Metabolomics Reveals Lysinibacillus capsici TT41-Induced Metabolic Shifts Enhancing Drought Stress Tolerance in Kimchi Cabbage (Brassica rapa L. subsp. pekinensis)
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Drought Tolerance Screening of Isolates
2.3. Plant Growth Conditions and Treatments
2.4. Validation of Drought Resistance Enhancement in Plants by Lactic Acid
2.5. Phenotype and Endogenous Change Measurement
2.6. Metabolite Profiling
2.7. Statistical Analysis
3. Results and Discussion
3.1. Selection of Drought Tolerance Microbial Resource
3.2. Plant Drought Tolerance Effect of the Microbial Resource
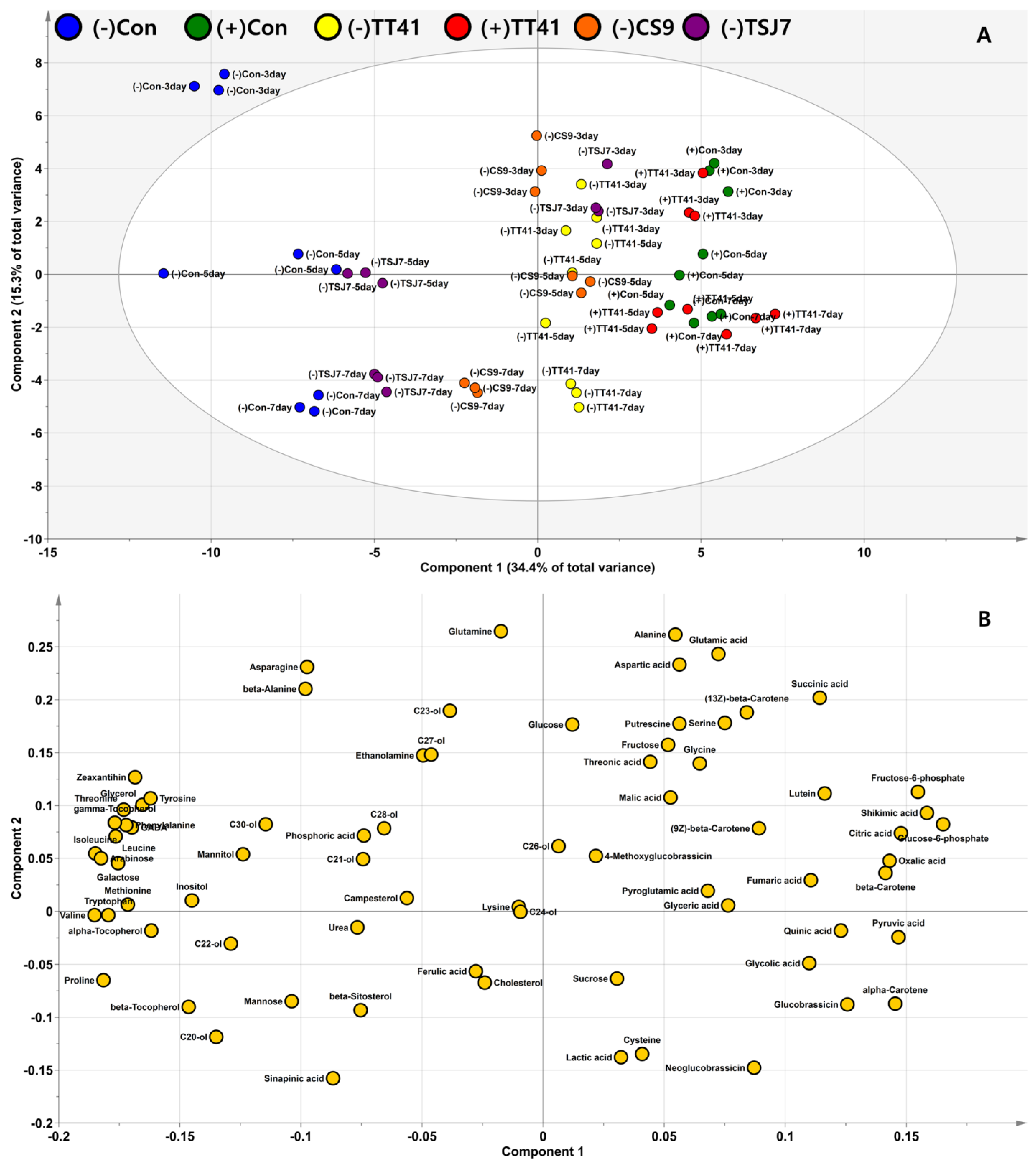
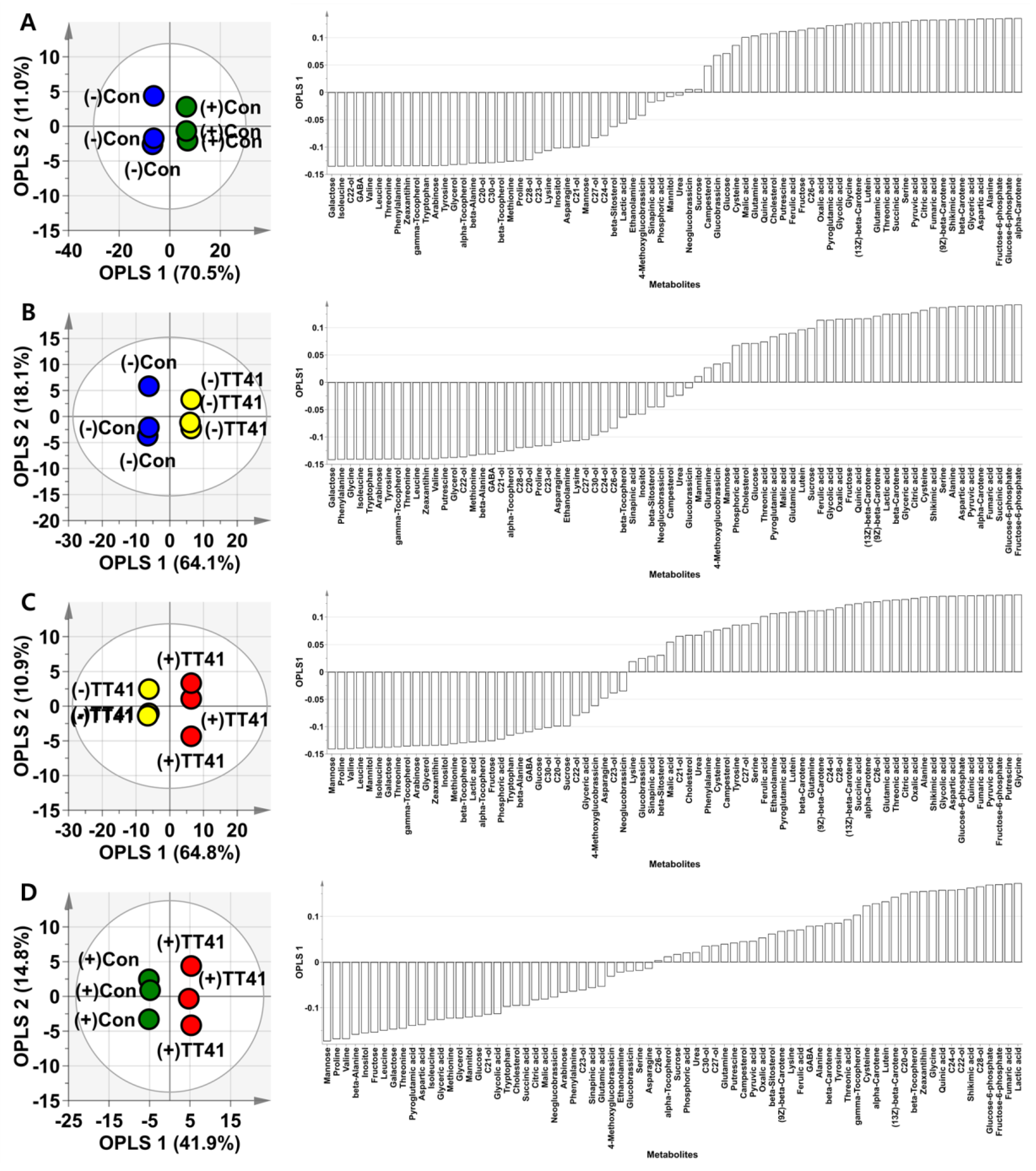
3.3. Effect of Microbial Resource on Plants’ Endogenous Metabolome
3.4. Metabolite Profiling of Lysinibacillus sp. TT41 Strain under Drought Stress
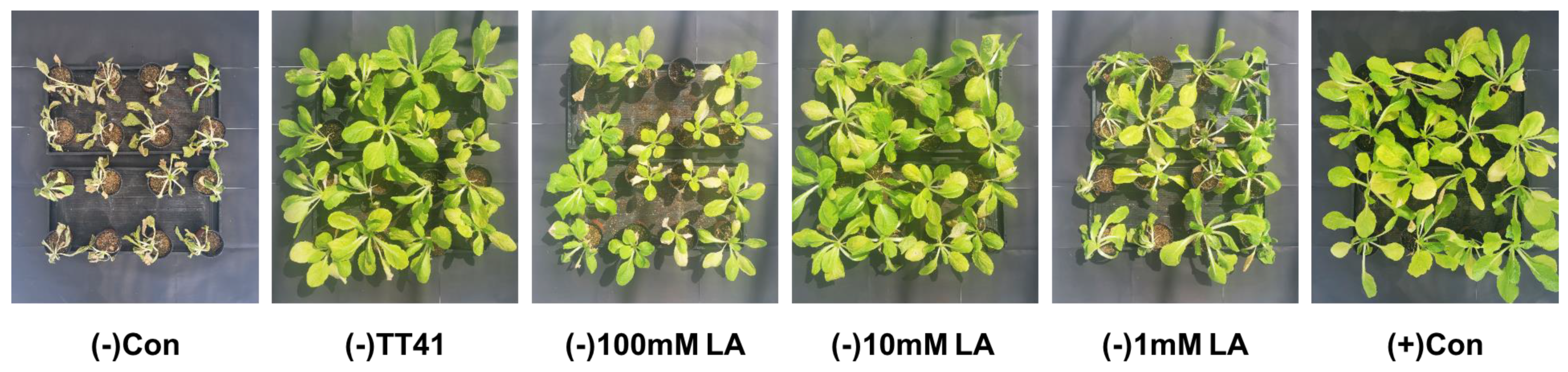
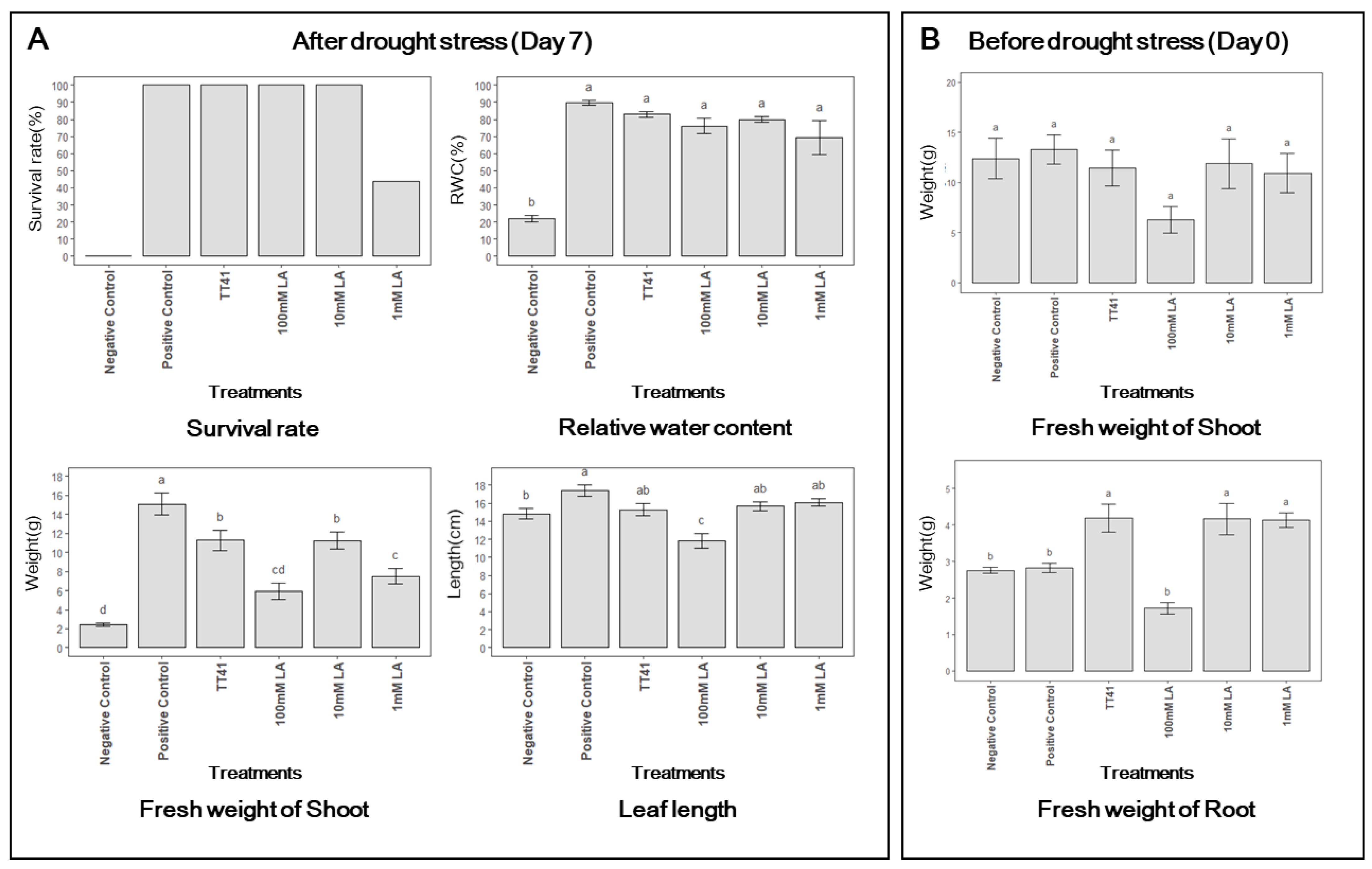
3.5. Effect of Lactic Acid Treatment on Kimchi Cabbage under Drought Stress
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mata, A.T.; Jorge, T.F.; Pires, M.V.; Antonio, C. Drought stress tolerance in plants: Insights from metabolomics. In Drought Stress Tolerance in Plants; Hossain, M.A., Wani, S.H., Bhattacharjee, S., Burritt, D.J., Tran, L.-S.P., Eds.; Springer International Publishing: Cham, Switzerland, 2016; Volume 2, pp. 187–216. [Google Scholar]
- Singh, A.; Mazahar, S.; Chapadgaonkar, S.S.; Giri, P.; Shourie, A. Phyto-microbiome to mitigate abiotic stress in crop plants. Front. Microbiol. 2023, 14, 1210890. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chai, B. Fate of antibiotic resistance genes and changes in bacterial community with increasing breeding scale of layer manure. Front. Microbiol. 2022, 13, 857046. [Google Scholar] [CrossRef] [PubMed]
