Discrimination of Lipogenic or Glucogenic Diet Effects in Early-Lactation Dairy Cows Using Plasma Metabolite Abundances and Ratios in Combination with Machine Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Design
2.2. Characteristics of Lipogenic and Glucogenic Diets
2.3. Measure of Cow Routine Indicators
2.4. Collection of Blood Samples
2.5. LC–MS Measurements
2.6. Data Processing
2.6.1. Metabolite Abundance Data
2.6.2. Ratios of Metabolite Abundances
2.7. Statistical Analysis
2.7.1. Exploratory Analysis
2.7.2. Discrimination and Classification Analysis of Plasma Metabolite Profiles
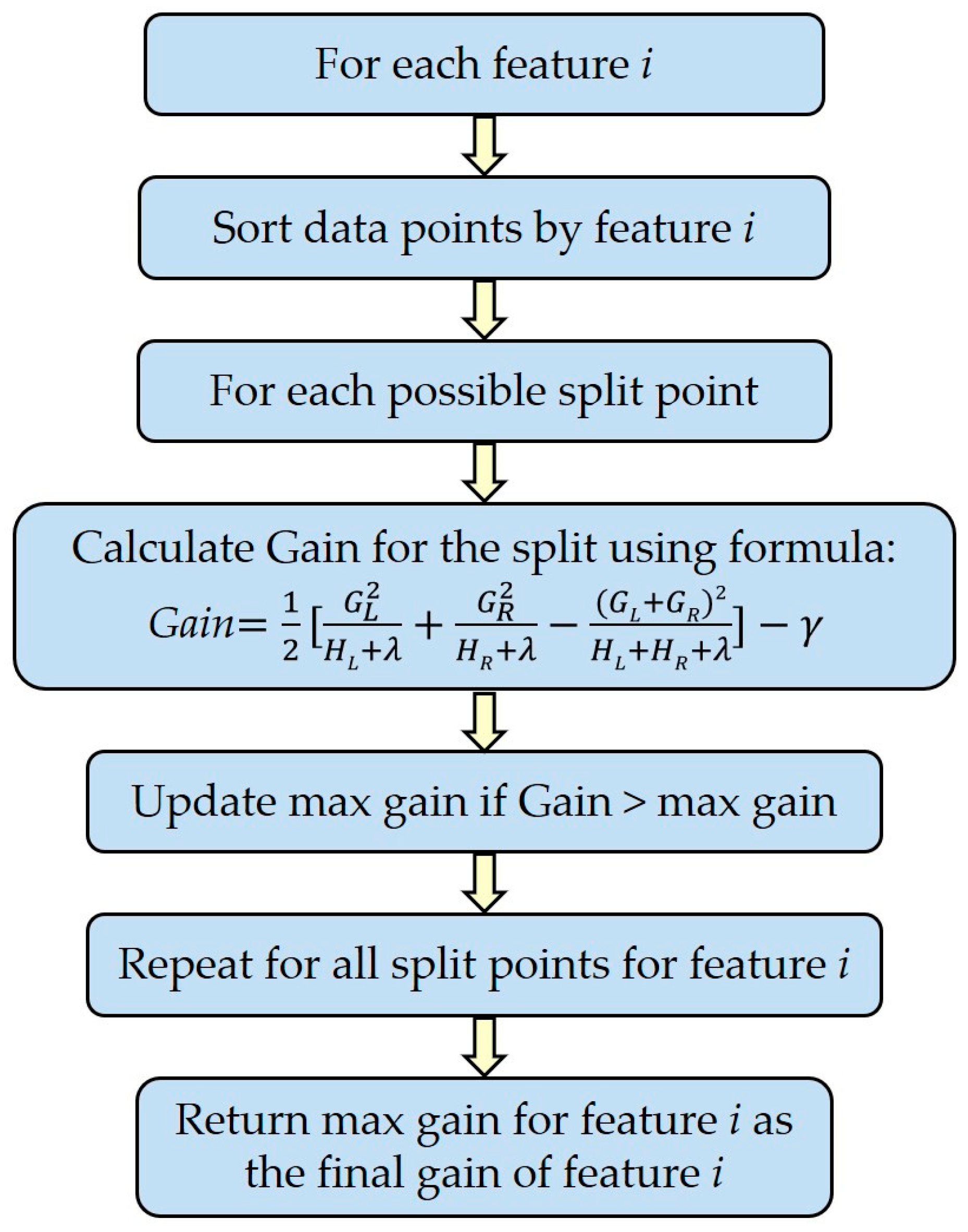
2.7.3. Variable Importance for Classification
2.7.4. Testing Metabolite Abundance and Metabolite Ratios between Dietary Intervention and Weeks
2.7.5. Software
3. Results and Discussion
3.1. Information of Cow Routine Indicators
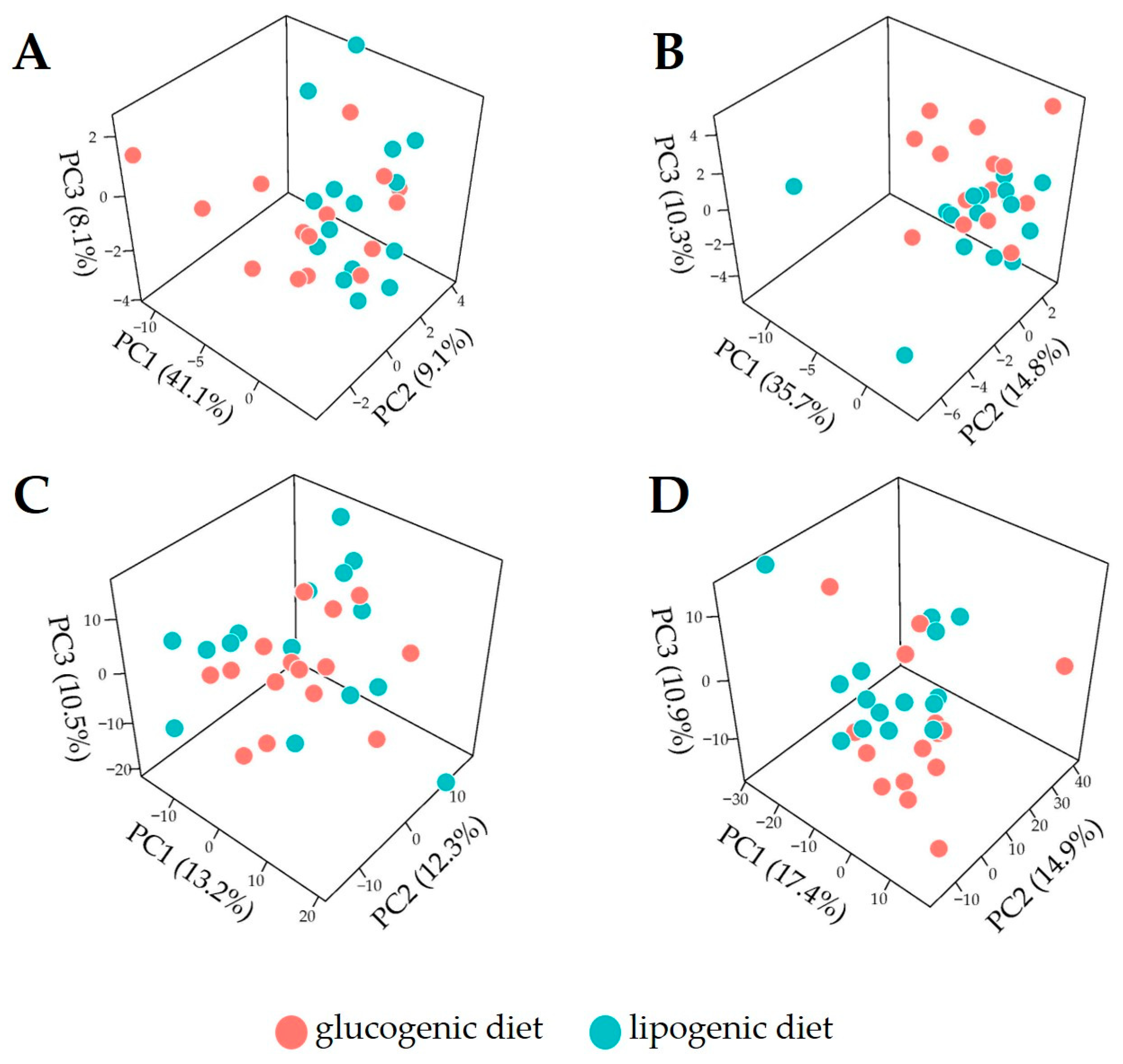
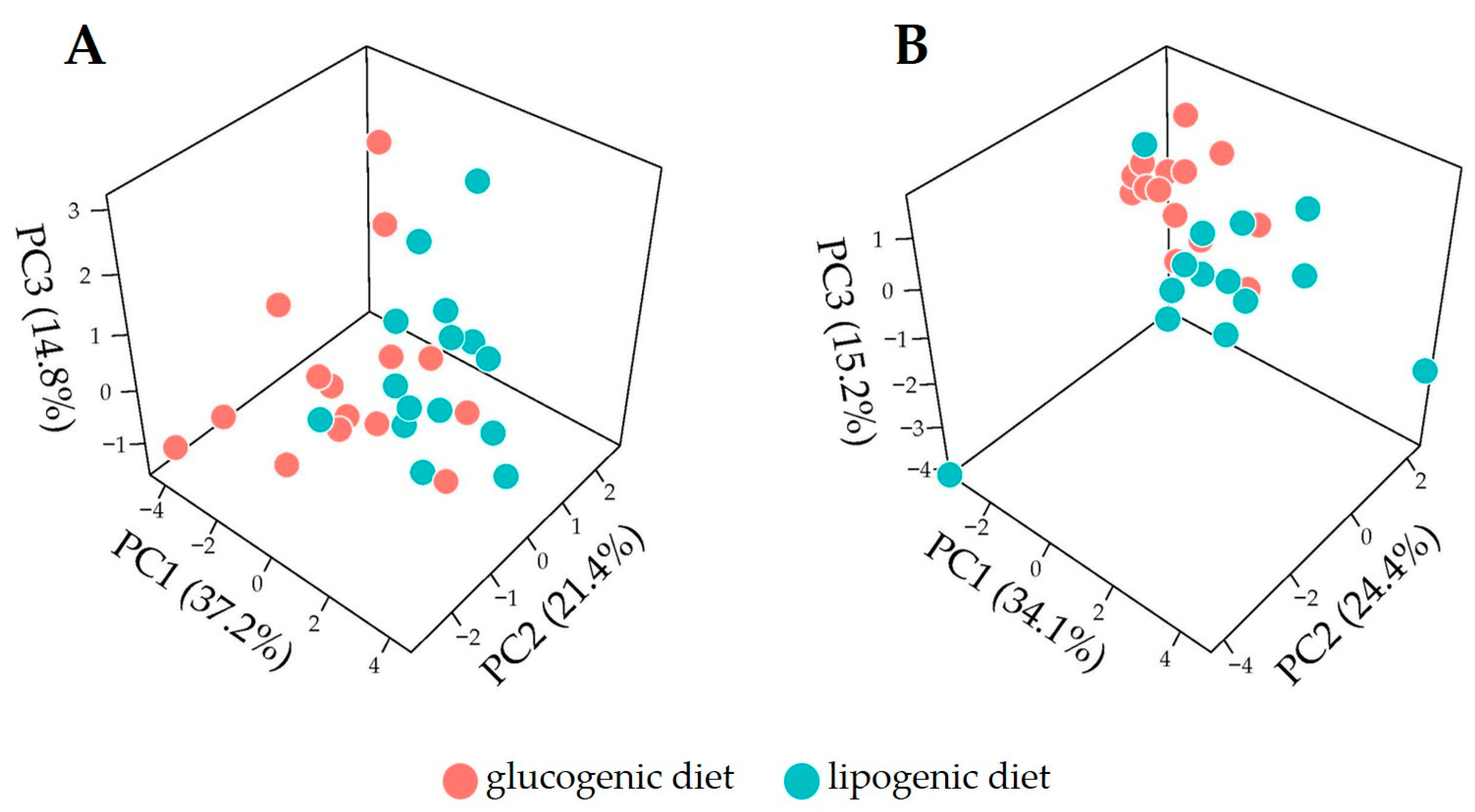
3.2. Overview of Metabolome Profiles for Cow Plasma
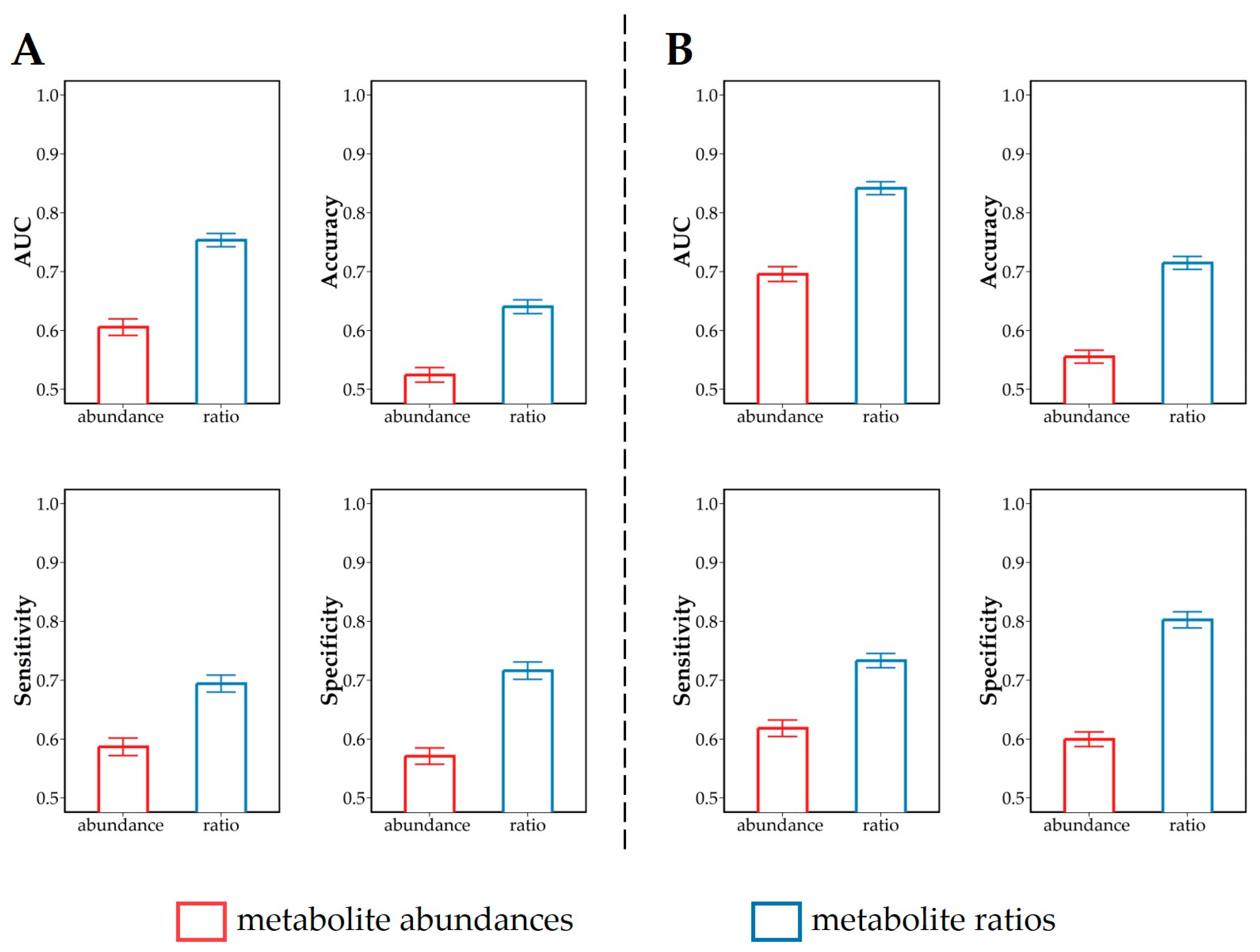
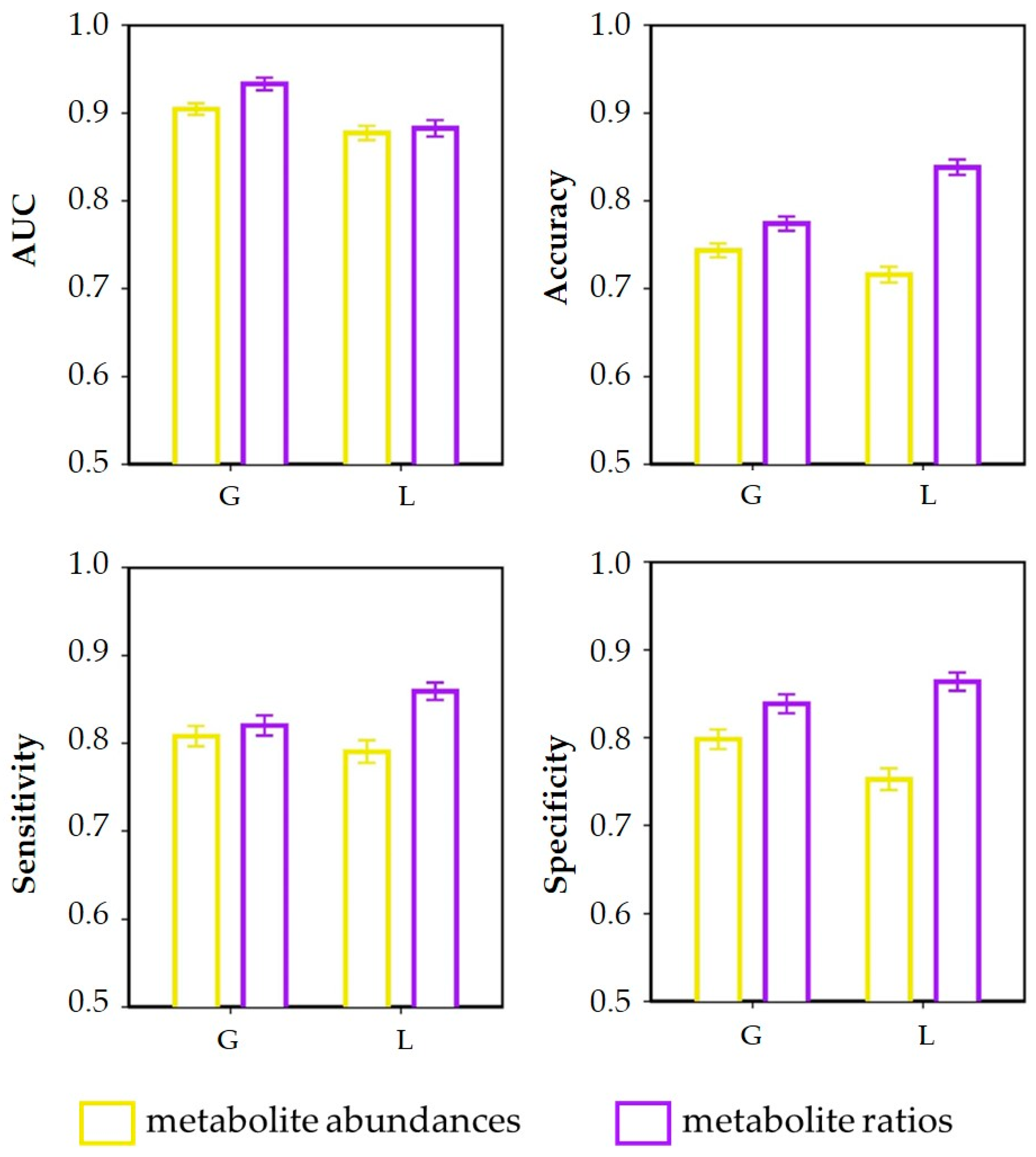
3.3. Discrimination of Dietary Intervention
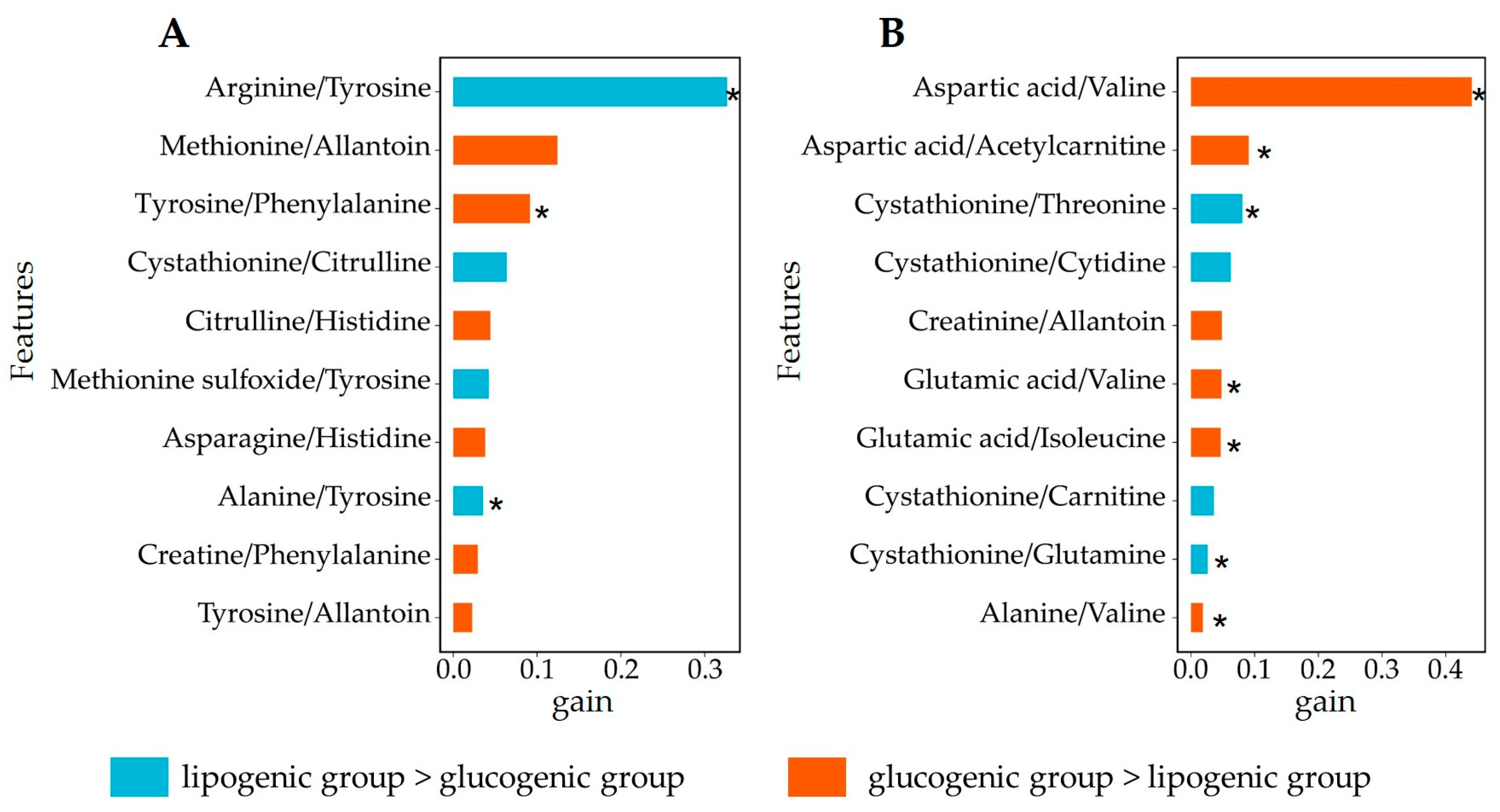
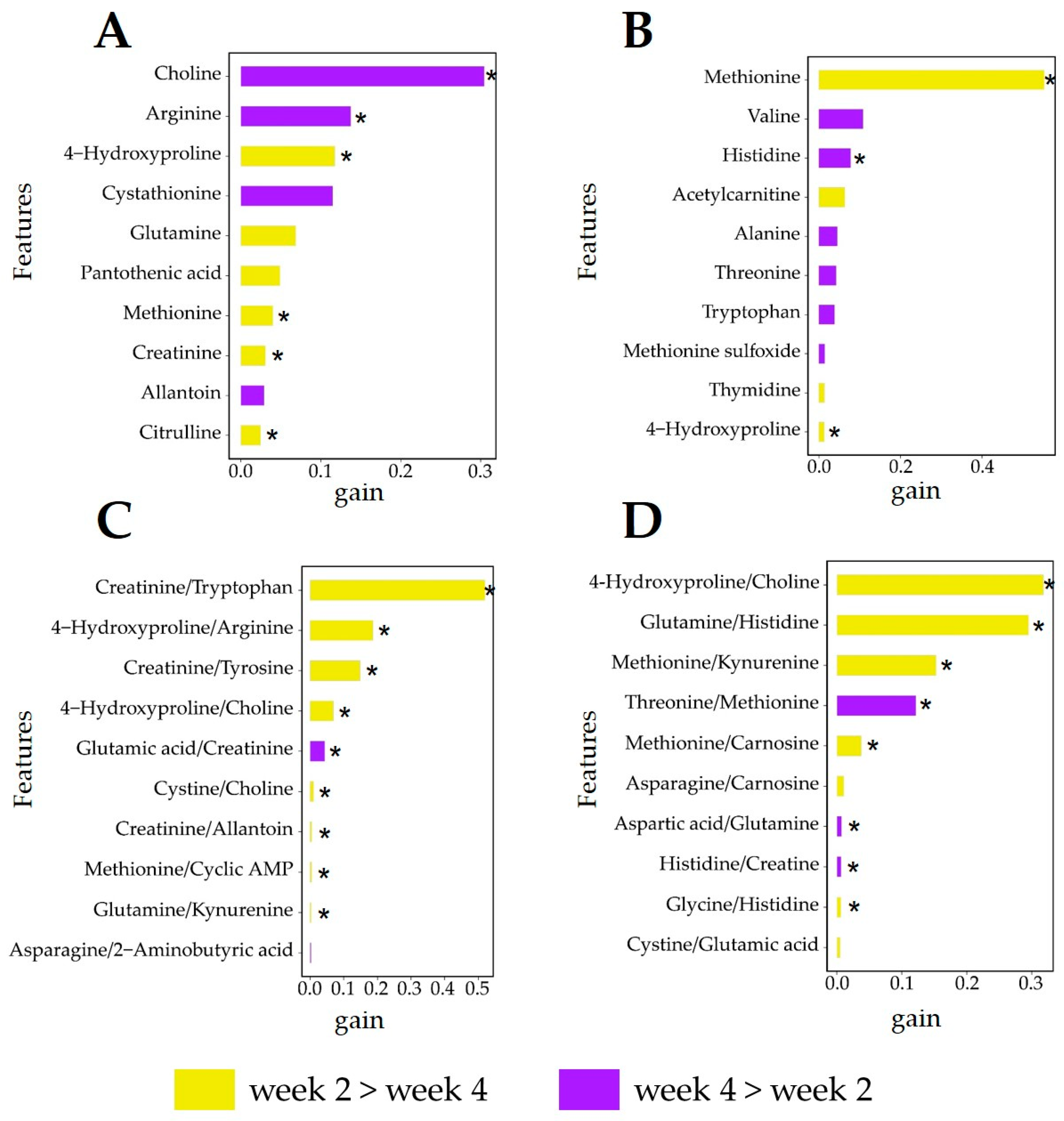
3.4. Analysis of Relevant Metabolite Features Discriminating between Glucogenic and Lipogenic Diets
