Comparative Impact of Organic Grass-Fed and Conventional Cattle-Feeding Systems on Beef and Human Postprandial Metabolomics—A Randomized Clinical Trial
Abstract
1. Introduction
2. Materials and Methods
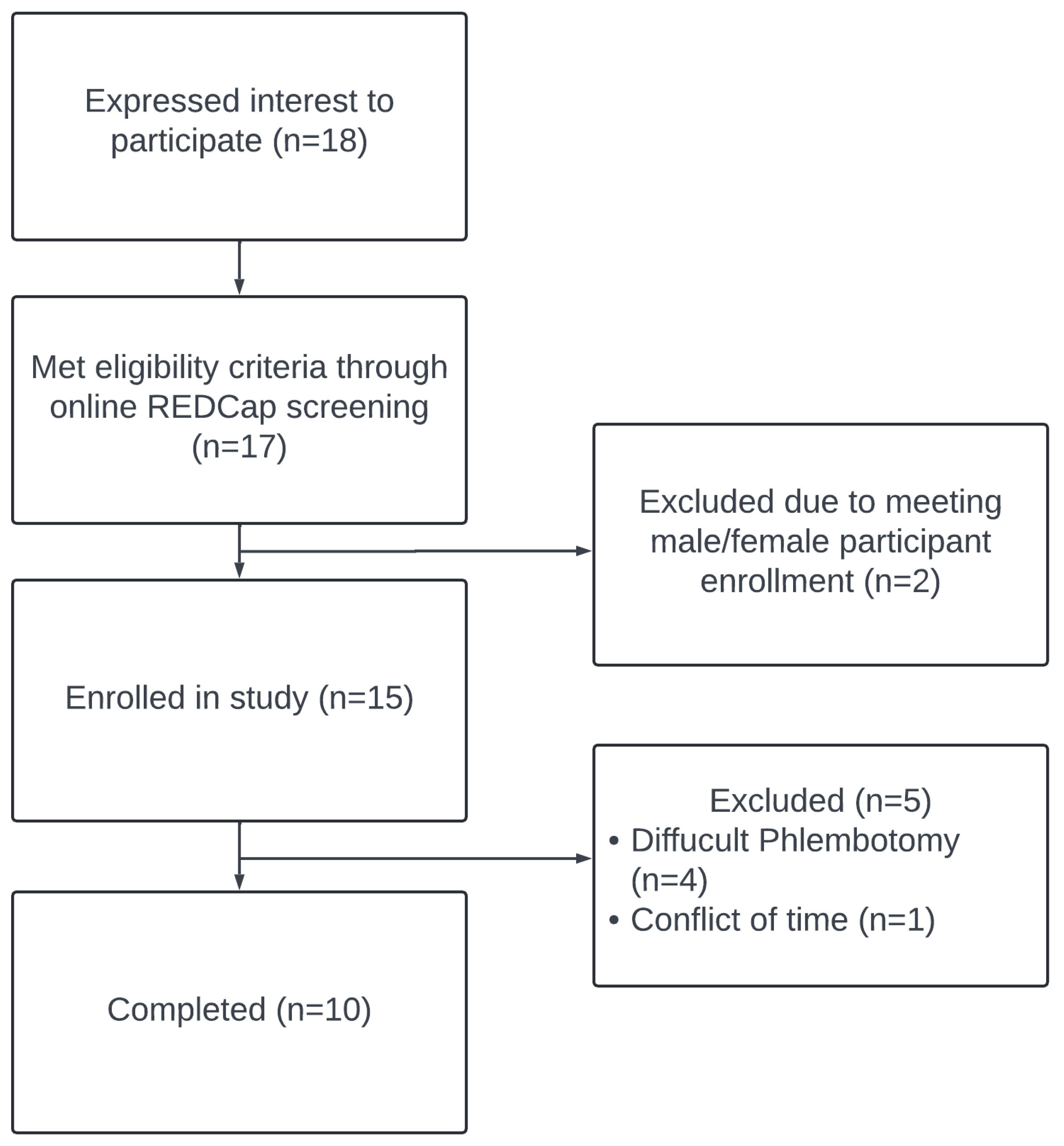
2.1. Study Population
2.2. Experimental Design
2.3. Dietary Intervention and Blood Sample Collection
2.4. Steak Preparation
2.5. Whole Beef Steak Metabolite Extraction
2.6. Plasma Metabolite Extraction
2.7. Untargeted Metabolomic Analysis
2.8. Statistical Analysis
3. Results
3.1. Participant Characteristics
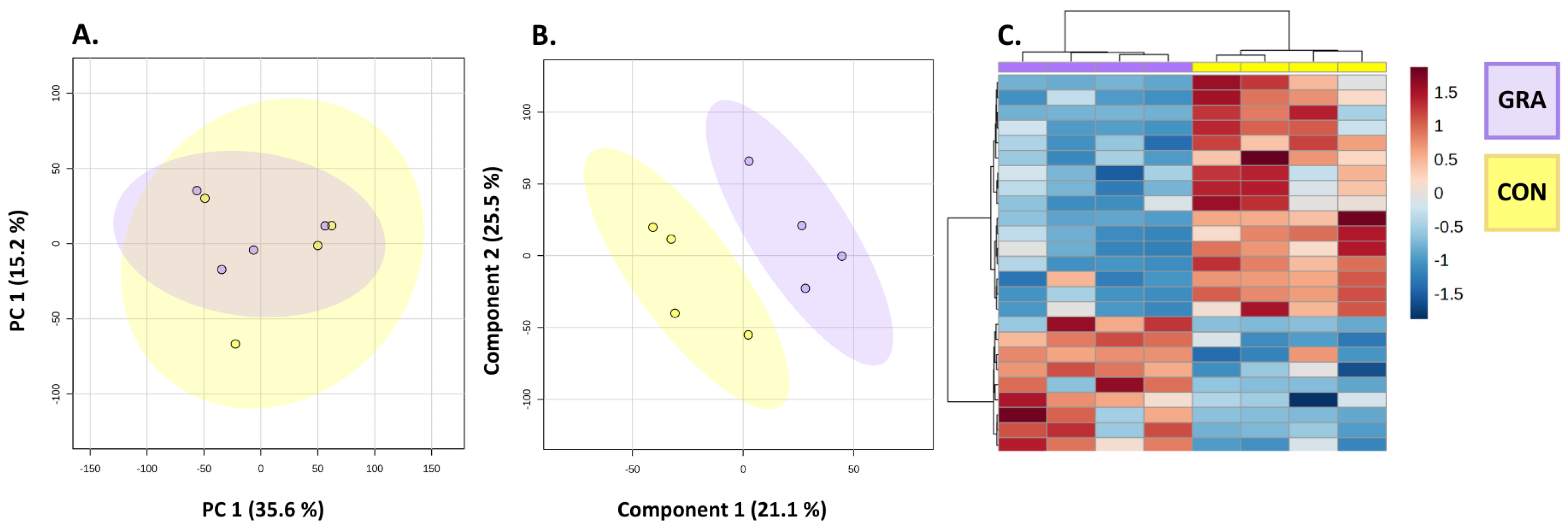
3.2. Whole Beef Steak Metabolomic Profiles
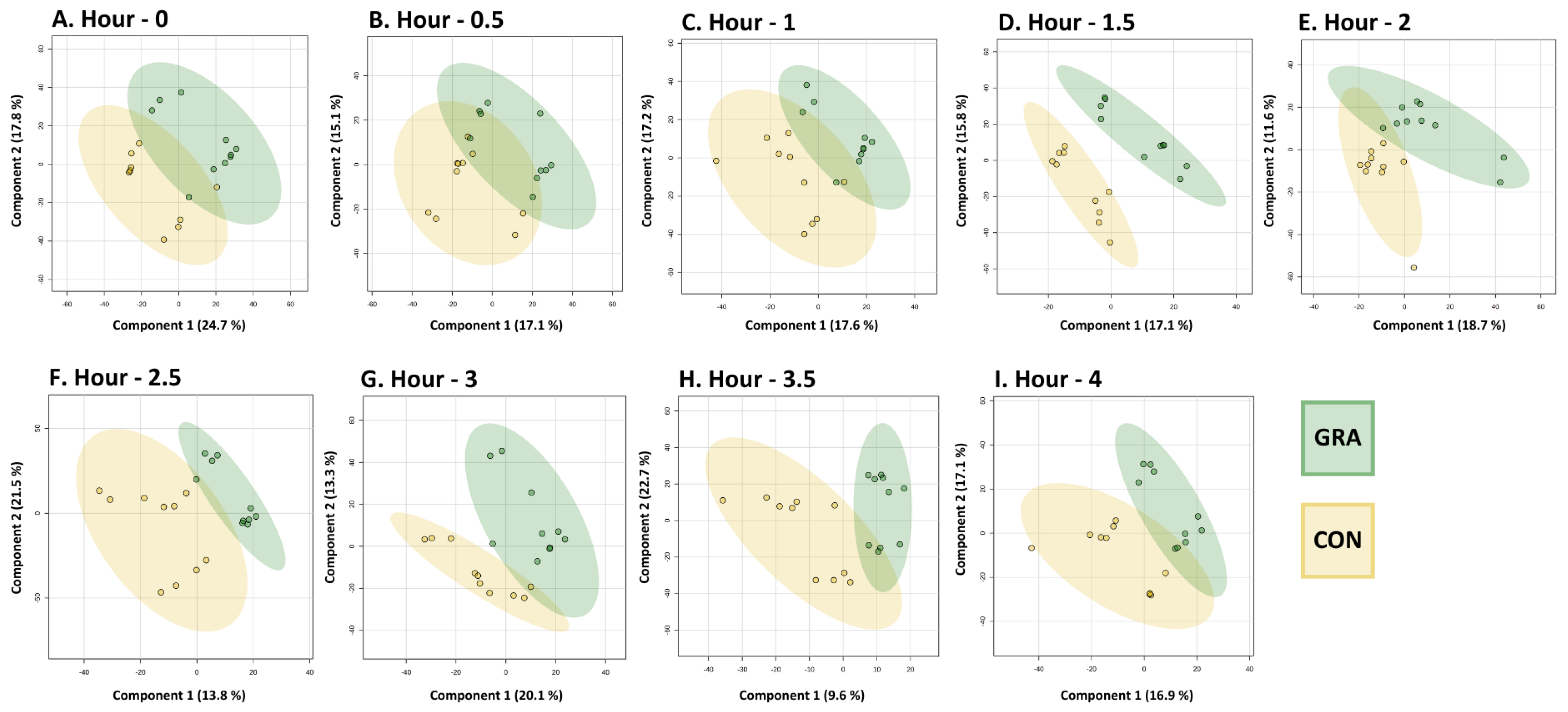