- Naureen, Z.; Rehman, N.U.; Hussain, H.; Hussain, J.; Gilani, S.A.; Al Housni, S.K.; Mabood, F.; Khan, A.L.; Farooq, S.; Abbas, G.; et al. Exploring the potentials of Lysinibacillus sphaericus ZA9 for plant growth promotion and biocontrol activities against phytopathogenic fungi. Front. Microbiol. 2017, 8, 1477. [Google Scholar] [CrossRef] [PubMed]
- Jha, C.K.; Saraf, M. Plant growth promoting rhizobacteria (PGPR): A review. J. Agric. Res. Deve. 2015, 5, 108–119. [Google Scholar]
- Ojiambo, P.S.; Scherm, H. Biological and application-oriented factors influencing plant disease suppression by biological control: A meta-analytical review. Phytopathology 2006, 96, 1168–1174. [Google Scholar] [CrossRef] [PubMed]
- Dayan, F.E.; Cantrell, C.L.; Duke, S.O. Natural products in crop protection. Bioorg. Med. Chem. 2009, 17, 4022–4034. [Google Scholar] [CrossRef] [PubMed]
- Mashabela, M.D.; Piater, L.A.; Dubery, I.A.; Tugizimana, F.; Mhlongo, M.I. Rhizosphere Tripartite Interactions and PGPR-Mediated Metabolic Reprogramming towards ISR and Plant Priming: A Metabolomics Review. Biology 2022, 11, 346. [Google Scholar] [CrossRef]
- Carlson, R.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A.; Labuschagne, N. Differential Metabolic Reprogramming in Paenibacillus alvei-Primed Sorghum bicolor Seedlings in Response to Fusarium pseudograminearum Infection. Metabolites 2019, 9, 150. [Google Scholar] [CrossRef]
- Kang, D.; Kirienko, D.R.; Webster, P.; Fisher, A.L.; Kirienko, N.V. Pyoverdine, a siderophore from Pseudomonas aeruginosa, translocates into C. elegans, removes iron, and activates a distinct host response. Virulence 2018, 9, 804–817. [Google Scholar] [CrossRef]