3.5. Comparison of Plasma Metabolite Features with Respect to Lactation Week
3.6. Analysis of Relevant Metabolite Features Discriminating between Lactation Weeks
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Pathway | Total | Hits | Raw p | Impact |
|---|---|---|---|---|
| Arginine biosynthesis | 14 | 5 | 1.49 × 10−5 | 0.42 |
| Histidine metabolism | 16 | 4 | 5.51 × 10−4 | 0.31 |
| Alanine, aspartate, and glutamate metabolism | 28 | 5 | 5.59 × 10−4 | 0.53 |
| Glycine, serine, and threonine metabolism | 34 | 5 | 1.41 × 10−3 | 0.30 |
| Arginine and proline metabolism | 38 | 5 | 2.36 × 10−3 | 0.29 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | 4 | 2 | 3.77 × 10−3 | 1.00 |
| D-Glutamine and D-glutamate metabolism | 5 | 2 | 6.17 × 10−3 | 1.00 |
| Cysteine and methionine metabolism | 33 | 4 | 9.03 × 10−3 | 0.32 |
| Pantothenate and CoA biosynthesis | 19 | 3 | 1.16 × 10−2 | 0.01 |
| beta-Alanine metabolism | 21 | 3 | 1.53 × 10−2 | 0.06 |
| Phenylalanine metabolism | 12 | 2 | 3.64 × 10−2 | 0.36 |
| Glyoxylate and dicarboxylate metabolism | 32 | 3 | 4.70 × 10−2 | 0.11 |
| Pyrimidine metabolism | 38 | 3 | 7.19 × 10−2 | 0.01 |
| Glutathione metabolism | 28 | 2 | 1.61 × 10−1 | 0.11 |
| Tryptophan metabolism | 41 | 2 | 2.86 × 10−1 | 0.24 |
| Nicotinate and nicotinamide metabolism | 13 | 1 | 2.89 × 10−1 | 0.19 |