3.3. Human Plasma Metabolomic Profiles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. Food Balance Sheets 2010–2022 Global, Regional and Country Trends; FAOSTAT Analytical Brief 91; Food and Agriculture Organization of the United Nations: Rome, Italy, 2024. [Google Scholar]
- Hawley, A.L.; Liang, X.; Børsheim, E.; Wolfe, R.R.; Salisbury, L.; Hendy, E.; Wu, H.; Walker, S.; Tacinelli, A.M.; Baum, J.I. The Potential Role of Beef and Nutrients Found in Beef on Outcomes of Wellbeing in Healthy Adults 50 Years of Age and Older: A Systematic Review of Randomized Controlled Trials. Meat Sci. 2022, 189, 108830. [Google Scholar] [CrossRef]
- Agarwal, S.; Fulgoni, V.L. Beef Consumption Is Associated with Higher Intakes and Adequacy of Key Nutrients in Older Adults Age 60+ Years: National Health and Nutrition Examination Survey 2011–2018 Analysis. Nutrients 2024, 16, 1779. [Google Scholar] [CrossRef] [PubMed]
- Leheska, J.M.; Thompson, L.D.; Howe, J.C.; Hentges, E.; Boyce, J.; Brooks, J.C.; Shriver, B.; Hoover, L.; Miller, M.F. Effects of Conventional and Grass-Feeding Systems on the Nutrient Composition of Beef. J. Anim. Sci. 2008, 86, 3575–3585. [Google Scholar] [CrossRef] [PubMed]
- Nogoy, K.M.C.; Sun, B.; Shin, S.; Lee, Y.; Li, X.Z.; Choi, S.H.; Park, S. Fatty Acid Composition of Grain- And Grass-Fed Beef and Their Nutritional Value and Health Implication. Food Sci. Anim. Resour. 2022, 42, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Regulations.Gov. Available online: https://www.regulations.gov/document/FSIS-2016-0021-4653 (accessed on 21 July 2024).