- Akram, W.; Aslam, H.; Ahmad, S.R.; Anjum, T.; Yasin, N.A.; Khan, W.U.; Ahmad, A.; Guo, J.; Wu, T.; Luo, W.; et al. Bacillus megaterium strain A12 ameliorates salinity stress in tomato plants through multiple mechanisms. J. Plant Interact. 2019, 14, 506–518. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, V.K.; Tripathi, V.; Singh, P.P.; Singh, A.K. Plant growth-promoting rhizobacteria (PGPR): Perspective in agriculture under biotic and abiotic stress. In Crop Improvement through Microbial Biotechnology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 333–342. [Google Scholar]
- Khan, N.; Bano, A.; Rahman, M.A.; Guo, J.; Kang, Z.; Babar, M.A. Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in Chickpea (Cicer arietinum L.) induced by PGPR and PGRs. Sci. Rep. 2019, 9, 2097. [Google Scholar] [CrossRef] [PubMed]
- Rozier, C.; Hamzaoui, J.; Lemoine, D.; Czarnes, S.; Legendre, L. Field-based assessment of the mechanism of maize yield enhancement by Azospirillum lipoferum CRT1. Sci. Rep. 2017, 7, 7416. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Metabolic profiling of PGPR-treated tomato plants reveal priming-related adaptations of secondary metabolites and aromatic amino acids. Metabolites 2020, 10, 210. [Google Scholar] [CrossRef] [PubMed]
- Fred, E.B.; Waksman, S.A. Laboratory Manual of General Microbiology, with Special Reference to the Microorganisms of the Soil, 1st ed.; McGraw-Hill Inc.: New York, NY, USA, 1928. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Patane, C.; Cosentino, S.L.; Romano, D.; Toscano, S. Relative water content, proline, and antioxidant enzymes in leaves of long shelf-life tomatoes under drought stress and rewatering. Plants 2022, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Sun, W.; Zhao, M. Controlled formation of emulsion gels stabilized by salted myofibrillar protein under malondialdehyde (MDA)-induced oxidative stress. J. Agric. Food Chem. 2015, 63, 3766–3777. