiol. 2022, 13, 832151. [Google Scholar] [CrossRef]
- Marrelli, M.; Conforti, F.; Araniti, F.; Statti, G.A. Effects of saponins on lipid metabolism: A review of potential health benefits in the treatment of obesity. Molecules 2016, 21, 1404. [Google Scholar] [CrossRef]
- Hughes, K.; Le, T.B.; Van Der Smissen, P.; Tyteca, D.; Mingeot-Leclercq, M.P.; Quetin-Leclercq, J. The antileishmanial activity of eugenol associated with lipid storage reduction rather than membrane properties alterations. Molecules 2023, 28, 3871. [Google Scholar] [CrossRef]
- Creighton, J.V.; de Souza Gonçalves, L.; Artioli, G.G.; Tan, D.; Elliott-Sale, K.J.; Turner, M.D.; Doig, C.L.; Sale, C. Physiological roles of carnosine in myocardial function and health. Adv. Nutr. 2022, 13, 1914–1929. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.S.; Prasad, S.K.; Shivamallu, C.; Prasad, K.S.; Syed, A.; Reddy, P.; Cull, C.A.; Amachawadi, R.G. Genistein: A potent anti-breast cancer agent. Curr Issues Mol. Biol. 2021, 43, 1502–1517. [Google Scholar] [CrossRef]
- Wert-Carvajal, C.; Reneaux, M.; Tchumatchenko, T.; Clopath, C. Dopamine and serotonin interplay for valence-based spatial learning. Cell Rep. 2022, 39, 110645. [Google Scholar] [CrossRef]
- Marshall, A.; McGrath, J.W.; Graham, R.; McMullan, G. Food for thought-The link between Clostridioides difficile metabolism and pathogenesis. PLoS Pathog. 2023, 19, e1011034. [Google Scholar] [CrossRef] [PubMed]
- Vicentini, F.A.; Keenan, C.M.; Wallace, L.E.; Woods, C.; Cavin, J.B.; Flockton, A.R.; Macklin, W.B.; Belkind-Gerson, J.; Hirota, S.A.; Sharkey, K.A. Intestinal microbiota shapes gut physiology and regulates enteric neurons and glia. Microbiome 2021, 9, 210. [Google Scholar] [CrossRef]
- Yin, W.; Zhang, S.-Q.; Pang, W.-L.; Chen, X.-J.; Wen, J.; Hou, J.; Wang, C.; Song, L.Y.; Qiu, Z.M.; Liang, P.T. Tang-Ping-San decoction remodel intestinal flora and barrier to ameliorate type 2 diabetes mellitus in rodent model. Diabetes Metab. Syndr. Obes. Targets Ther. 2022, 15, 2563–2581. [Google Scholar] [CrossRef] [PubMed]
- Ju, M.; Liu, Y.; Li, M.; Cheng, M.; Zhang, Y.; Deng, G.; Kang, X.; Liu, H. Baicalin improves intestinal microecology and abnormal metabolism induced by high-fat diet. Eur. J. Pharmacol. 2019, 857, 172457. [Google Scholar] [CrossRef]
- Moon, H.D.; Eunjung, L.; Mi-Jin, O.; Yoonsook, K.; Ho-Young, P. High-glucose or -fructose diet cause changes of the gut microbiota and metabolic disorders in mice without body weight change. Nutrients 2018, 10, 761. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Wang, B.; Kong, Q.; Li, X.; Zhao, J.; Zhang, H.; Chen, W.; Wang, G.A. High-Fat Diet Increases Gut Microbiota Biodiversity and Energy Expenditure Due to Nutrient Difference. Nutrients 2020, 12, 3197. [Google Scholar] [CrossRef]
