Application of Clinical Blood Metabogram to Type 2 Diabetes Mellitus
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Mass Spectrometry Analysis of Blood Samples
2.3. Template Design for CBM
2.4. CBM for Participants in the Study Cohort
2.5. Adjustment of Mass Peak Intensity Due to Ionic Inconsistency in Samples
2.6. Cluster Analysis
2.7. Diagnostic Parameters
3. Results
3.1. Studied Subjects
3.2. CBM Data
3.3. Statistical Data and Diagnostic Parameters
3.4. Cluster Analysis
4. Discussion
- CBM is a new method for diagnosing DM that can complement the FPG level and OGTT;
- CBM makes it possible to identify the metabotypes (molecular phenotypes) of diabetic patients and the deviations in blood metabolites associated with these metabotypes;
- CBM provides precise measurement of diabetic changes in blood metabolome, both in volume and severity;
- Treatment for DM can be tailored to the patient’s needs by means of the CBM’s ability to measure and interpret variations in the blood metabolome. The effectiveness of treatment can be assessed and adjusted by tracking changes in the metabogram components and their return to normal Z-scores. The same applies to assessing lifestyle, selecting an effective diet, physical activity, and other factors affecting health.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials; Board on Health Care Services; Board on Health Sciences Policy; Institute of Medicine. Evolution of Translational Omics: Lessons Learned and the Path Forward; Micheel, C.M., Sharyl, N.J., Omenn, G.S., Eds.; National Academies Press (US): Washington, DC, USA, 2012; ISBN 9780309224185. [Google Scholar]
- Beger, R.D.; Dunn, W.; Schmidt, M.A.; Gross, S.S.; Kirwan, J.A.; Cascante, M.; Brennan, L.; Wishart, D.S.; Oresic, M.; Hankemeier, T.; et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics 2016, 12, 149. [Google Scholar] [CrossRef]
- Tebani, A.; Gummesson, A.; Zhong, W.; Koistinen, I.S.; Lakshmikanth, T.; Olsson, L.M.; Boulund, F.; Neiman, M.; Stenlund, H.; Hellström, C.; et al. Integration of molecular profiles in a longitudinal wellness profiling cohort. Nat. Commun. 2020, 11, 4487. [Google Scholar] [CrossRef]
- Gurke, R.; Bendes, A.; Bowes, J.; Koehm, M.; Twyman, R.M.; Barton, A.; Elewaut, D.; Goodyear, C.; Hahnefeld, L.; Hillenbrand, R.; et al. Omics and Multi-Omics Analysis for the Early Identification and Improved Outcome of Patients with Psoriatic Arthritis. Biomedicines 2022, 10, 2387. [Google Scholar] [CrossRef]
- Zheng, M.; Piermarocchi, C.; Mias, G.I. Temporal response characterization across individual multiomics profiles of prediabetic and diabetic subjects. Sci. Rep. 2022, 12, 12098. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef]
- Chen, R.; Mias, G.I.; Li-Pook-Than, J.; Jiang, L.; Lam, H.Y.K.; Chen, R.; Miriami, E.; Karczewski, K.J.; Hariharan, M.; Dewey, F.E.; et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 2012, 148, 1293–1307. [Google Scholar] [CrossRef]
- Schüssler-Fiorenza Rose, S.M.; Contrepois, K.; Moneghetti, K.J.; Zhou, W.; Mishra, T.; Mataraso, S.; Dagan-Rosenfeld, O.; Ganz, A.B.; Dunn, J.; Hornburg, D.; et al. A longitudinal big data approach for precision health. Nat. Med. 2019, 25, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Hall, H.; Perelman, D.; Breschi, A.; Limcaoco, P.; Kellogg, R.; McLaughlin, T.; Snyder, M. Glucotypes reveal new patterns of glucose dysregulation. PLoS Biol. 2018, 16, e2005143. [Google Scholar] [CrossRef] [PubMed]
- Ahadi, S.; Zhou, W.; Schüssler-Fiorenza Rose, S.M.; Sailani, M.R.; Contrepois, K.; Avina, M.; Ashland, M.; Brunet, A.; Snyder, M. Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat. Med. 2020, 26, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Hood, L.; Price, N.D. Promoting Wellness & Demystifying Disease: The 100K Project. Clin. Omi. 2015, 1, 20–23. [Google Scholar] [CrossRef]
- Price, N.D.; Magis, A.T.; Earls, J.C.; Glusman, G.; Levy, R.; Lausted, C.; McDonald, D.T.; Kusebauch, U.; Moss, C.L.; Zhou, Y.; et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat. Biotechnol. 2017, 35, 747–756. [Google Scholar] [CrossRef]
- Oldoni, E.; Saunders, G.; Bietrix, F.; Garcia Bermejo, M.L.; Niehues, A.; ’t Hoen, P.A.C.; Nordlund, J.; Hajduch, M.; Scherer, A.; Kivinen, K.; et al. Tackling the translational challenges of multi-omics research in the realm of European personalised medicine: A workshop report. Front. Mol. Biosci. 2022, 9, 974799. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Asmis, R.; Hawkins, G.A.; Howard, T.D.; Cox, L.A. The Need for Multi-Omics Biomarker Signatures in Precision Medicine. Int. J. Mol. Sci. 2019, 20, 4781. [Google Scholar] [CrossRef] [PubMed]
- Krassowski, M.; Das, V.; Sahu, S.K.; Misra, B.B. State of the Field in Multi-Omics Research: From Computational Needs to Data Mining and Sharing. Front. Genet. 2020, 11, 610798. [Google Scholar] [CrossRef] [PubMed]
- EATRIS ERIC. EATRIS-Plus Project Quality Initiative. Available online: https://eatris.eu/eatris-quality-initiative (accessed on 12 January 2023).