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Wu, Z.; Zhang, L.; Li, J.; Zhou, J.; Jun, Z.; Zhu, B. Monitoring of peanut leaves chlorophyll content based on drone-based multispectral image feature extraction. Comput Electron Agric. 2021, 187, 106292. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. [34] Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1987; Volume 148, pp. 350–382. [Google Scholar]
- Baek, S.-A.; Kim, K.W.; Kim, J.O.; Kim, T.J.; Ahn, S.K.; Choi, J.; Kim, J.; Ahn, J.; Kim, J.K. Metabolic profiling reveals an increase in stress-related metabolites in Arabidopsis thaliana exposed to honeybees. J. Appl. Biol. Chem. 2021, 64, 141–151. [Google Scholar] [CrossRef]
- Jung, J.W.; Oh, S.-D.; Park, S.-Y.; Jang, Y.; Lee, S.-K.; Yun, D.-W.; Chang, A.; Park, S.U.; Ha, S.-H.; Kim, J.K. Metabolic profiling and antioxidant properties of hybrid soybeans with different seed coat colors, obtained by crossing β-carotene-enhanced (Glycine max) and wild (Glycine soja) soybeans. Plant Biotechnol. Rep. 2022, 16, 449–463. [Google Scholar] [CrossRef]
- Baek, S.A.; Jung, Y.H.; Lim, S.H.; Park, S.U.; Kim, J.K. Metabolic Profiling in Chinese Cabbage (Brassica rapa L. subsp. pekinensis) Cultivars Reveals that Glucosinolate Content Is Correlated with Carotenoid Content. J. Agric. Food. Chem. 2016, 64, 4426–4434. [Google Scholar] [CrossRef]
- Kim, T.J.; Lee, K.B.; Baek, S.-A.; Choi, J.; Ha, S.-H.; Lim, S.-H.; Park, S.-Y.; Yeo, Y.; Park, S.U.; Kim, J.K. Determination of lipophilic metabolites for species discrimination and quality assessment of nine leafy vegetables. J. Korean Soc. Appl. Biol. Chem. 2015, 58, 909–918. [Google Scholar] [CrossRef]