- Redondo-Cuenca, A.; García-Alonso, A.; Rodríguez-Arcos, R.; Castro, I.; Alba, C.; Rodríguez, J.M.; Goñi, I. Nutritional composition of green asparagus (Asparagus officinalis L.), edible part and by-products, and assessment of their effect on the growth of human gut-associated bacteria. Food Res. Int. 2023, 163, 112284. [Google Scholar] [CrossRef]
- Sergeev, I.N.; Aljutaily, T.; Walton, G.; Huarte, E. Effects of Synbiotic Supplement on Human Gut Microbiota, Body Composition and Weight Loss in Obesity. Nutrients 2020, 12, 222. [Google Scholar] [CrossRef] [PubMed]
- Di Modica, M.; Gargari, G.; Regondi, V.; Bonizzi, A.; Arioli, S.; Belmonte, B.; De Cecco, L.; Fasano, E.; Bianchi, F.; Bertolotti, A.; et al. Gut microbiota conditions the therapeutic efficacy of trastuzumab in her2-positive breast cancer. Cancer Res. 2021, 81, 2195–2206. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, Y. Gut microbiota derived metabolites in cardiovascular health and disease. Protein Cell. 2018, 9, 416–431. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, H.; Zhao, X.; Zhou, R.; Liu, H.; Sun, Y.; Fan, Y.; Shi, Y.; Qiao, S.; Liu, S.; et al. Multi-omics study reveals that statin therapy is associated with restoration of gut microbiota homeostasis and improvement in outcomes in patients with acute coronary syndrome. Theranostics 2021, 11, 5778–5793. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, Y.; Lu, F.; Mohammed, S.A.D.; Liu, H.; Ding, S.; Liu, S.M. Mechanism of action of shenerjiangzhi formulation on hyperlipidemia induced by consumption of a high-fat diet in rats using network pharmacology and analyses of the gut microbiota. Front. Pharmacol. 2022, 13, 745074. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.X.; Jiang, X.M.; Wang, H.W.; Ge, L.; Lai, Y.T.; Jiang, X.Y.; Chen, F.; Huang, P.P. Probiotic supplements alleviate gestational diabetes mellitus by restoring the diversity of gut microbiota: A study based on 16S rRNA sequencing. J. Microbiol. 2021, 59, 827–839. [Google Scholar] [CrossRef] [PubMed]
| ID | CpdName | Score | KEGGID | HMDBID | Microbe | Rtmed | Mzmed | Exp Mean | Ctrl Mean | VIP | p-Value | Fold Change |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FT0338 | Eugenol | 0.99059 | C10453 | HMDB0005809 | NA | 246.5537 | 128.0419 | 1.36 × 10−2 | 2.18 × 10−5 | 1.253382 | 0.017216 | 624.381 |
| FT6284 | Fasciculic acid B | 0.90713 | NA | HMDB0036438 | NA | 426.4757 | 795.4509 | 1.5 × 10−5 | 6.05 × 10−8 | 1.425796 | 0.006844 | 247.4448 |
| FT0799 | Homocarnosine | 0.78974 | C00884 | HMDB0000745 | YES | 318.4291 | 174.1119 | 1.13 × 10−3 | 1.29 × 10−5 | 1.240513 | 0.013296 | 87.37652 |
| FT4987 | Moclobemide | 0.80195 | NA | HMDB0015302 | NA | 266.6079 | 479.2484 | 2.9 × 10−4 | 9.02 × 10−5 | 0.399071 | 0.011479 | 32.17849 |
| FT1164 | Glycylleucine | 0.58063 | C02155 | HMDB0000759 | NA | 84.13725 | 203.0521 | 2.41 × 10−3 | 9.7 × 10−5 | 1.198751 | 0.005106 | 24.86357 |
| FT6289 | Soyasaponin I | 0.87364 | C08983 | HMDB0034649 | NA | 407.55 | 797.4662 | 7.55 × 10−4 | 4.02 × 10−5 | 1.02781 | 0.000364 | 18.79097 |