- Schreier, J.; Feeney, R.; Keeling, P. Diagnostics Reform and Harmonization of Clinical Laboratory Testing. J. Mol. Diagn. 2019, 21, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Spitzenberger, F.; Patel, J.; Gebuhr, I.; Kruttwig, K.; Safi, A.; Meisel, C. Laboratory-Developed Tests: Design of a Regulatory Strategy in Compliance with the International State-of-the-Art and the Regulation (EU) 2017/746 (EU IVDR [In Vitro Diagnostic Medical Device Regulation]). Ther. Innov. Regul. Sci. 2022, 56, 47–64. [Google Scholar] [CrossRef]
- Graden, K.C.; Bennett, S.A.; Delaney, S.R.; Gill, H.E.; Willrich, M.A.V. A High-Level Overview of the Regulations Surrounding a Clinical Laboratory and Upcoming Regulatory Challenges for Laboratory Developed Tests. Lab Med. 2021, 52, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Genzen, J.R. Regulation of Laboratory-Developed Tests. Am. J. Clin. Pathol. 2019, 152, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Centers for Medicare and Medicaid Services. Background Document on CLIA Oversight of LDTs. Available online: https://www.cms.gov/Regulations-and-Guidance/Legislation/CLIA/Downloads/LDT-and-CLIA_FAQs.pdf (accessed on 15 January 2023).
- Nightingale Health Plc. Next-Generation Health Risk Management for Informed Decision-Making. Available online: https://pro.nightingalehealth.com (accessed on 15 January 2023).
- Ajinomoto Group. AminoIndex®|The amino acid profile as a marker for cancer screening. Available online: https://www.ajinomoto.com/innovation/action/aminoindex (accessed on 15 January 2023).
- Lokhov, P.G.; Balashova, E.E.; Trifonova, O.P.; Maslov, D.L.; Grigoriev, A.I.; Ponomarenko, E.A.; Archakov, A.I. Mass Spectrometric Blood Metabogram: Acquisition, Characterization, and Prospects for Application. Int. J. Mol. Sci. 2023, 24, 1736. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Balashova, E.E.; Trifonova, O.P.; Maslov, D.L.; Plotnikova, O.A.; Sharafetdinov, K.K.; Nikityuk, D.B.; Tutelyan, V.A.; Ponomarenko, E.A.; Archakov, A.I. Clinical Blood Metabogram: Application to Overweight and Obese Patients. Metabolites 2023, 13, 798. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Balashova, E.E.; Maslov, D.L.; Trifonova, O.P.; Lisitsa, A.V.; Markova, Y.M.; Stetsenko, V.V.; Polyanina, A.S.; Sheveleva, S.A.; Sharafetdinov, K.K.; et al. Linking Clinical Blood Metabogram and Gut Microbiota. Metabolites 2023, 13, 1095. [Google Scholar] [CrossRef]