- Van Elswyk, M.E.; Mcneill, S.H. Impact of Grass/Forage Feeding versus Grain Finishing on Beef Nutrients and Sensory Quality: The U.S. Experience. Meat Sci. 2013, 96, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Siriwardhana, N.; Kalupahana, N.S.; Moustaid-Moussa, N. Health Benefits of N-3 Polyunsaturated Fatty Acids. Eicosapentaenoic Acid and Docosahexaenoic Acid. Adv. Food. Nutr. Res. 2012, 65, 211–222. [Google Scholar] [CrossRef]
- López-Miranda, J.; Pérez-Martinez, P.; Pérez-Jiménez, F. Health Benefits of Monounsaturated Fatty Acids. Improv. Fat Content Foods 2006, 71–106. [Google Scholar] [CrossRef]
- Apaoblaza, A.; Gerrard, S.D.; Matarneh, S.K.; Wicks, J.C.; Kirkpatrick, L.; England, E.M.; Scheffler, T.L.; Duckett, S.K.; Shi, H.; Silva, S.L.; et al. Muscle from Grass- and Grain-Fed Cattle Differs Energetically. Meat Sci. 2020, 161, 107996. [Google Scholar] [CrossRef]
- Gómez, J.F.M.; Cônsolo, N.R.B.; Antonelo, D.S.; Beline, M.; Gagaoua, M.; Higuera-Padilla, A.; Colnago, L.A.; Gerrard, D.E.; Silva, S.L. Impact of Cattle Feeding Strategy on the Beef Metabolome. Metabolites 2022, 12, 640. [Google Scholar] [CrossRef]
- Kaur, L.; Elamurugan, A.; Chian, F.M.; Zhu, X.; Boland, M. Protein and Lipid Digestibility of Pasture-Raised and Grain-Finished Beef: An In Vitro Comparison. Foods 2023, 12, 1239. [Google Scholar] [CrossRef]
- Dillon, E.L. Nutritionally Essential Amino Acids and Metabolic Signaling in Aging. Amino Acids 2013, 45, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Duckett, S.K.; Neel, J.P.S.; Fontenot, J.P.; Clapham, W.M. Effects of Winter Stocker Growth Rate and Finishing System on: III. Tissue Proximate, Fatty Acid, Vitamin, and Cholesterol Content. J. Anim. Sci. 2009, 87, 2961–2970. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.S. The Role of B Vitamins in Preventing and Treating Cognitive Impairment and Decline. Adv. Nutr. 2012, 3, 801–812. [Google Scholar] [CrossRef]
- Uebanso, T.; Shimohata, T.; Mawatari, K.; Takahashi, A. Functional Roles of B-Vitamins in the Gut and Gut Microbiome. Mol. Nutr. Food Res. 2020, 64, 2000426. [Google Scholar] [CrossRef]
- Berry, S.E.; Valdes, A.M.; Drew, D.A.; Asnicar, F.; Mazidi, M.; Wolf, J.; Capdevila, J.; Hadjigeorgiou, G.; Davies, R.; Al Khatib, H.; et al. Human Postprandial Responses to Food and Potential for Precision Nutrition. Nat. Med. 2020, 26, 964–973. [Google Scholar] [CrossRef]
- Neacsu, M.; Vaughan, N.J.; Multari, S.; Haljas, E.; Scobbie, L.; Duncan, G.J.; Cantlay, L.; Fyfe, C.; Anderson, S.; Horgan, G.; et al. Hemp and Buckwheat Are Valuable Sources of Dietary Amino Acids, Beneficially Modulating Gastrointestinal Hormones and Promoting Satiety in Healthy Volunteers. Eur. J. Nutr. 2022, 61, 1057–1072. [Google Scholar] [CrossRef]
- Ross, A.B.; Svelander, C.; Undeland, I.; Pinto, R.; Sandberg, A.-S. Herring and Beef Meals Lead to Differences in Plasma 2-Aminoadipic Acid, b-Alanine, 4-Hydroxyproline, Cetoleic Acid, and Docosahexaenoic Acid Concentrations in Overweight Men 1–3. J. Nutr. Genom. 2015, 145, 2456–2463. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.S. The Emerging Role of Branched-Chain Amino Acids in Insulin Resistance and Metabolism. Nutrients 2016, 8, 405. [Google Scholar] [CrossRef]