| Glycerophospholipid metabolism | 36 | 1 | 6.14 × 10−1 | 0.03 |
| Tyrosine metabolism | 42 | 1 | 6.71 × 10−1 | 0.14 |
| Primary bile acid biosynthesis | 46 | 1 | 7.05 × 10−1 | 0.02 |
References
- Leduc, A.; Souchet, S.; Gelé, M.; Le Provost, F.; Boutinaud, M. Effect of Feed Restriction on Dairy Cow Milk Production: A Review. J. Anim. Sci. 2021, 99, skab130. [Google Scholar] [CrossRef]
- Kuhla, B.; Nürnberg, G.; Albrecht, D.; Görs, S.; Hammon, H.M.; Metges, C.C. Involvement of Skeletal Muscle Protein, Glycogen, and Fat Metabolism in the Adaptation on Early Lactation of Dairy Cows. J. Proteome Res. 2011, 10, 4252–4262. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.S.; Almstetter, M.F.; Nürnberger, N.; Sigl, G.; Gronwald, W.; Wiedemann, S.; Dettmer, K.; Oefner, P.J. Correlations between Milk and Plasma Levels of Amino and Carboxylic Acids in Dairy Cows. J. Proteome Res. 2013, 12, 5223–5232. [Google Scholar] [CrossRef] [PubMed]
- van Knegsel, A.T.M.; van den Brand, H.; Dijkstra, J.; Tamminga, S.; Kemp, B. Effect of Dietary Energy Source on Energy Balance, Production, Metabolic Disorders and Reproduction in Lactating Dairy Cattle. Reprod. Nutr. Dev. 2005, 45, 665–688. [Google Scholar] [CrossRef]
- Vossebeld, F.; van Knegsel, A.T.M.; Saccenti, E. Phenotyping Metabolic Status of Dairy Cows Using Clustering of Time Profiles of Energy Balance Peripartum. J. Dairy Sci. 2022, 105, 4565–4580. [Google Scholar] [CrossRef] [PubMed]