- Alberti, K.G.; Zimmet, P.Z. Definition, Diagnosis and Classification of Diabetes Mellitus and its Complications Part 1: Diagnosis and Classification of Diabetes Mellitus. Deabet Med. 1998, 15, 539–553. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Trifonova, O.P.; Maslov, D.L.; Balashova, E.E.; Archakov, A.I.; Shestakova, E.A.; Shestakova, M.V.; Dedov, I.I. Diagnosing impaired glucose tolerance using direct infusion mass spectrometry of blood plasma. PLoS ONE 2014, 9, e105343. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Kharybin, O.N.; Archakov, A.I. Diagnosis of lung cancer based on direct-infusion electrospray mass spectrometry of blood plasma metabolites. Int. J. Mass Spectrom. 2011, 309, 200–205. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38 (Suppl. 2), W71–W77. [Google Scholar] [CrossRef]
- Safiri, S.; Karamzad, N.; Kaufman, J.S.; Bell, A.W.; Nejadghaderi, S.A.; Sullman, M.J.M.; Moradi-Lakeh, M.; Collins, G.; Kolahi, A.-A. Prevalence, Deaths and Disability-Adjusted-Life-Years (DALYs) Due to Type 2 Diabetes and Its Attributable Risk Factors in 204 Countries and Territories, 1990-2019: Results From the Global Burden of Disease Study 2019. Front. Endocrinol. 2022, 13, 838027. [Google Scholar] [CrossRef]
- Zhu, R.; Zhou, S.; Xia, L.; Bao, X. Incidence, Morbidity and years Lived With Disability due to Type 2 Diabetes Mellitus in 204 Countries and Territories: Trends From 1990 to 2019. Front. Endocrinol. 2022, 13, 905538. [Google Scholar] [CrossRef]
- Kahanovitz, L.; Sluss, P.M.; Russell, S.J. Type 1 Diabetes—A Clinical Perspective. Point Care 2017, 16, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martín, C. Pathophysiology of Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef] [PubMed]
- Sagel, J.; Colwell, J.A. Shock During Oral Glucose Tolerance Testing. JAMA 1973, 226, 667–668. [Google Scholar] [CrossRef] [PubMed]
- Mcdonald, G.W.; Fisher, G.F.; Burnham, C. Reproducibility of the Oral Glucose Tolerance Test. Diabetes 1965, 14, 473–480. [Google Scholar] [CrossRef]
- Balion, C.M.; Raina, P.S.; Gerstein, H.C.; Santaguida, P.L.; Morrison, K.M.; Booker, L.; Hunt, D.L. Reproducibility of impaired glucose tolerance (IGT) and impaired fasting glucose (IFG) classification: A systematic review. Clin. Chem. Lab. Med. 2007, 45, 1180–1185. [Google Scholar] [CrossRef]
- Ko, G.T.C.; Chan, J.C.N.; Woo, J.; Lau, E.; Yeung, V.T.F.; Chow, C.C.; Cockram, C.S. The reproducibility and usefulness of the oral glucose tolerance test in screening for diabetes and other cardiovascular risk factors. Ann. Clin. Biochem. 1998, 35, 62–67. [Google Scholar] [CrossRef]