- Lee, H.J.; Jang, H.B.; Kim, W.H.; Park, K.J.; Kim, K.Y.; Park, S.I.; Lee, H.J. 2-Aminoadipic Acid (2-AAA) as a Potential Biomarker for Insulin Resistance in Childhood Obesity. Sci. Rep. 2019, 9, 13610. [Google Scholar] [CrossRef]
- Westerterp-Plantenga, M.S.; Lemmens, S.G.; Westerterp, K.R. Dietary Protein—Its Role in Satiety, Energetics, Weight Loss and Health. Br. J. Nutr. 2012, 108, S105–S112. [Google Scholar] [CrossRef]
- Lépine, G.; Tremblay-Franco, M.; Bouder, S.; Dimina, L.; Fouillet, H.; Mariotti, F.; Polakof, S. Investigating the Postprandial Metabolome after Challenge Tests to Assess Metabolic Flexibility and Dysregulations Associated with Cardiometabolic Diseases. Nutrients 2022, 14, 472. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yan, S.; Bakker, B.M.; Hoefsloot, H.; Chawes, B.; Horner, D.; Rasmussen, M.A.; Smilde, A.K.; Acar, E. Analyzing Postprandial Metabolomics Data Using Multiway Models: A Simulation Study. BMC Bioinform. 2024, 25, 94. [Google Scholar] [CrossRef]
- Pimentel, G.; Burton, K.J.; Pralong, F.P.; Vionnet, N.; Portmann, R.; Vergères, G. The Postprandial Metabolome—A Source of Nutritional Biomarkers of Health. Curr. Opin. Food Sci. 2017, 16, 67–73. [Google Scholar] [CrossRef]
- Valdés, A.; Álvarez-Rivera, G.; Socas-Rodríguez, B.; Herrero, M.; Ibáñez, E.; Cifuentes, A. Foodomics: Analytical Opportunities and Challenges. Anal. Chem. 2022, 94, 366–381. [Google Scholar] [CrossRef]
- O’Gorman, A.; Brennan, L. The Role of Metabolomics in Determination of New Dietary Biomarkers. Proc. Nutr. Soc. 2017, 76, 295–302. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond Biomarkers and towards Mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- López-López, Á.; López-Gonzálvez, Á.; Barker-Tejeda, T.C.; Barbas, C. A Review of Validated Biomarkers Obtained through Metabolomics. Expert Rev. Mol. Diagn. 2018, 18, 557–575. [Google Scholar] [CrossRef]
- Liang, Q.; Liu, H.; Zhang, T.; Jiang, Y.; Zhang, A.H. Untargeted Lipidomics Study of Coronary Artery Disease by FUPLC-Q-TOF-MS. Anal. Methods 2016, 8, 1229–1234. [Google Scholar] [CrossRef]
- Jones, D.P.; Park, Y.; Ziegler, T.R. Nutritional Metabolomics: Progress in Addressing Complexity in Diet and Health. Annu. Rev. Nutr. 2012, 32, 183. [Google Scholar] [CrossRef]
- Harris, P.A.; Taylor, R.; Minor, B.L.; Elliott, V.; Fernandez, M.; O’Neal, L.; McLeod, L.; Delacqua, G.; Delacqua, F.; Kirby, J.; et al. The REDCap Consortium: Building an International Community of Software Platform Partners. J. Biomed. Inform. 2019, 95, 103208. [Google Scholar] [CrossRef]
- Chamberlin, M.L.; Peach, J.T.; Wilson, S.M.G.; Miller, Z.T.; Bothner, B.; Walk, S.T.; Yeoman, C.J.; Miles, M.P. Polyphenol-Rich Aronia Melanocarpa Fruit Beneficially Impact Cholesterol, Glucose, and Serum and Gut Metabolites: A Randomized Clinical Trial. Foods 2024, 13, 2768. [Google Scholar] [CrossRef] [PubMed]
- Smilde, A.K.; Jansen, J.J.; Hoefsloot, H.C.J.; Lamers, R.-J.A.N.; Van Der Greef, J.; Timmerman, M.E. ANOVA-Simultaneous Component Analysis (ASCA): A New Tool for Analyzing Designed Metabolomics Data. Bioinform. Orig. Pap. 2005, 21, 3043–3048. [Google Scholar] [CrossRef] [PubMed]
- Nueda, M.J.; Conesa, A.; Westerhuis, J.A.; Hoefsloot, H.C.J.; Smilde, A.K.; Taló, M.; Ferrer, A. Discovering Gene Expression Patterns in Time Course Microarray Experiments by ANOVA-SCA. Bioinformatics 2007, 23, 1792–1800. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting Network Activity from High Throughput Metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef]