- de Vries, M.J.; Veerkamp, R.F. Energy Balance of Dairy Cattle in Relation to Milk Production Variables and Fertility. J. Dairy Sci. 2000, 83, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, S.; Derno, M.; Schäff, C.T.; Börner, S.; Kautzsch, U.; Kuhla, B.; Hammon, H.M.; Tuchscherer, A.; Röntgen, M. Comparative Analyses of Estimated and Calorimetrically Determined Energy Balance in High-Yielding Dairy Cows. J. Dairy Sci. 2019, 102, 4002–4013. [Google Scholar] [CrossRef] [PubMed]
- Churakov, M.; Karlsson, J.; Edvardsson Rasmussen, A.; Holtenius, K. Milk Fatty Acids as Indicators of Negative Energy Balance of Dairy Cows in Early Lactation. Animal 2021, 15, 100253. [Google Scholar] [CrossRef]
- Pires, J.A.A.; Larsen, T.; Leroux, C. Milk Metabolites and Fatty Acids as Noninvasive Biomarkers of Metabolic Status and Energy Balance in Early-Lactation Cows. J. Dairy Sci. 2022, 105, 201–220. [Google Scholar] [CrossRef]
- van Knegsel, A.T.M.; Remmelink, G.J.; Jorjong, S.; Fievez, V.; Kemp, B. Effect of Dry Period Length and Dietary Energy Source on Energy Balance, Milk Yield, and Milk Composition of Dairy Cows. J. Dairy Sci. 2014, 97, 1499–1512. [Google Scholar] [CrossRef]