- Chai, J.H.; Ma, S.; Heng, D.; Yoong, J.; Lim, W.-Y.; Toh, S.-A.; Loh, T.P. Impact of analytical and biological variations on classification of diabetes using fasting plasma glucose, oral glucose tolerance test and HbA1c. Sci. Rep. 2017, 7, 13721. [Google Scholar] [CrossRef] [PubMed]
- Coskun, C.; Inal, B.B.; Emre, H.O.; Baz, S.; Gumus, A.; Sonmez, D.; Orhan, B.; Duz, M.E.; Serin, E.; Koldas, M. Evaluation of biological variations in glucose and glycated hemoglobin levels in healthy individuals. Turk. J. Biochem. 2018, 43, 495–501. [Google Scholar] [CrossRef]
- Fraser, C.G.; Williams, P. Short-term biological variation of plasma analytes in renal disease. Clin. Chem. 1983, 29, 508–510. [Google Scholar] [CrossRef] [PubMed]
- Duong, K.N.C.; Tan, C.J.; Rattanasiri, S.; Thakkinstian, A.; Anothaisintawee, T.; Chaiyakunapruk, N. Comparison of diagnostic accuracy for diabetes diagnosis: A systematic review and network meta-analysis. Front. Med. 2023, 10, 1016381. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.Z. Individuality of clinical biochemical patterns in preventive health maintenance. J. Occup. Med. 1967, 9, 567–570. [Google Scholar] [PubMed]
- Kroll, M.H. Biological variation of glucose and insulin includes a deterministic chaotic component. Biosystems 1999, 50, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Harris, E.K.; Kanofsky, P.; Shakarji, G.; Cotlove, E. Biological and analytic components of variation in long-term studies of serum constituents in normal subjects. II. Estimating biological components of variation. Clin. Chem. 1970, 16, 1022–1027. [Google Scholar] [CrossRef] [PubMed]
- Fraser, C.G. Biological variation in clinical chemistry. An update: Collated data, 1988–1991. Arch. Pathol. Lab. Med. 1992, 116, 916–923. [Google Scholar]
- Leite, N.C.; Salles, G.F.; Araujo, A.L.E.; Villela-Nogueira, C.A.; Cardoso, C.R.L. Prevalence and associated factors of non-alcoholic fatty liver disease in patients with type-2 diabetes mellitus. Liver Int. 2009, 29, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Targher, G.; Bertolini, L.; Padovani, R.; Rodella, S.; Tessari, R.; Zenari, L.; Day, C.; Arcaro, G. Prevalence of nonalcoholic fatty liver disease and its association with cardiovascular disease among type 2 diabetic patients. Diabetes Care 2007, 30, 1212–1218. [Google Scholar] [CrossRef]
- Silverman, J.F.; O’Brien, K.F.; Long, S.; Leggett, N.; Khazanie, P.G.; Pories, W.J.; Norris, H.T.; Caro, J.F. Liver pathology in morbidly obese patients with and without diabetes. Am. J. Gastroenterol. 1990, 85, 1349–1355. [Google Scholar]