- Welhaven, H.D.; Vahidi, G.; Walk, S.T.; Bothner, B.; Martin, S.A.; Heveran, C.M.; June, R.K. The Cortical Bone Metabolome of C57BL/6J Mice Is Sexually Dimorphic. JBMR Plus 2022, 6, e10654. [Google Scholar] [CrossRef]
- Wolfe, R.R. Branched-Chain Amino Acids and Muscle Protein Synthesis in Humans: Myth or Reality? J. Int. Soc. Sports Nutr. 2017, 14, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jorge-Smeding, E.; Polakof, S.; Bonnet, M.; Durand, S.; Centeno, D.; Pétéra, M.; Taussat, S.; Cantalapiedra-Hijar, G. Untargeted Metabolomics Confirms the Association between Plasma Branched Chain Amino Acids and Residual Feed Intake in Beef Heifers. PLoS ONE 2022, 17, e0277458. [Google Scholar] [CrossRef]
- Ye, Z.; Wang, S.; Zhang, C.; Zhao, Y. Coordinated Modulation of Energy Metabolism and Inflammation by Branched-Chain Amino Acids and Fatty Acids. Front. Endocrinol. 2020, 11, 617. [Google Scholar] [CrossRef]
- Mann, G.; Mora, S.; Madu, G.; Adegoke, O.A.J. Branched-Chain Amino Acids: Catabolism in Skeletal Muscle and Implications for Muscle and Whole-Body Metabolism. Front. Physiol. 2021, 12, 702826. [Google Scholar] [CrossRef]
- Nie, C.; He, T.; Zhang, W.; Zhang, G.; Ma, X. Branched Chain Amino Acids: Beyond Nutrition Metabolism. Int. J. Mol. Sci. 2018, 19, 954. [Google Scholar] [CrossRef]
- Polizel, G.H.G.; Cançado, F.A.C.Q.; Dias, E.F.F.; Fernandes, A.C.; Cracco, R.C.; Carmona, B.T.; Castellar, H.H.; Poleti, M.D.; de Almeida Santana, M.H. Effects of Different Prenatal Nutrition Strategies on the Liver Metabolome of Bulls and Its Correlation with Body and Liver Weight. Metabolites 2022, 12, 441. [Google Scholar] [CrossRef] [PubMed]
- Pekala, J.; Patkowska-Sokola, B.; Bodkowski, R.; Jamroz, D.; Nowakowski, P.; Lochynski, S.; Librowski, T. L-Carnitine--Metabolic Functions and Meaning in Humans Life. Curr. Drug Metab. 2011, 12, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.; Provenza, F.D.; Leroy, F.; Van Vliet, S.; Hamlin, M.; Elliot, C.; Garrett, K.; Marshall, C.J.; Gregorini, P. Connecting Plant, Animal, and Human Health Using Untargeted Metabolomics. Res. Sq. 2024. preprint. [Google Scholar] [CrossRef]
- Shah, R.V.; Steffen, L.M.; Nayor, M.; Reis, J.P.; Jacobs, D.R.; Allen, N.B.; Lloyd-Jones, D.; Meyer, K.; Cole, J.; Piaggi, P.; et al. Dietary Metabolic Signatures and Cardiometabolic Risk. Eur. Heart J. 2023, 44, 557. [Google Scholar] [CrossRef]
- Schnuck, J.K.; Sunderland, K.L.; Kuennen, M.R.; Vaughan, R.A. Characterization of the Metabolic Effect of β-Alanine on Markers of Oxidative Metabolism and Mitochondrial Biogenesis in Skeletal Muscle. J. Exerc. Nutrition Biochem. 2016, 20, 34. [Google Scholar] [CrossRef]
- Fernandes, A.C.; Polizel, G.H.G.; Cracco, R.C.; Cançado, F.A.C.Q.; Baldin, G.C.; Poleti, M.D.; Ferraz, J.B.S.; de Santana, M.H.A. Metabolomics Changes in Meat and Subcutaneous Fat of Male Cattle Submitted to Fetal Programming. Metabolites 2024, 14, 9. [Google Scholar] [CrossRef]
- Oppermann, H.; Heinrich, M.; Birkemeyer, C.; Meixensberger, J.; Gaunitz, F. The Proton-Coupled Oligopeptide Transporters PEPT2, PHT1 and PHT2 Mediate the Uptake of Carnosine in Glioblastoma Cells. Amino Acids 2019, 51, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Yin, Q.; Zhao, H.; Cao, Y.; Cai, C.; Yao, J. Metabolomics for the Effect of Biotin and Nicotinamide on Transition Dairy Cows. J. Agric. Food Chem. 2018, 66, 5723–5732. [Google Scholar] [CrossRef] [PubMed]