- Xu, W.; van Knegsel, A.; Saccenti, E.; van Hoeij, R.; Kemp, B.; Vervoort, J. Metabolomics of Milk Reflects a Negative Energy Balance in Cows. J. Proteome Res. 2020, 19, 2942–2949. [Google Scholar] [CrossRef] [PubMed]
- Wehrens, R.; Hageman, J.A.; van Eeuwijk, F.; Kooke, R.; Flood, P.J.; Wijnker, E.; Keurentjes, J.J.B.; Lommen, A.; van Eekelen, H.D.L.M.; Hall, R.D.; et al. Improved Batch Correction in Untargeted MS-Based Metabolomics. Metabolomics 2016, 12, 88. [Google Scholar] [CrossRef]
- Eckhart, A.D.; Beebe, K.; Milburn, M. Metabolomics as a Key Integrator for “Omic” Advancement of Personalized Medicine and Future Therapies. Clin. Transl. Sci. 2012, 5, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-Generation Machine Learning for Biological Networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef]
- Rosato, A.; Tenori, L.; Cascante, M.; De Atauri Carulla, P.R.; Martins Dos Santos, V.A.P.; Saccenti, E. From Correlation to Causation: Analysis of Metabolomics Data Using Systems Biology Approaches. Metabolomics 2018, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Giannuzzi, D.; Mota, L.F.M.; Pegolo, S.; Tagliapietra, F.; Schiavon, S.; Gallo, L.; Marsan, P.A.; Trevisi, E.; Cecchinato, A. Prediction of Detailed Blood Metabolic Profile Using Milk Infrared Spectra and Machine Learning Methods in Dairy Cattle. J. Dairy Sci. 2023, 106, 3321–3344. [Google Scholar] [CrossRef] [PubMed]