| FT2334 | Genistein | 0.99973 | C06563 | HMDB0003217 | YES | 341.9878 | 271.0593 | 4.5 × 10−4 | 3.1 × 10−5 | 1.352531 | 0.000707 | 14.54516 |
| FT1837 | L-Glutamine | 0.91368 | C00064 | HMDB0000641 | YES | 241.1555 | 244.1284 | 2.97 × 10−4 | 2.35 × 10−5 | 1.37705 | 0.023164 | 12.63435 |
| FT4728 | Phlorizin | 0.69916 | C01604 | HMDB0036634 | YES | 407.3074 | 448.243 | 5.23 × 10−5 | 4.62 × 10−6 | 1.351436 | 0.007354 | 11.31761 |
| FT1557 | Altretamine | 0.73286 | NA | HMDB0014631 | NA | 417.2145 | 228.1952 | 2.19 × 10−3 | 2.16 × 10−4 | 1.198833 | 0.027437 | 10.12372 |
| FT0340 | Dopamine | 0.96353 | C03758 | HMDB0000073 | YES | 296.1454 | 332.2169 | 2.17 × 10−4 | 4.6 × 10−5 | 1.336598 | 0.037817 | 4.722736 |
| FT0495 | 1,2,3,4-Tetrahydro-beta-carboline | 0.99943 | NA | HMDB0012488 | NA | 287.099 | 144.0803 | 6.98 × 10−3 | 2.68 × 10−3 | 1.166997 | 0.046653 | 2.606954 |
| ID | CpdName | Score | KEGGID | HMDBID | Microbe | Rtmed | Mzmed | Exp Mean | Ctrl Mean | VIP | p-Value | Fold Change |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FT1381 | Isocitric acid | 0.96879 | C00311 | HMDB0000193 | YES | 94.38789 | 353.072 | 0.020077 | 6.85 × 10−7 | 1.280033 | 0.045768 | 29325.62 |
| FT0754 | DL-O-Phosphoserine | 0.64298 | C01005 | HMDB0001721 | YES | 54.73537 | 259.0221 | 0.003051 | 7.57 × 10−5 | 1.06287 | 0.008156 | 40.28009 |
| FT1176 | Aspalathin | 0.96254 | NA | HMDB0030851 | NA | 331.2802 | 331.118 | 0.001473 | 3.99 × 10−5 | 1.246585 | 0.021972 | 36.96136 |
| FT0090 | Malic acid | 0.94492 | C00149 | HMDB0000156 | YES | 68.6133 | 133.0143 | 0.026847 | 8.41 × 10−4 | 1.154805 | 0.021612 | 31.92659 |
| FT0234 | Rosmarinic acid | 0.86928 | C01850 | HMDB0003572 | YES | 63.7528 | 179.0562 | 0.044586 | 1.57 × 10−3 | 1.276599 | 0.025802 | 28.32409 |
| FT2818 | Phenytoin | 0.89564 | C07443 | HMDB0014397 | NA | 94.07186 | 503.1612 | 0.000834 | 4.9 × 10−5 | 1.278964 | 0.002793 | 17.00339 |
| FT1535 | Ornithine | 0.7267 | C00077 | HMDB0000214 | YES | 236.4346 | 515.1625 | 0.000537 | 4.45 × 10−5 | 1.14777 | 0.032421 | 12.06561 |
| FT0207 | L-Leucine | 0.99821 | C00123 | HMDB0000687 | YES | 317.1682 | 172.0978 | 0.001018 | 8.7 × 10−5 | 1.137649 | 0.019915 | 11.69737 |
| FT1500 | (-)-Epigallocatechin | 0.99869 | C12136 | HMDB0038361 | YES | 376.3322 | 253.0502 | 0.000785 | 8.39 × 10−5 | 1.181254 | 0.010312 | 9.351916 |
| FT0017 | Succinic acid | 0.93795 | C00042 | HMDB0000254 | YES | 70.84436 | 101.0245 | 0.000561 | 1.22 × 10−4 | 1.122522 | 0.042969 | 4.60625 |
| FT0132 | Cortisone | 0.61173 | C00762 | HMDB0002802 | YES | 331.8418 | 147.045 | 0.011588 | 2.63 × 10−3 | 1.233049 | 0.000266 | 4.406416 |
| FT2017 | Citric acid | 0.99132 | C00158 | HMDB0000094 | YES | 72.08018 | 405.0281 | 0.000482 | 1.31 × 10−4 | 1.035968 | 0.029749 | 3.676637 |
| FT0078 | Genipin | 0.83861 | C09780 | HMDB0035830 | NA | 314.9441 | 129.0557 | 0.001566 | 5.69 × 10−4 | 1.150752 | 0.000124 | 2.751451 |
| HFF Diet vs. Normal Diet | ||||
