Combined Metabolomics and Biochemical Analyses of Serum and Milk Revealed Parity-Related Metabolic Differences in Sanhe Dairy Cattle
Abstract
:1. Introduction
2. Material and Methods
2.1. Animals, Housing, and Experimental Design
2.2. Diet and Feed Ingredients
2.3. Blood Variables
2.4. Milk Production and Composition
2.5. Metabolomics Analysis by UPLC-MS/MS
2.6. Metabolomics Data Processing and Analysis
2.7. Statistical Analysis of Data
3. Results
3.1. Lactation Performance and Milk Traits
3.2. Serum Biochemical Parameters and Free Amino Acid Levels
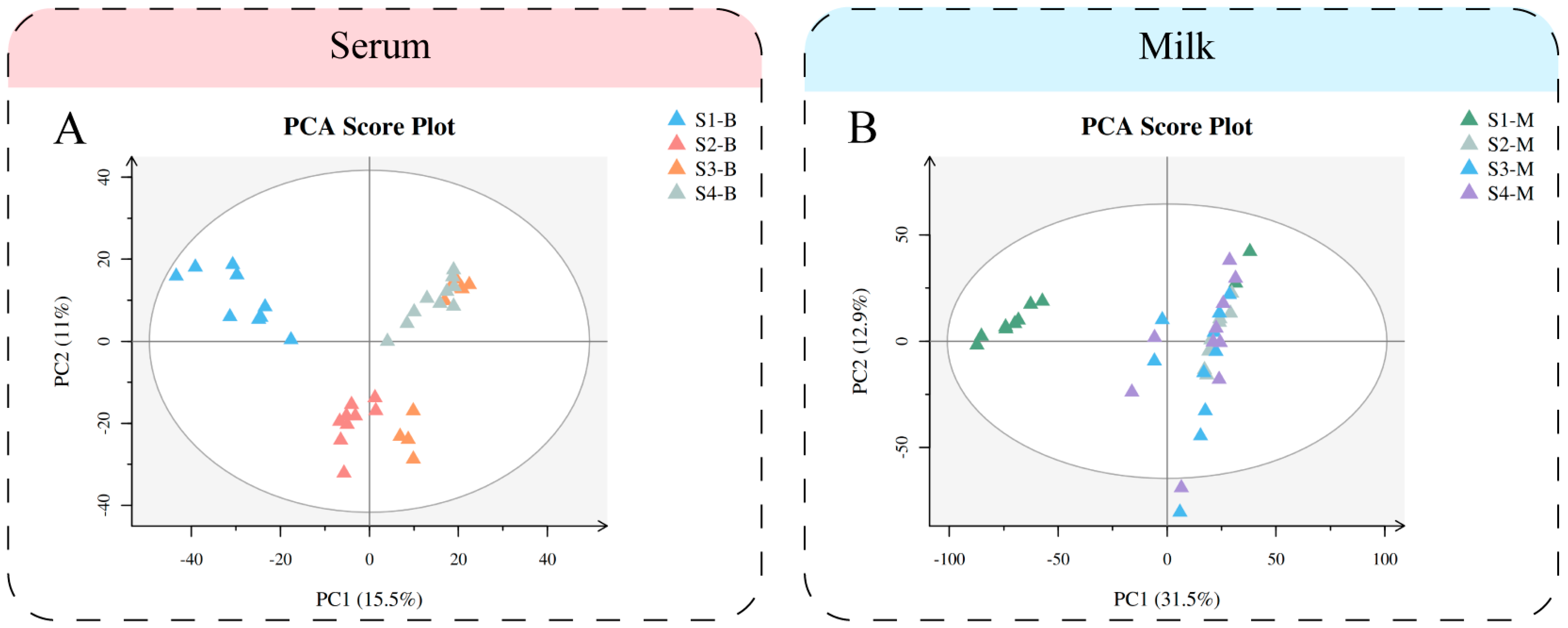
3.3. Metabolomics Multivariate Analysis of Serum and Milk
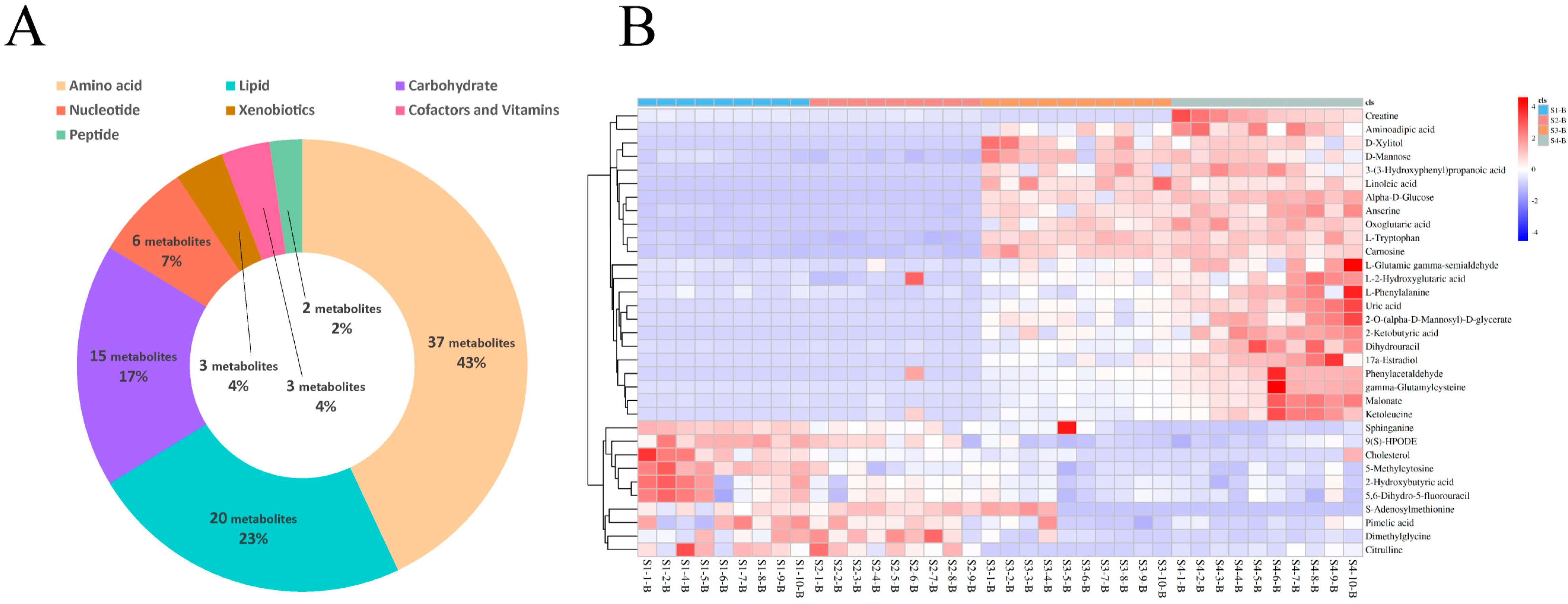
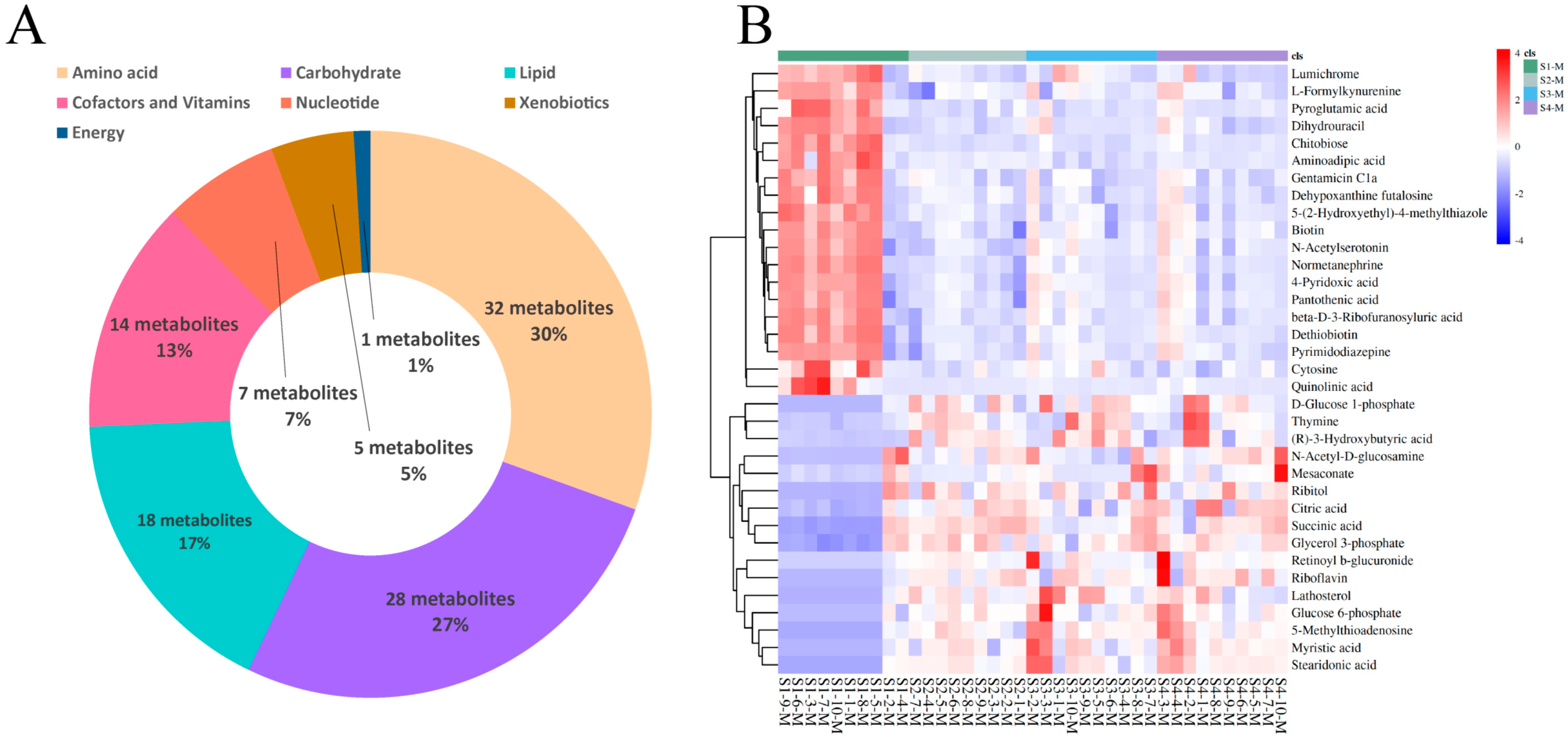
3.4. Differential Metabolite Identification in Serum and Milk