- Kim, T.J.; Hyeon, H.; Park, N.I.; Yi, T.G.; Lim, S.H.; Park, S.Y.; Ha, S.H.; Kim, J.K. A high-throughput platform for interpretation of metabolite profile data from pepper (Capsicum) fruits of 13 phenotypes associated with different fruit maturity states. Food Chem. 2020, 331, 127286. [Google Scholar] [CrossRef]
- Kutmon, M.; van Iersel, M.P.; Bohler, A.; Kelder, T.; Nunes, N.; Pico, A.R.; Evelo, C.T. PathVisio 3: An extendable pathway analysis toolbox. PLoS Comput. Biol. 2015, 11, e1004085. [Google Scholar] [CrossRef]
- Zhou, G.; Yang, L.-T.; Li, Y.-R.; Zou, C.-L.; Huang, L.-P.; Qiu, L.-H.; Huang, X.; Srivastava, M.K. Proteomic Analysis of Osmotic Stress-Responsive Proteins in Sugarcane Leaves. Plant Mol. Biol. Rep. 2012, 30, 349–359. [Google Scholar] [CrossRef]
- Carlson, R.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A.; Hassen, A.I.; Labuschagne, N. Rhizobacteria-induced systemic tolerance against drought stress in Sorghum bicolor (L.) Moench. Microbiol. Res. 2020, 232, 126388. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Kumar Patel, M.; Kumar, N.; Bajpai, A.B.; Siddique, K.H.M. Metabolomics and Molecular Approaches Reveal Drought Stress Tolerance in Plants. Int. J. Mol. Sci. 2021, 22, 9108. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Cheng, B.; Yong, B.; Liu, T.; Peng, Y.; Zhang, X.; Ma, X.; Huang, L.; Liu, W.; Nie, G. Metabolomics and physiological analyses reveal β-sitosterol as an important plant growth regulator inducing tolerance to water stress in white clover. Planta 2019, 250, 2033–2046. [Google Scholar] [CrossRef] [PubMed]