| Name | Fold Change | p-Value | FDR p-Value | Bonferroni |
| Akkermansia muciniphila ATCC BAA-835; NR_042817.1 | 151.15 | 3.61 × 10−23 | 2.36 × 10−20 | 4.73 × 10−20 |
| Romboutsia timonensis; NR_144740.1 | 104.99 | 3.03 × 10−18 | 1.32 × 10−15 | 3.96 × 10−15 |
| Akkermansia muciniphila; NR_074436.1 | 96.59 | 1.09 × 10−15 | 3.55 × 10−13 | 1.42 × 10−12 |
| Akkermansia glycaniphila; NR_152695.1 | 74.95 | 2.29 × 10−12 | 4.99 × 10−10 | 2.99 × 10−9 |
| Romboutsia ilealis; NR_125597.1 | 59.95 | 1.96 × 10−11 | 3.67 × 10−9 | 2.57 × 10−8 |
| Shigella boydii; NR_104901.1 | 25.39 | 1.02 × 10−6 | 9.57 × 10−5 | 1.34 × 10−3 |
| Luteolibacter gellanilyticus; NR_158117.1 | 20.04 | 1.54 × 10−6 | 1.26 × 10−4 | 2.01 × 10−3 |
| Paraclostridium benzoelyticum; NR_148815.1 | 19.27 | 2.20 × 10−6 | 1.69 × 10−4 | 2.87 × 10−4 |
| Parabacteroides goldsteinii DSM 19448 = WAL 12034; NR_113076.1 | 10.45 | 2.03 × 10−28 | 2.66 × 10−25 | 2.66 × 10−25 |
| Eubacterium coprostanoligenes; NR_104907.1 | 8.62 | 2.27 × 10−5 | 1.45 × 10−3 | 0.03 |
| Clostridium scindens; NR_028785.1 | 4.31 | 1.66 × 10−7 | 1.67 × 10−5 | 2.17 × 10−4 |
| Parabacteroides chongii; NR_165699.1 | 4.29 | 5.04 × 10−6 | 3.47 × 10−4 | 6.60 × 10−3 |
| Lactobacillus faecis; NR_114391.1 | −2.78 | 4.25 × 10−8 | 6.18 × 10−6 | 5.56 × 10−5 |
| Lactobacillus murinus; NR_112689.1 | −3.44 | 5.44 × 10−10 | 8.89 × 10−8 | 7.11 × 10−7 |
| Lactobacilpodemedemi; NR_112752.1 | −4.53 | 3.33 × 10−6 | 2.42 × 10−4 | 4.35 × 10−4 |
| HFF Diet + SFBS vs. Normal Diet | ||||
| Name | Fold Change | p-Value | FDR p-Value | Bonferroni |
| Romboutsia timonensis; NR_144740.1 | 35.93 | 5.54 × 10−19 | 1.81 × 10−16 | 7.25 × 10−16 |
| Akkermansia muciniphila ATCC BAA-835; NR_042817.1 | 31.89 | 2.03 × 10−13 | 2.41 × 10−11 | 2.66 × 10−10 |
| Romboutsia ilealis; NR_125597.1 | 29.72 | 2.55 × 10−10 | 1.85 × 10−8 | 3.34 × 10−7 |
| Akkermansia muciniphila; NR_074436.1 | 30.81 | 1.23 × 10−8 | 6.70 × 10−7 | 1.61 × 10−5 |
| Paraclostridium benzoelyticum; NR_148815.1 | 24.82 | 2.88 × 10−7 | 1.18 × 10−5 | 3.77 × 10−5 |
| Akkermansia glycaniphila; NR_152695.1 | 19.69 | 4.64 × 10−6 | 1.52 × 10−4 | 6.08 × 10−3 |
| Eubacterium coprostanoligenes; NR_104907.1 | 15.2 | 4.06 × 10−8 | 1.97 × 10−6 | 5.31 × 10−5 |
| Parabacteroides goldsteinii DSM 19448 = WAL 12034; NR_113076.1 | 9.48 | 9.00 × 10−24 | 9.36 × 10−21 | 1.18 × 10−20 |
| Lactobacillus faecis; NR_114391.1 | −2.66 | 1.71 × 10−6 | 6.21 × 10−5 | 2.23 × 10−3 |
| Lactobacillus murinus; NR_112689.1 | −3.11 | 1.38 × 10−7 | 6.02 × 10−6 | 1.81 × 10−4 |
| Mucispirillum schaedleri; NR_042896.1 | −3.89 | 1.94 × 10−5 | 5.78 × 10−4 | 0.03 |
| HFF Diet vs. Normal Diet | ||||||
| Pathway | MetaCyc ID | Min. Solution | Confidence | Coverage | Fold Change | Metabolite |
| tetrahydro pteridine recycling | PWY-8099 | TRUE | 0.5 | 1 | 17.68 | tetrahydrobiopterin |
| glutathionylspermidine biosynthesis | PWY-4121 | TRUE | 1 | 1 | 13.15 | glutathionylspermidine |