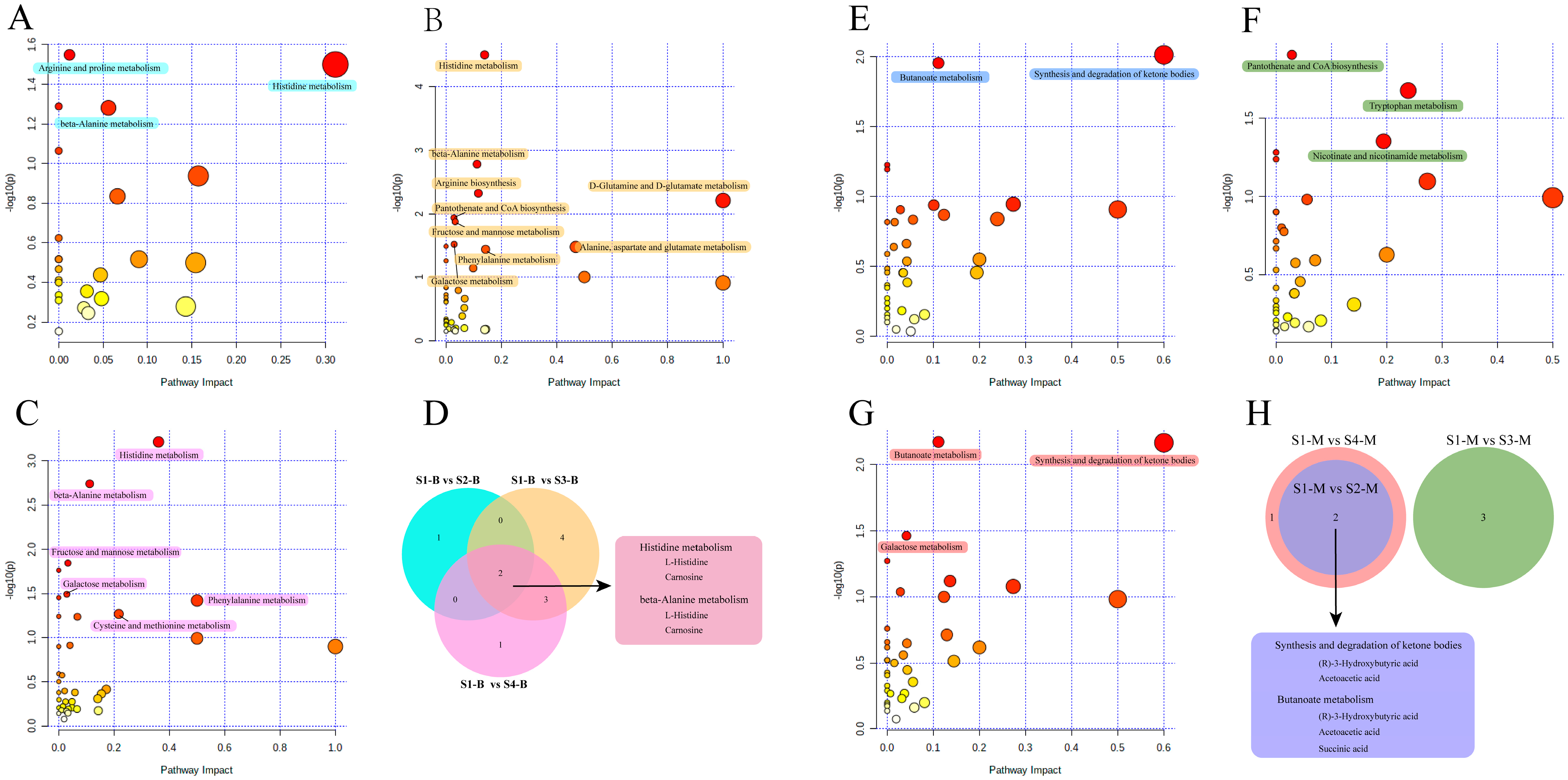
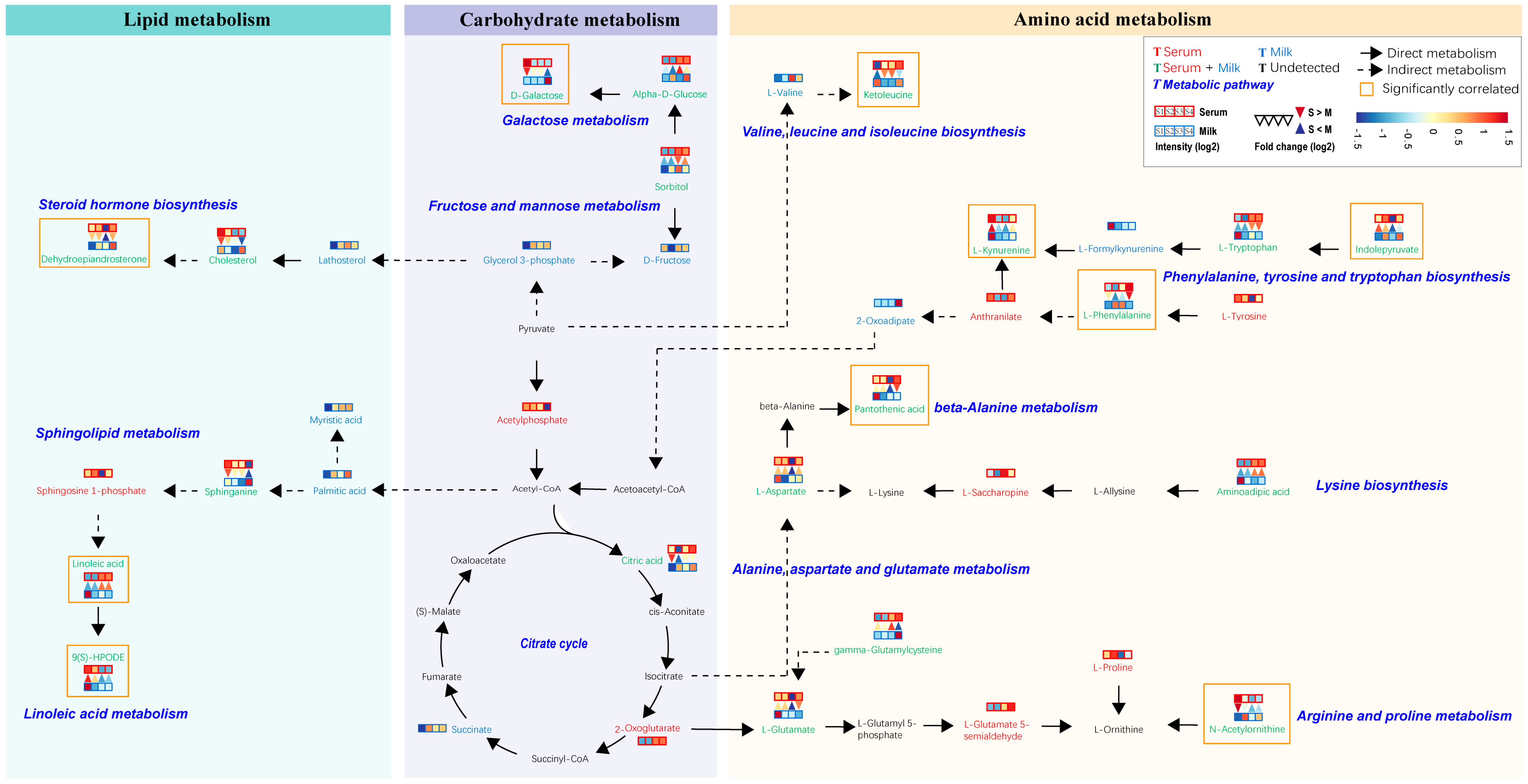
3.5. Pathway Analysis of Serum and Milk
3.6. Interaction Analysis of Serum and Milk Metabolites
4. Discussion
4.1. Characteristics of Serum and Milk from Sanhe Dairy Cattle with Parities of 1–4
4.2. Association and Transformation of Amino Acids, Carbohydrates, and Lipids between Serum and Milk Samples from Sanhe Dairy Cattle with Parities of 1–4
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pacífico, C.; Stauder, A.; Reisinger, N.; Schwartz-Zimmermann, H.E.; Zebeli, Q. Distinct serum metabolomic signatures of multiparous and primiparous dairy cows switched from a moderate to high-grain diet during early lactation. Metabolomics 2020, 16, 96. [Google Scholar] [CrossRef] [PubMed]