- Pantoja-Guerra, M.; Burkett-Cadena, M.; Cadena, J.; Dunlap, C.A.; Ramirez, C.A. Lysinibacillus spp.: An IAA-producing endospore forming-bacteria that promotes plant growth. Antonie Van Leeuwenhoek 2023, 116, 615–630. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Mpovo, C.L.; Aaqil Khan, M.; Shaffique, S.; Ninson, D.; Bilal, S.; Khan, M.; Kwon, E.H.; Kang, S.M.; Yun, B.W.; et al. Synergistic Effect of Melatonin and Lysinibacillus fusiformis L. (PLT16) to Mitigate Drought Stress via Regulation of Hormonal, Antioxidants System, and Physio-Molecular Responses in Soybean Plants. Int. J. Mol. Sci. 2023, 24, 8489. [Google Scholar] [CrossRef]
- Raman, J.; Kim, J.S.; Choi, K.R.; Eun, H.; Yang, D.; Ko, Y.J.; Kim, S.J. Application of Lactic Acid Bacteria (LAB) in Sustainable Agriculture: Advantages and Limitations. Int. J. Mol. Sci. 2022, 23, 7784. [Google Scholar] [CrossRef]
- Chen, W.; He, P.; Zhang, H.; Duan, H.; Shao, L.; Lü, F. The low dose of L-lactic acid as loess soil amendment enhances wheat rhizosphere microecosystem. Rhizosphere 2023, 28, 100784. [Google Scholar] [CrossRef]
- Lee, S.-M.; Kim, S.-K.; Lee, N.; Ahn, C.-Y.; Ryu, C.-M. d-Lactic acid secreted by Chlorella fusca primes pattern-triggered immunity against Pseudomonas syringae in Arabidopsis. Plant J. 2020, 102, 761–778. [Google Scholar] [CrossRef]



| Treatment | Leaf Number | Leaf Length (cm) | Leaf Width (cm) | Fresh Weight (g) | Leaf Relative Water Content (%) | Chlorophyll Content (mg/gFW) | Malondialdehyde Content (mg/gFW) |
|---|---|---|---|---|---|---|---|
| (+)Con | 13.00 ± 0.82 a | 19.08 ± 2.35 a | 9.27 ± 1.08 a | 35.67 ± 9.75 a | 84.7 ± 7.50 a | 31.34 ± 1.20 ab | 6.88 ± 1.03 ab |
| (−)Con | 7.00 ± 0.00 c | 16.40 ± 0.30 a | 6.44 ± 0.21 c | 7.87 ± 0.82 b | 40.29 ± 2.83 d | 23.64 ± 2.53 b | 10.04 ± 0.64 ab |