- Petersen, A.-K.; Krumsiek, J.; Wägele, B.; Theis, F.J.; Wichmann, H.-E.; Gieger, C.; Suhre, K. On the Hypothesis-Free Testing of Metabolite Ratios in Genome-Wide and Metabolome-Wide Association Studies. BMC Bioinform. 2012, 13, 120. [Google Scholar] [CrossRef]
- Di Cesare, F.; Luchinat, C.; Tenori, L.; Saccenti, E. Age- and Sex-Dependent Changes of Free Circulating Blood Metabolite and Lipid Abundances, Correlations, and Ratios. J. Gerontol. Ser. A 2022, 77, 918–926. [Google Scholar] [CrossRef]
- Jahagirdar, S.; Suarez-Diez, M.; Saccenti, E. Simulation and Reconstruction of Metabolite–Metabolite Association Networks Using a Metabolic Dynamic Model and Correlation Based Algorithms. J. Proteome Res. 2019, 18, 1099–1113. [Google Scholar] [CrossRef]
- Klein, M.S.; Buttchereit, N.; Miemczyk, S.P.; Immervoll, A.-K.; Louis, C.; Wiedemann, S.; Junge, W.; Thaller, G.; Oefner, P.J.; Gronwald, W. NMR Metabolomic Analysis of Dairy Cows Reveals Milk Glycerophosphocholine to Phosphocholine Ratio as Prognostic Biomarker for Risk of Ketosis. J. Proteome Res. 2012, 11, 1373–1381. [Google Scholar] [CrossRef]
- Sustainable Development Goals. United Nations Development Programme. Available online: https://www.undp.org/sustainable-development-goals (accessed on 4 April 2024).