| phenylmercury acetate degradation | P641-PWY | TRUE | 1 | 1 | 13 | catechol |
| uracil degradation III | PWY0-1471 | TRUE | 0.53 | 0.67 | 12 | 3-hydroxypropanoate |
| sulfoquinovose degradation I | PWY-7446 | TRUE | 1 | 0.8 | 11.78 | dihydroxypropane-1-sulfonate |
| L-arginine degradation II (AST pathway) | AST-PWY | TRUE | 1 | 1 | 11.59 | glutamate |
| putrescine degradation IV | PWY-2 | TRUE | 0.56 | 0.67 | 10.07 | 4-aminobutanoate (GABA) |
| adenine and adenosine salvage V | PWY-6611 | TRUE | 0.52 | 0.67 | 8.4 | ribosyl hypoxanthine monophosphate |
| enterobacterial common antigen biosynthesis | ECASYN-PWY | TRUE | 1 | 0.67 | 7.7 | enterobacterial common antigen |
| cinnamate and 3-hydroxycinnamate degradation to 2-hydroxypentadienoate | PWY-6690 | TRUE | 0.5 | 1 | 7.22 | 2-hydroxypenta-2,4-dienoate |
| histamine biosynthesis | PWY-6173 | TRUE | 1 | 1 | 6.84 | histamine |
| aerobactin biosynthesis | AEROBACTINSYN-PWY | TRUE | 1 | 1 | 6.33 | aerobactin |
| aminopropanol phosphate biosynthesis I | PWY-5443 | TRUE | 1 | 1 | 6.05 | 1-amino-2-propanol O-2-phosphate |
| HFF Diet + SFBS vs. Normal Diet | ||||||
| Pathway | MetaCyc ID | Min. Solution | Confidence | Coverage | Fold Change | Metabolite |
| glutathionylspermidine biosynthesis | PWY-4121 | TRUE | 1 | 1 | 81.6 | glutathionylspermidine |
| sulfoquinovose degradation I | PWY-7446 | TRUE | 1 | 0.8 | 73.07 | dihydroxy propane-1-sulfonate |
| L-arginine degradation II (AST pathway) | AST-PWY | TRUE | 1 | 1 | 64.36 | glutamate |
| uracil degradation III | PWY0-1471 | TRUE | 0.53 | 0.67 | 62.66 | 3-hydroxy propanoate |
| putrescine degradation IV | PWY-2 | TRUE | 0.54 | 0.67 | 53.42 | 4-aminobutanoate (GABA) |
| enterobacterial common antigen biosynthesis | ECASYN-PWY | TRUE | 1 | 0.67 | 39.35 | enterobacterial common antigen |
| cinnamate and 3-hydroxycinnamate degradation to 2-hydroxypentadienoate | PWY-6690 | TRUE | 0.52 | 1 | 38.31 | 2-hydroxypenta-2,4-dienoate |
| aerobactin biosynthesis | AEROBACTINSYN-PWY | TRUE | 1 | 1 | 3.76 | aerobactin |
| Adenine and adenosine salvage V | PWY-6611 | TRUE | 0.55 | 1 | 35.9 | ribosyl hypoxanthine monophosphate |
| phenylmercury acetate degradation | P641-PWY | TRUE | 1 | 1 | 33.05 | catechol |
| tetrahydro pteridine recycling | PWY-8099 | TRUE | 0.52 | 1 | 13.51 | tetrahydrobiopterin |
| aminopropanol phosphate biosynthesis I | PWY-5443 | TRUE | 1 | 1 | 8.7 | 1-amino-2-propanol O-2-phosphate |
| histamine biosynthesis | PWY-6173 | TRUE | 1 | 1 | 4.97 | histamine |
| Post-SFBS Group vs. Pre-SFBS Group | ||||
|---|---|---|---|---|
| Name | Fold Change | p-Value | FDR p-Value | Bonferroni |
| Phascolarctobacterium succinatutens; NR_112902.1 | 3.37 | 8.37 × 10−5 | 2.35 × 10−4 | 6.34 × 10−4 |
| Ruminococcus faecis JCM 15917; NR_116747.1 | 2.11 | 3.27 × 10−5 | 3.54 × 10−3 | 0.02 |
| Phascolarctobacterium faecium; NR_026111.1 | 1.81 | 2.23 × 10−6 | 3.38 × 10−4 | 1.69 × 10−3 |
| Fusicatenibacter saccharivorans; NR_114326.1 | 1.68 | 1.69 × 10−5 | 2.14 × 10−3 | 0.01 |