- Bugianesi, E.; McCullough, A.J.; Marchesini, G. Insulin resistance: A metabolic pathway to chronic liver disease. Hepatology 2005, 42, 987–1000. [Google Scholar] [CrossRef] [PubMed]
- Marchesini, G.; Brizi, M.; Morselli-Labate, A.M.; Bianchi, G.; Bugianesi, E.; McCullough, A.J.; Forlani, G.; Melchionda, N. Association of nonalcoholic fatty liver disease with insulin resistance. Am. J. Med. 1999, 107, 450–455. [Google Scholar] [CrossRef]
- Efe, O.; Verma, A.; Waikar, S.S. Urinary oxalate as a potential mediator of kidney disease in diabetes mellitus and obesity. Curr. Opin. Nephrol. Hypertens. 2019, 28, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Nikiforova, V.J.; Giesbertz, P.; Wiemer, J.; Bethan, B.; Looser, R.; Liebenberg, V.; Ruiz Noppinger, P.; Daniel, H.; Rein, D. Glyoxylate, a new marker metabolite of type 2 diabetes. J. Diabetes Res. 2014, 2014, 685204. [Google Scholar] [CrossRef]
- Padberg, I.; Peter, E.; González-Maldonado, S.; Witt, H.; Mueller, M.; Weis, T.; Bethan, B.; Liebenberg, V.; Wiemer, J.; Katus, H.A.; et al. A new metabolomic signature in type-2 diabetes mellitus and its pathophysiology. PLoS ONE 2014, 9, e85082. [Google Scholar] [CrossRef]
- Laffel, L. Ketone bodies: A review of physiology, pathophysiology and application of monitoring to diabetes. Diabetes Metab. Res. Rev. 1999, 15, 412–426. [Google Scholar] [CrossRef]
- Guasch-Ferré, M.; Hruby, A.; Toledo, E.; Clish, C.B.; Martínez-González, M.A.; Salas-Salvadó, J.; Hu, F.B. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care 2016, 39, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Alqudah, A.; Wedyan, M.; Qnais, E.; Jawarneh, H.; McClements, L. Plasma Amino Acids Metabolomics’ Important in Glucose Management in Type 2 Diabetes. Front. Pharmacol. 2021, 12, 695418. [Google Scholar] [CrossRef] [PubMed]
- Tessaro, F.H.G.; Ayala, T.S.; Martins, J.O. Lipid Mediators Are Critical in Resolving Inflammation: A Review of the Emerging Roles of Eicosanoids in Diabetes Mellitus. Biomed Res. Int. 2015, 2015, 568408. [Google Scholar] [CrossRef]
- Tuomisto, K.; Palmu, J.; Long, T.; Watrous, J.D.; Mercader, K.; Lagerborg, K.A.; Andres, A.; Salmi, M.; Jalkanen, S.; Vasan, R.S.; et al. A plasma metabolite score of three eicosanoids predicts incident type 2 diabetes: A prospective study in three independent cohorts. BMJ Open Diabetes Res. Care 2022, 10, e002519. [Google Scholar] [CrossRef]