- Van Es, A.J.H. Feed Evaluation for Dairy Cows. Livest. Prod. Sci. 1975, 2, 95–107. [Google Scholar] [CrossRef]
- Tamminga, S.; Van Straalen, W.M.; Subnel, A.P.J.; Meijer, R.G.M.; Steg, A.; Wever, C.J.G.; Blok, M.C. The Dutch Protein Evaluation System: The DVE/OEB-System. Livest. Prod. Sci. 1994, 40, 139–155. [Google Scholar] [CrossRef]
- Chen, J.; Gross, J.J.; van Dorland, H.A.; Remmelink, G.J.; Bruckmaier, R.M.; Kemp, B.; van Knegsel, A.T.M. Effects of Dry Period Length and Dietary Energy Source on Metabolic Status and Hepatic Gene Expression of Dairy Cows in Early Lactation. J. Dairy Sci. 2015, 98, 1033–1045. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Nishiumi, S.; Kobayashi, T.; Azuma, T.; Yoshida, M. LC–MS/MS-Based Metabolome Analysis Detected Changes in the Metabolic Profiles of Small and Large Intestinal Adenomatous Polyps in Apc Min/+ Mice. Metabolomics 2016, 12, 68. [Google Scholar] [CrossRef]
- Matsubara, A.; Izumi, Y.; Nishiumi, S.; Suzuki, M.; Azuma, T.; Fukusaki, E.; Bamba, T.; Yoshida, M. Supercritical Fluid Extraction as a Preparation Method for Mass Spectrometry of Dried Blood Spots. J. Chromatogr. B 2014, 969, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Vervoort, J.; Saccenti, E.; van Hoeij, R.; Kemp, B.; van Knegsel, A. Milk Metabolomics Data Reveal the Energy Balance of Individual Dairy Cows in Early Lactation. Sci. Rep. 2018, 8, 15828. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses; R Packages: Marseille, France, 2020. [Google Scholar]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T.; et al. Xgboost: Extreme Gradient Boosting; R Packages: Marseille, France, 2023. [Google Scholar]
- Søreide, K. Receiver-Operating Characteristic Curve Analysis in Diagnostic, Prognostic and Predictive Biomarker Research. J. Clin. Pathol. 2009, 62, 1–5. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R.; Hastie, T. A Working Guide to Boosted Regression Trees. J. Anim. Ecol. 2008, 77, 802–813. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining; Association for Computing Machinery: New York, NY, USA, 2016. [Google Scholar]
- Husson, F.; Le, S.; Pagès, J. Exploratory Multivariate Analysis by Example Using R; Chapman and Hall/CRC: Boca Raton, FL, USA, 2017; ISBN 978-0-429-22543-7. [Google Scholar]
- van Knegsel, A.T.M.; van den Brand, H.; Dijkstra, J.; Kemp, B. Effects of Dietary Energy Source on Energy Balance, Metabolites and Reproduction Variables in Dairy Cows in Early Lactation. Theriogenology 2007, 68, S274–S280. [Google Scholar] [CrossRef]
- Xiao, F.; Guo, F. Impacts of Essential Amino Acids on Energy Balance. Mol. Metab. 2022, 57, 101393. [Google Scholar] [CrossRef]
- Xu, W.; Vervoort, J.; Saccenti, E.; Kemp, B.; van Hoeij, R.J.; van Knegsel, A.T.M. Relationship between Energy Balance and Metabolic Profiles in Plasma and Milk of Dairy Cows in Early Lactation. J. Dairy Sci. 2020, 103, 4795–4805. [Google Scholar] [CrossRef] [PubMed]
- Thams, P.; Capito, K. L-Arginine Stimulation of Glucose-Induced Insulin Secretion through Membrane Depolarization and Independent of Nitric Oxide. Eur. J. Endocrinol. 1999, 140, 87–93. [Google Scholar] [CrossRef] [PubMed]
- National Academies of Sciences, Engineering, and Medicine; Division on Earth and Life Studies; Board on Agriculture and Natural Resources; Committee on Nutrient Requirements of Dairy Cattle. Nutrient Requirements of Dairy Cattle: Eighth Revised Edition; National Academies Press: Washington, DC, USA, 2021; ISBN 978-0-309-67777-6. [Google Scholar]
- Giesecke, D.; Ehrentreich, L.; Stangassinger, M.; Ahrens, F. Mammary and Renal Excretion of Purine Metabolites in Relation to Energy Intake and Milk Yield in Dairy Cows. J. Dairy Sci. 1994, 77, 2376–2381. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, G.N.; Larson, B.L. Conversion of Phenylalanine to Tyrosine in the Bovine Mammary Secretory Cell. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 1968, 165, 121–126. [Google Scholar] [CrossRef]
- Guinard, J.; Rulquin, H. Effect of Graded Levels of Duodenal Infusions of Casein on Mammary Uptake in Lactating Cows. 2. Individual Amino Acids. J. Dairy Sci. 1994, 77, 3304–3315. [Google Scholar] [CrossRef] [PubMed]
- Whitton, P.D.; Rodrigues, L.M.; Hems, D.A. Stimulation by Vasopressin, Angiotensin and Oxytocin of Gluconeogenesis in Hepatocyte Suspensions. Biochem. J. 1978, 176, 893–898. [Google Scholar] [CrossRef]