| Eubacterium rectale ATCC 33656; NR_074634.1 | −1.79 | 1.33 × 10−16 | 1.01 × 10−13 | 1.01 × 10−13 |
| Roseburia intestinalis L1-82; NR_027557.1 | −2 | 4.31 × 10−5 | 4.08 × 10−3 | 0.03 |
| Clostridium saudiense; NR_144696.1 | −3.34 | 1.96 × 10−6 | 3.38 × 10−4 | 1.49 × 10−3 |
| Post-SFBS vs. Pre-SFBS | ||||||
|---|---|---|---|---|---|---|
| Pathway | MetaCyc ID | Min. Solution | Confidence | Coverage | Fold Change | Metabolite |
| sulfoacetaldehyde degradation III | PWY-6718 | TRUE | 1 | 1 | 2.87 | isethionate |
| L-arginine degradation XII | PWY-7523 | TRUE | 0.77 | 0.75 | 2.34 | 4-aminobutanoate |
| cellulose and hemicellulose degradation (cellulolosome) | PWY-6784 | TRUE | 1 | 0.73 | 2.34 | polysacharide |
| vanillin and vanillate degradation II | PWY-7098 | TRUE | 1 | 0.5 | 2.34 | protocatechuate |
| 1,4-dihydroxy-6-naphthoate biosynthesis II | PWY-7371 | TRUE | 0.55 | 0.6 | 2.11 | dihydroxyacetone |
| aminopropanol phosphate biosynthesis I | PWY-5443 | TRUE | 1 | 0.5 | 1.87 | 1-amino-2-propanol O-2-phosphate |
| gallate degradation I | GALLATE-DEGRADATION-II-PWY | TRUE | 1 | 0.75 | 1.84 | oxaloacetate |
| retinol biosynthesis | PWY-6857 | TRUE | 0.72 | 0.62 | 1.7 | trans-retinol |
| benzoate degradation I (aerobic) | PWY-2503 | TRUE | 1 | 0.5 | 1.67 | catechol |
| 4-methyl catechol degradation (ortho cleavage) | PWY-6185 | TRUE | 0.58 | 0.54 | 1.67 | acetyl-CoA |
| methiin metabolism | PWY-7614 | TRUE | 0.64 | 0.54 | 1.64 | pyruvate |
| pyruvate fermentation to propanoate I | P108-PWY | TRUE | 1 | 0.57 | 1.57 | propanoate |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.-Y.; Huang, C.-K.; Hong, W.-S.; Liu, Y.-H.; Shih, M.-C.; Lin, J.-C. Influence of Symbiotic Fermentation Broth on Regulating Metabolism with Gut Microbiota and Metabolite Profiles Is Estimated Using a Third-Generation Sequencing Platform. Metabolites 2023, 13, 999. https://doi.org/10.3390/metabo13090999
Wu C-Y, Huang C-K, Hong W-S, Liu Y-H, Shih M-C, Lin J-C. Influence of Symbiotic Fermentation Broth on Regulating Metabolism with Gut Microbiota and Metabolite Profiles Is Estimated Using a Third-Generation Sequencing Platform. Metabolites. 2023; 13(9):999. https://doi.org/10.3390/metabo13090999
Chicago/Turabian StyleWu, Chih-Yin, Chun-Kai Huang, Wei-Sheng Hong, Yin-Hsiu Liu, Ming-Chi Shih, and Jung-Chun Lin. 2023. "Influence of Symbiotic Fermentation Broth on Regulating Metabolism with Gut Microbiota and Metabolite Profiles Is Estimated Using a Third-Generation Sequencing Platform" Metabolites 13, no. 9: 999. https://doi.org/10.3390/metabo13090999
APA StyleWu, C.-Y., Huang, C.-K., Hong, W.-S., Liu, Y.-H., Shih, M.-C., & Lin, J.-C. (2023). Influence of Symbiotic Fermentation Broth on Regulating Metabolism with Gut Microbiota and Metabolite Profiles Is Estimated Using a Third-Generation Sequencing Platform. Metabolites, 13(9), 999. https://doi.org/10.3390/metabo13090999