- Luo, P.; Wang, M.-H. Eicosanoids, β-cell function, and diabetes. Prostaglandins Other Lipid Mediat. 2011, 95, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.C.; Shulman, G.I. Roles of Diacylglycerols and Ceramides in Hepatic Insulin Resistance. Trends Pharmacol. Sci. 2017, 38, 649–665. [Google Scholar] [CrossRef]
- Luukkonen, P.K.; Zhou, Y.; Sädevirta, S.; Leivonen, M.; Arola, J.; Orešič, M.; Hyötyläinen, T.; Yki-Järvinen, H. Hepatic ceramides dissociate steatosis and insulin resistance in patients with non-alcoholic fatty liver disease. J. Hepatol. 2016, 64, 1167–1175. [Google Scholar] [CrossRef]
- Albareda, M.; de Leiva, A.; Corcoy, R. Reproducibility of diabetes mellitus diagnosis (WHO 1999 criteria) in women. Acta Diabetol. 2004, 41, 14–17. [Google Scholar] [CrossRef]
- Hwang, J.L.; Weiss, R.E. Steroid-induced diabetes: A clinical and molecular approach to understanding and treatment. Diabetes Metab. Res. Rev. 2014, 30, 96–102. [Google Scholar] [CrossRef]
- Tamez-Pérez, H.E.; Quintanilla-Flores, D.L.; Rodríguez-Gutiérrez, R.; González-González, J.G.; Tamez-Peña, A.L. Steroid hyperglycemia: Prevalence, early detection and therapeutic recommendations: A narrative review. World J. Diabetes 2015, 6, 1073–1081. [Google Scholar] [CrossRef]
- I S Sobczak, A.; A Blindauer, C.; J Stewart, A. Changes in Plasma Free Fatty Acids Associated with Type-2 Diabetes. Nutrients 2019, 11, 2022. [Google Scholar] [CrossRef]
- Hui, D.Y. Phospholipase A(2) enzymes in metabolic and cardiovascular diseases. Curr. Opin. Lipidol. 2012, 23, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Prunonosa Cervera, I.; Gabriel, B.M.; Aldiss, P.; Morton, N.M. The phospholipase A2 family’s role in metabolic diseases: Focus on skeletal muscle. Physiol. Rep. 2021, 9, e14662. [Google Scholar] [CrossRef] [PubMed]
- Inouye, M.; Mio, T.; Sumino, K. Dicarboxylic acids as markers of fatty acid peroxidation in diabetes. Atherosclerosis 2000, 148, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Vangipurapu, J.; Stancáková, A.; Smith, U.; Kuusisto, J.; Laakso, M. Nine Amino Acids Are Associated With Decreased Insulin Secretion and Elevated Glucose Levels in a 7.4-Year Follow-up Study of 5,181 Finnish Men. Diabetes 2019, 68, 1353–1358. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wang, Y.; Liang, X.; Zou, L.; Ong, C.N.; Yuan, J.-M.; Koh, W.-P.; Pan, A. Serum Amino Acids in Association with Prevalent and Incident Type 2 Diabetes in A Chinese Population. Metabolites 2019, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar] [CrossRef]
- Chen, T.; Ni, Y.; Ma, X.; Bao, Y.; Liu, J.; Huang, F.; Hu, C.; Xie, G.; Zhao, A.; Jia, W.; et al. Branched-chain and aromatic amino acid profiles and diabetes risk in Chinese populations. Sci. Rep. 2016, 6, 20594. [Google Scholar] [CrossRef]