- Siri-Tarino, P.W.; Sun, Q.; Hu, F.B.; Krauss, R.M. Saturated Fatty Acids and Risk of Coronary Heart Disease: Modulation by Replacement Nutrients. Curr. Atheroscler. Rep. 2010, 12, 384–390. [Google Scholar] [CrossRef]
- Ruiz-Núñez, B.; Dijck-Brouwer, D.A.J.; Muskiet, F.A.J. The Relation of Saturated Fatty Acids with Low-Grade Inflammation and Cardiovascular Disease. J. Nutr. Biochem. 2016, 36, 1–20. [Google Scholar] [CrossRef]
- Legrand, P.; Rioux, V. The Complex and Important Cellular and Metabolic Functions of Saturated Fatty Acids. Lipids 2010, 45, 941–946. [Google Scholar] [CrossRef]
- Harrison, S.; Brassard, D.; Lemieux, S.; Lamarche, B. Consumption and Sources of Saturated Fatty Acids According to the 2019 Canada Food Guide: Data from the 2015 Canadian Community Health Survey. Nutrients 2019, 11, 1964. [Google Scholar] [CrossRef]
- Yamamura, S.; Iguchi, M.; Nishiyama, A.; Niwa, M.; Koyama, H.; Hirata, Y. Sesquiterpenes from Acorus Calamus L. Tetrahedron 1971, 27, 5419–5431. [Google Scholar] [CrossRef]
- Jain, D.K.; Gupta, S.; Jain, R.; Jain, N. Nilesh Anti-Inflammatory Activity of 80% Ethanolic Extract of Acorus Calamus Linn. Leaves in Albino Rats. Res. J. Pharm. Technol. 2010, 3, 882–884. [Google Scholar]
- Wang, X.; Zhang, H.; Quinn, P.J. Production of L-Valine from Metabolically Engineered Corynebacterium Glutamicum. Appl. Microbiol. Biotechnol. 2018, 102, 4319–4330. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Ma, Q.; Liu, X.; Fan, X.; Men, J.; Wu, H.; Jiang, S.; Tian, D.; Xiong, B.; Xie, X. High-Yield Production of L-Valine in Engineered Escherichia Coli by a Novel Two-Stage Fermentation. Metab. Eng. 2020, 62, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Yang, P.; Fu, Z.; Wang, Y.; Zhou, Y.; Ye, Z.; Gong, Y.; Huang, A.; Sun, L.; Zhao, Y.; et al. High L-Valine Concentrations Associate with Increased Oxidative Stress and Newly-Diagnosed Type 2 Diabetes Mellitus: A Cross-Sectional Study. Diabetes Metab. Syndr. Obes. Targets Ther. 2022, 15, 499–509. [Google Scholar] [CrossRef]
- Kandola, K.; Bowman, A.; Birch-Machin, M.A. Oxidative Stress—A Key Emerging Impact Factor in Health, Ageing, Lifestyle and Aesthetics. Int. J. Cosmet. Sci. 2015, 37, 1–8. [Google Scholar] [CrossRef]
- Arya, F.; Egger, S.; Colquhoun, D.; Sullivan, D.; Pal, S.; Egger, G. Differences in Postprandial Inflammatory Responses to a “modern” v. Traditional Meat Meal: A Preliminary Study. Br. J. Nutr. 2010, 104, 724–728. [Google Scholar] [CrossRef]
- Grant, S.S.; Magruder, K.P.; Friedman, B.H. Controlling for Caffeine in Cardiovascular Research: A Critical Review. Int. J. Psychophysiol. 2018, 133, 193–201. [Google Scholar] [CrossRef]
- Sato, S.; Parr, E.B.; Devlin, B.L.; Hawley, J.A.; Sassone-Corsi, P. Human Metabolomics Reveal Daily Variations under Nutritional Challenges Specific to Serum and Skeletal Muscle. Mol. Metab. 2018, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Caesar, L.K.; Kellogg, J.J.; Kvalheim, O.M.; Cech, N.B. Opportunities and Limitations for Untargeted Mass Spectrometry Metabolomics to Identify Biologically Active Constituents in Complex Natural Product Mixtures. J. Nat. Prod. 2019, 82, 469. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.J.; Yen, C.H.; Wu, I.W.; Liu, M.H.; Cheng, H.Y.; Lin, Y.T.; Lee, C.C.; Hsu, K.H.; Sun, C.Y.; Chen, C.Y.; et al. The Association between Low Protein Diet and Body Composition, Muscle Function, Inflammation, and Amino Acid-Based Metabolic Profile in Chronic Kidney Disease Stage 3–5 Patients. Clin. Nutr. ESPEN 2021, 46, 405–415. [Google Scholar] [CrossRef] [PubMed]