| (−)TT41 | 9.34 ± 1.25 bc | 17.34 ± 1.32 a | 7.44 ± 0.38 bc | 15.10 ± 2.41 b | 76.79 ± 7.00 ab | 31.54 ± 1.73 ab | 7.59 ± 1.04 ab |
| (+)TT41 | 13.00 ± 0.82 a | 19.00 ± 1.19 a | 8.74 ± 0.53 ab | 32.37 ± 6.1 a | 93.36 ± 5.76 a | 35.84 ± 3.11 ab | 6.28 ± 0.61 b |
| (−)TSJ7 | 9.75 ± 1.09 b | 17.55 ± 0.46 a | 7.50 ± 0.44 bc | 13.03 ± 0.8 b | 55.19 ± 11.99 cd | 36.49 ± 4.19 a | 10.89 ± 2.31 a |
| (−)CS9 | 8.67 ± 0.48 bc | 15.54 ± 0.72 a | 7.00 ± 0.46 c | 10.74 ± 2.5 b | 63.78 ± 5.01 bc | 33.64 ± 9.99 ab | 7.64 ± 0.72 ab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, T.J.; Hwang, Y.J.; Park, Y.J.; Lee, J.S.; Kim, J.K.; Lee, M.-H. Metabolomics Reveals Lysinibacillus capsici TT41-Induced Metabolic Shifts Enhancing Drought Stress Tolerance in Kimchi Cabbage (Brassica rapa L. subsp. pekinensis). Metabolites 2024, 14, 87. https://doi.org/10.3390/metabo14020087
Kim TJ, Hwang YJ, Park YJ, Lee JS, Kim JK, Lee M-H. Metabolomics Reveals Lysinibacillus capsici TT41-Induced Metabolic Shifts Enhancing Drought Stress Tolerance in Kimchi Cabbage (Brassica rapa L. subsp. pekinensis). Metabolites. 2024; 14(2):87. https://doi.org/10.3390/metabo14020087
Chicago/Turabian StyleKim, Tae Jin, Ye Ji Hwang, Young Jin Park, Jong Sung Lee, Jae Kwang Kim, and Mi-Hwa Lee. 2024. "Metabolomics Reveals Lysinibacillus capsici TT41-Induced Metabolic Shifts Enhancing Drought Stress Tolerance in Kimchi Cabbage (Brassica rapa L. subsp. pekinensis)" Metabolites 14, no. 2: 87. https://doi.org/10.3390/metabo14020087
APA StyleKim, T. J., Hwang, Y. J., Park, Y. J., Lee, J. S., Kim, J. K., & Lee, M.-H. (2024). Metabolomics Reveals Lysinibacillus capsici TT41-Induced Metabolic Shifts Enhancing Drought Stress Tolerance in Kimchi Cabbage (Brassica rapa L. subsp. pekinensis). Metabolites, 14(2), 87. https://doi.org/10.3390/metabo14020087