- Diamanti, K.; Cavalli, M.; Pan, G.; Pereira, M.J.; Kumar, C.; Skrtic, S.; Grabherr, M.; Risérus, U.; Eriksson, J.W.; Komorowski, J.; et al. Intra- and inter-individual metabolic profiling highlights carnitine and lysophosphatidylcholine pathways as key molecular defects in type 2 diabetes. Sci. Rep. 2019, 9, 9653. [Google Scholar] [CrossRef]
- Coregliano-Ring, L.; Goia-Nishide, K.; Rangel, É.B. Hypokalemia in Diabetes Mellitus Setting. Medicina 2022, 58, 431. [Google Scholar] [CrossRef]
- Liamis, G.; Liberopoulos, E.; Barkas, F.; Elisaf, M. Diabetes mellitus and electrolyte disorders. World J. Clin. Cases 2014, 2, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Trifonova, O.P.; Maslov, D.L.; Balashova, E.E.; Lokhov, P.G. Evaluation of Dried Blood Spot Sampling for Clinical Metabolomics: Effects of Different Papers and Sample Storage Stability. Metabolites 2019, 9, 277. [Google Scholar] [CrossRef] [PubMed]



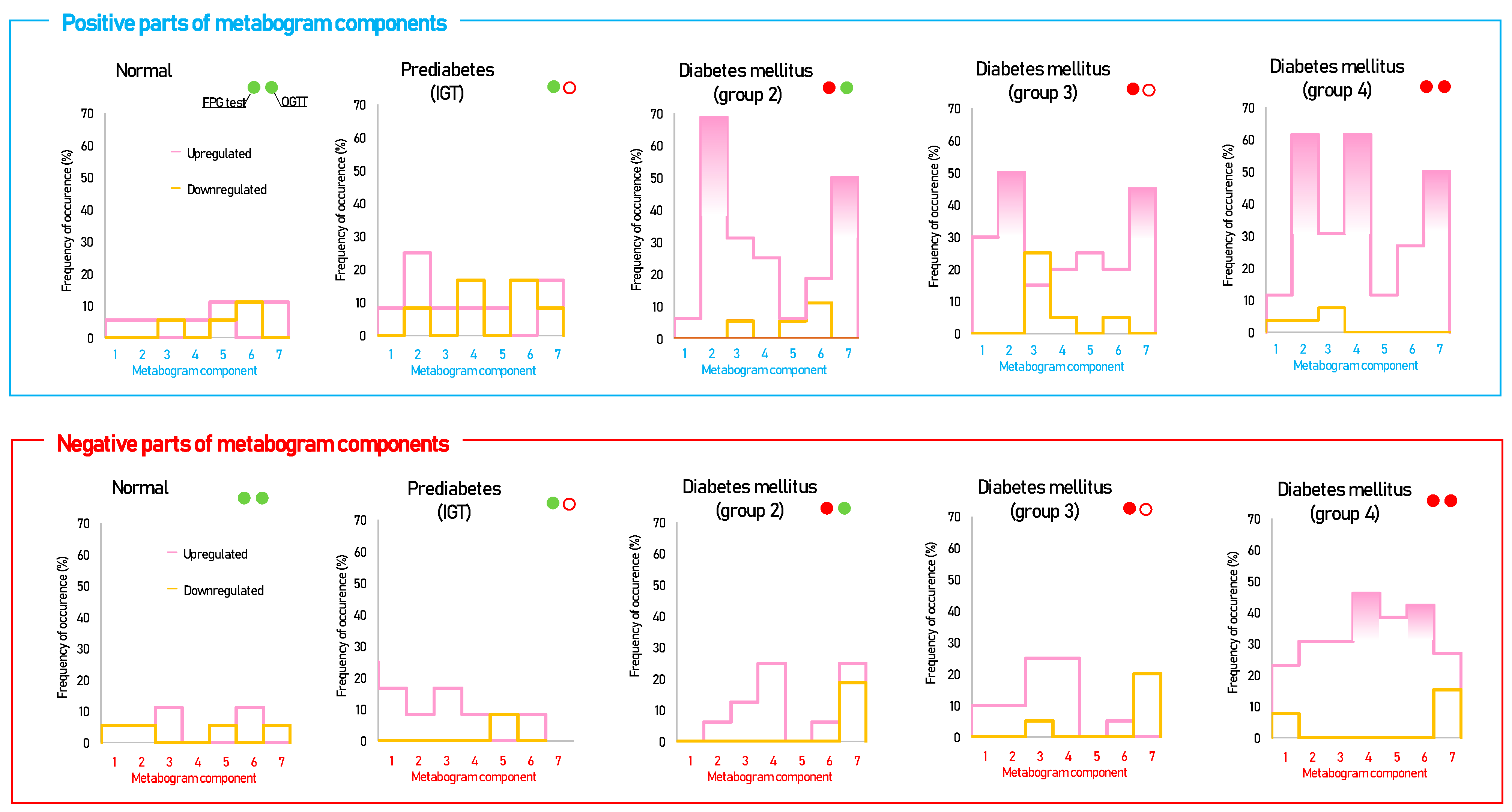


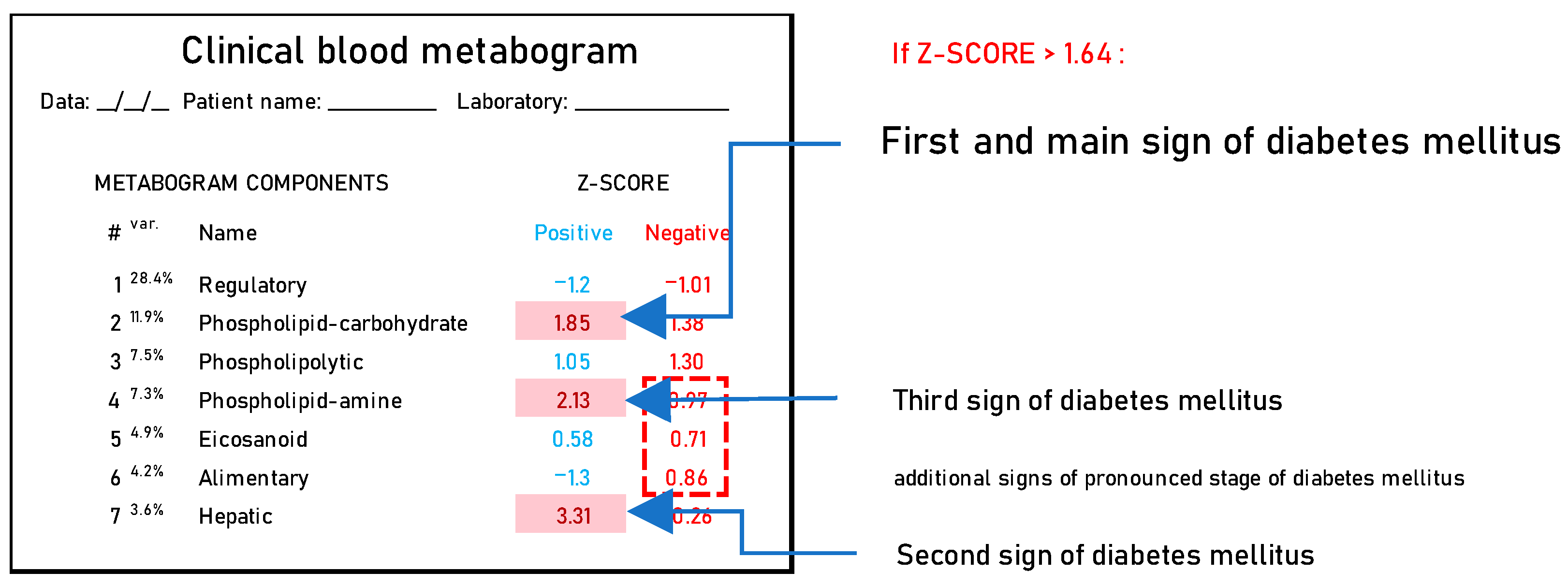
| Parameter | Groups | |||||
|---|---|---|---|---|---|---|
| Normal | Prediabetes | Type 2 Diabetes Mellitus | ||||
| Group 1 | Group 2 | Group 3 | Group 4 | |||
| Number | 18 | 12 | 2 | 16 | 20 | 26 |
| Gender (males/females) | 9/9 | 6/6 | 1/1 | 8/8 | 10/10 | 13/13 |
| Age (years) | 54.7 ± 13.2 1 | 63.1 ± 9.6 | 63, 81 | 48.8 ± 12.5 | 54.6 ± 12.0 | 58.8 ± 11.0 |
| BMI (kg/m2) | 33.4 ± 9.1 | 35.7 ± 10.1 | 28.7, 38.9 | 33.7 ±8.9 | 33.5 ± 6.2 | 33.6 ± 6.8 |
| FPG (mmol/L) | 5.3 ± 0.3 | 5.4 ± 0.2 | 5.1, 5.6 | 6.2 ± 0.4 | 7.1 ± 1.7 | 8.0 ± 2.3 |
| Glucose in OGTT (mmol/L) | 6.2 ± 0.9 | 9.3 ± 0.9 | 14.1, 11.4 | 6.2 ± 1.3 | 9.2 ± 0.9 | 14.3 ± 2.3 |
| HbA1c (%) [mmol/mol] | 5.7 ± 0.4 | 6.1 ± 0.5 | 6.2, 6.0 | 5.9 ± 0.5 | 6.3 ± 0.6 | 7.2 ± 1.5 |
| LDL (mmol/L) | 3.5 ± 0.8 | 3.2 ± 1.0 | 2.7, 2.8 | 3.3 ±0.9 | 3.5 ± 1.1 | 3.6 ± 1.0 |
| HDL (mmol/L) | 1.2 ± 0.4 | 1.2 ± 0.4 | 0.7, 1.4 | 1.1 ±0.2 | 1.1 ± 0.3 | 1.0 ± 0.3 |
| Triglycerides (mmol/L) | 1.3 ± 0.5 | 1.3 ± 0.4 | 2.1, 0.8 | 1.4 ±0.6 | 2.5 ± 2.9 | 2.7 ±2.1 |
| Cholesterol (mmol/L) | 5.2 ± 0.9 | 4.9 ± 1.2 | 4.7, 4.5 | 5.1 ± 0.9 | 5.6 ± 1.1 | 5.7 ± 1.4 |
| Uric acid (µmol/L) | 386 ± 82 | 392 ± 97 | 380, 266 | 353 ± 94 | 395 ± 97 | 356 ± 88 |
| Insulin (µU/mL) | 15.2 ± 22.6 | 13.4 ± 7.7 | 12.1, 5.8 | 15.9 ± 13.4 | 26.9 ± 31.8 | 25.1 ± 33.3 |
| HOMA-IR | 3.6 ± 5.4 | 3.2 ± 1.8 | 2.7, 1.4 | 4.3 ± 3.4 | 8.6 ± 9.8 | 8.3 ± 9.5 |
| Metabogram Component | t-Test (p-Value) | |||||
|---|---|---|---|---|---|---|