- Zang, Y.; Samii, S.S.; Phipps, Z.C.; Tager, L.R.; McFadden, J.W.; Krause, K.M. Comparative Effects of Multiple Sources of Rumen-Protected Methionine on Milk Production and Serum Amino Acid Levels in Mid-Lactation Dairy Cows. Prof. Anim. Sci. 2017, 33, 692–699. [Google Scholar] [CrossRef]
- Weber, C.; Hametner, C.; Tuchscherer, A.; Losand, B.; Kanitz, E.; Otten, W.; Sauerwein, H.; Bruckmaier, R.M.; Becker, F.; Kanitz, W.; et al. Hepatic Gene Expression Involved in Glucose and Lipid Metabolism in Transition Cows: Effects of Fat Mobilization during Early Lactation in Relation to Milk Performance and Metabolic Changes. J. Dairy Sci. 2013, 96, 5670–5681. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jian, J. Exponential Convergence and Lagrange Stability for Impulsive Cohen–Grossberg Neural Networks with Time-Varying Delays. J. Comput. Appl. Math. 2015, 277, 23–35. [Google Scholar] [CrossRef]
- Bene, J.; Szabo, A.; Komlósi, K.; Melegh, B. Mass Spectrometric Analysis of L-Carnitine and Its Esters: Potential Biomarkers of Disturbances in Carnitine Homeostasis. Curr. Mol. Med. 2020, 20, 336–354. [Google Scholar] [CrossRef]
- Krumsiek, J.; Stückler, F.; Suhre, K.; Gieger, C.; Spector, T.D.; Soranzo, N.; Kastenmüller, G.; Theis, F.J. Network-Based Metabolite Ratios for an Improved Functional Characterization of Genome-Wide Association Study Results. bioRxiv 2016, 048512. [Google Scholar]
- Wang, F.; Rico, J.E.; Fontoura, A.B.P.; Gervais, R.; McFadden, J.W. Short Communication: Effects of Dietary Deoiled Soy Lecithin Supplementation on Circulating Choline and Choline Metabolites, and the Plasma Phospholipid Profile in Holstein Cows Fed Palm Fat. J. Dairy Sci. 2021, 104, 1838–1845. [Google Scholar] [CrossRef] [PubMed]
- de Veth, M.J.; Artegoitia, V.M.; Campagna, S.R.; Lapierre, H.; Harte, F.; Girard, C.L. Choline Absorption and Evaluation of Bioavailability Markers When Supplementing Choline to Lactating Dairy Cows. J. Dairy Sci. 2016, 99, 9732–9744. [Google Scholar] [CrossRef]
- Orozco, J.S.; Hertz-Picciotto, I.; Abbeduto, L.; Slupsky, C.M. Metabolomics Analysis of Children with Autism, Idiopathic-Developmental Delays, and Down Syndrome. Transl. Psychiatry 2019, 9, 243. [Google Scholar] [CrossRef] [PubMed]
- Castillo, C.; Hernández, J.; Valverde, I.; Pereira, V.; Sotillo, J.; Alonso, M.L.; Benedito, J.L. Plasma Malonaldehyde (MDA) and Total Antioxidant Status (TAS) during Lactation in Dairy Cows. Res. Vet. Sci. 2006, 80, 133–139. [Google Scholar] [CrossRef]
- Clare, C.E.; Brassington, A.H.; Kwong, W.Y.; Sinclair, K.D. One-Carbon Metabolism: Linking Nutritional Biochemistry to Epigenetic Programming of Long-Term Development. Annu. Rev. Anim. Biosci. 2019, 7, 263–287. [Google Scholar] [CrossRef]
- Cole, L.K.; Vance, J.E.; Vance, D.E. Phosphatidylcholine Biosynthesis and Lipoprotein Metabolism. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2012, 1821, 754–761. [Google Scholar] [CrossRef]
- Zeisel, S.H. Choline: Critical Role During Fetal Development and Dietary Requirements in Adults. Annu. Rev. Nutr. 2006, 26, 229–250. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Jahagirdar, S.; Bakker, W.; Lute, C.; Kemp, B.; Knegsel, A.v.; Saccenti, E. Discrimination of Lipogenic or Glucogenic Diet Effects in Early-Lactation Dairy Cows Using Plasma Metabolite Abundances and Ratios in Combination with Machine Learning. Metabolites 2024, 14, 230. https://doi.org/10.3390/metabo14040230
Wang X, Jahagirdar S, Bakker W, Lute C, Kemp B, Knegsel Av, Saccenti E. Discrimination of Lipogenic or Glucogenic Diet Effects in Early-Lactation Dairy Cows Using Plasma Metabolite Abundances and Ratios in Combination with Machine Learning. Metabolites. 2024; 14(4):230. https://doi.org/10.3390/metabo14040230
Chicago/Turabian StyleWang, Xiaodan, Sanjeevan Jahagirdar, Wouter Bakker, Carolien Lute, Bas Kemp, Ariette van Knegsel, and Edoardo Saccenti. 2024. "Discrimination of Lipogenic or Glucogenic Diet Effects in Early-Lactation Dairy Cows Using Plasma Metabolite Abundances and Ratios in Combination with Machine Learning" Metabolites 14, no. 4: 230. https://doi.org/10.3390/metabo14040230
APA StyleWang, X., Jahagirdar, S., Bakker, W., Lute, C., Kemp, B., Knegsel, A. v., & Saccenti, E. (2024). Discrimination of Lipogenic or Glucogenic Diet Effects in Early-Lactation Dairy Cows Using Plasma Metabolite Abundances and Ratios in Combination with Machine Learning. Metabolites, 14(4), 230. https://doi.org/10.3390/metabo14040230