| Participants | Age (Years) | BMI (kg/m2) | WC (cm) | VAT (L) | Glucose (mmol/L) | Triglyceride (mmol/L) |
|---|---|---|---|---|---|---|
| All (n = 10) | 26.6 ± 5.8 | 24.44 ± 2.20 | 82.33 ± 8.32 | 0.78 ± 0.73 | 5.47 ± 0.81 | 0.98 ± 0.45 |
| Female (n = 5) | 24.8 ± 5.2 | 23.99 ± 2.38 | 76.77 ± 7.34 | 0.34 ± 0.09 | 5.52 ± 1.14 | 0.80 ± 0.22 |
| Male (n = 5) | 28.4 ± 6.3 | 24.89 ± 2.16 | 87.88 ± 4.97 * | 1.22 ± 0.81 * | 5.41 ± 0.32 | 1.15 ± 0.56 |
| Metabolite | Score 1 | Raw p-Value | CON vs. GRA 2 |
|---|---|---|---|
| 9-Hexadecenoylcholine | 39.2 | 3.2919 × 10−4 | CON > GRA |
| L-threonine | 28.6 | 0.0035 | CON < GRA |
| Keratan sulfate II | 38.4 | 0.0051 | CON > GRA |
| 3-hydroxy-5,8-tetradecadienoylcarnitine | 34.9 | 0.0236 | CON > GRA |
| Proionylcarnitine | 37.1 | 0.0021 | CON > GRA |
| Aspartyl-Serine | 34.3 | 0.0251 | CON < GRA |
| Glycyl–Phenylalanine | 37.1 | 0.0488 | CON < GRA |
| L-Tryptophan | 38.7 | 0.0419 | CON > GRA |
| Histamine-betaxanthin | 38.6 | 0.0366 | CON < GRA |
| Leucylphenylalanine | 40.1 | 0.0416 | CON < GRA |
| N-palmitoyl serine | 38.7 | 0.0276 | CON > GRA |
| LysoPC(18:1/0:0) | 37.9 | 0.0474 | CON < GRA |
| DIBOA trihexose | 37.9 | 0.0330 | CON > GRA |
| Metabolite | Score 1 | Leverage | SPE 2 |
|---|---|---|---|
| L-valine | 38.7 | 0.0017 | 2.7733 × 10−31 |
| Calamendiol | 46.5 | 0.0020 | 2.7733 × 10−31 |
| 5-Aminopentanal | 39.4 | 0.0020 | 2.7733 × 10−31 |
| 3-Amino-4,7-dihydroxy-8-methylcoumarin | 39.1 | 0.0015 | 6.9333 × 10−32 |
| 3-beta-D-glucopyranuronosyloxy-5-methylisoxazole | 39.1 | 0.0013 | 0 |
| PE(16:1(9Z)/20:5(5Z,8Z,11Z,14Z,17Z) | 37.8 | 0.0014 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spears, M.; Cooper, G.; Sather, B.; Bailey, M.; Boles, J.A.; Bothner, B.; Miles, M.P. Comparative Impact of Organic Grass-Fed and Conventional Cattle-Feeding Systems on Beef and Human Postprandial Metabolomics—A Randomized Clinical Trial. Metabolites 2024, 14, 533. https://doi.org/10.3390/metabo14100533
Spears M, Cooper G, Sather B, Bailey M, Boles JA, Bothner B, Miles MP. Comparative Impact of Organic Grass-Fed and Conventional Cattle-Feeding Systems on Beef and Human Postprandial Metabolomics—A Randomized Clinical Trial. Metabolites. 2024; 14(10):533. https://doi.org/10.3390/metabo14100533
Chicago/Turabian StyleSpears, Meghan, Gwendolyn Cooper, Brett Sather, Marguerite Bailey, Jane A. Boles, Brian Bothner, and Mary P. Miles. 2024. "Comparative Impact of Organic Grass-Fed and Conventional Cattle-Feeding Systems on Beef and Human Postprandial Metabolomics—A Randomized Clinical Trial" Metabolites 14, no. 10: 533. https://doi.org/10.3390/metabo14100533
APA StyleSpears, M., Cooper, G., Sather, B., Bailey, M., Boles, J. A., Bothner, B., & Miles, M. P. (2024). Comparative Impact of Organic Grass-Fed and Conventional Cattle-Feeding Systems on Beef and Human Postprandial Metabolomics—A Randomized Clinical Trial. Metabolites, 14(10), 533. https://doi.org/10.3390/metabo14100533