| Normal Subjects Versus Subjects with | ||||||
| Prediabetes 1 | Type 2 Diabetes Mellitus | |||||
| Group 2 | Group 3 | Group 4 | All Groups | |||
| Positive | 1 | 0.67 | 0.83 | 0.01 | 0.33 | 0.13 |
| 2 | 0.79 | 0.00001 | 0.008 | 0.0006 | 0.0008 | |
| 3 | 0.72 | 0.01 | 0.71 | 0.09 | 0.11 | |
| 4 | 0.39 | 0.11 | 0.04 | 0.0003 | 0.01 | |
| 5 | 0.64 | 0.73 | 0.04 | 0.09 | 0.15 | |
| 6 | 0.19 | 0.05 | 0.25 | 0.04 | 0.04 | |
| 7 | 0.93 | 0.0003 | 0.003 | 0.0006 | 0.0005 | |
| Negative | 1 | 0.36 | 0.10 | 0.80 | 0.05 | 0.17 |
| 2 | 0.65 | 0.52 | 0.70 | 0.02 | 0.13 | |
| 3 | 0.46 | 0.97 | 0.08 | 0.24 | 0.26 | |
| 4 | 0.34 | 0.006 | 0.17 | 0.001 | 0.004 | |
| 5 | 0.64 | 0.54 | 0.28 | 0.0001 | 0.01 | |
| 6 | 0.53 | 0.37 | 0.19 | 0.002 | 0.02 | |
| 7 | 0.83 | 0.92 | 0.03 | 0.95 | 0.53 | |
| Metabogram Component | |||||
|---|---|---|---|---|---|
| Prediabetes 1 | Type 2 Diabetes Mellitus | ||||
| Group 2 | Group 3 | Group 4 | |||
| Positive | 1 | ||||
| 2 | |||||
| 3 | |||||
| 4 | |||||
| 5 | |||||
| 6 | |||||
| 7 | |||||
| Negative | 1 | ||||
| 2 | |||||
| 3 | |||||
| 4 | |||||
| 5 | |||||
| 6 | |||||
| 7 | |||||
| Measured m/z | Calculated m/z | Delta Δm/z | Detected Ion | Elemental Formula |
|---|---|---|---|---|
| 128.963 | 128.9585 | 0.0045 | [M+K39]+ | C2H2O4 |
| 140.995 | 140.9949 | 0.0001 | [M+K39]+ | C4H6O3 |
| 142.990 | 142.9930 | −0.0030 | [M+K40]+ | C4H6O3 |
| 184.985 | 184.9847 | 0.0003 | [M+K39]+ | C5H6O5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lokhov, P.G.; Balashova, E.E.; Trifonova, O.P.; Maslov, D.L.; Shestakova, E.A.; Shestakova, M.V.; Dedov, I.I. Application of Clinical Blood Metabogram to Type 2 Diabetes Mellitus. Metabolites 2024, 14, 168. https://doi.org/10.3390/metabo14030168
Lokhov PG, Balashova EE, Trifonova OP, Maslov DL, Shestakova EA, Shestakova MV, Dedov II. Application of Clinical Blood Metabogram to Type 2 Diabetes Mellitus. Metabolites. 2024; 14(3):168. https://doi.org/10.3390/metabo14030168
Chicago/Turabian StyleLokhov, Petr G., Elena E. Balashova, Oxana P. Trifonova, Dmitry L. Maslov, Ekaterina A. Shestakova, Marina V. Shestakova, and Ivan I. Dedov. 2024. "Application of Clinical Blood Metabogram to Type 2 Diabetes Mellitus" Metabolites 14, no. 3: 168. https://doi.org/10.3390/metabo14030168
APA StyleLokhov, P. G., Balashova, E. E., Trifonova, O. P., Maslov, D. L., Shestakova, E. A., Shestakova, M. V., & Dedov, I. I. (2024). Application of Clinical Blood Metabogram to Type 2 Diabetes Mellitus. Metabolites, 14(3), 168. https://doi.org/10.3390/metabo14030168