- Janovick, N.A.; Drackley, J.K. Prepartum dietary management of energy intake affects postpartum intake and lactation performance by primiparous and multiparous holstein cows. J. Dairy Sci. 2010, 93, 3086–3102. [Google Scholar] [CrossRef] [PubMed]
- Neave, H.W.; Lomb, J.; von Keyserlingk, M.A.G.; Behnam-Shabahang, A.; Weary, D.M. Parity differences in the behavior of transition dairy cows. J. Dairy Sci. 2017, 100, 548–561. [Google Scholar] [CrossRef] [PubMed]
- Morales Piñeyrúa, J.T.; Fariña, S.R.; Mendoza, A. Effects of parity on productive, reproductive, metabolic and hormonal responses of holstein cows. Anim. Reprod. Sci. 2018, 191, 9–21. [Google Scholar] [CrossRef]
- Adriaens, I.; van den Brulle, I.; D’Anvers, L.; Statham, J.M.E.; Geerinckx, K.; De Vliegher, S.; Piepers, S.; Aernouts, B. Milk losses and dynamics during perturbations in dairy cows differ with parity and lactation stage. J. Dairy Sci. 2021, 104, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Honig, H.; Ofer, L.; Elbaz, M.; Kaim, M.; Shinder, D.; Gershon, E. Seasonal and parity effects on ghrelin levels throughout the estrous cycle in dairy cows. Gen. Comp. Endocrinol. 2016, 235, 64–69. [Google Scholar] [CrossRef]
- Bittante, G.; Cipolat-Gotet, C.; Malchiodi, F.; Sturaro, E.; Tagliapietra, F.; Schiavon, S.; Cecchinato, A. Effect of dairy farming system, herd, season, parity, and days in milk on modeling of the coagulation, curd firming, and syneresis of bovine milk. J. Dairy Sci. 2015, 98, 2759–2774. [Google Scholar] [CrossRef]
- Bittante, G.; Cecchinato, A.; Schiavon, S. Dairy system, parity, and lactation stage affect enteric methane production, yield, and intensity per kilogram of milk and cheese predicted from gas chromatography fatty acids. J. Dairy Sci. 2018, 101, 1752–1766. [Google Scholar] [CrossRef]
- Hu, L.; Ma, Y.; Liu, L.; Kang, L.; Brito, L.F.; Wang, D.; Wu, H.; Liu, A.; Wang, Y.; Xu, Q. Detection of functional polymorphisms in the hsp70 gene and association with cold stress response in inner-mongolia sanhe cattle. Cell Stress Chaperones 2019, 24, 409–418. [Google Scholar] [CrossRef]
- Freitas, P.H.F.; Wang, Y.; Yan, P.; Oliveira, H.R.; Schenkel, F.S.; Zhang, Y.; Xu, Q.; Brito, L.F. Genetic diversity and signatures of selection for thermal stress in cattle and other two bos species adapted to divergent climatic conditions. Front. Genet. 2021, 12, 604823. [Google Scholar] [CrossRef]
- Xu, Q.; Wang, Y.C.; Liu, R.; Brito, L.F.; Kang, L.; Yu, Y.; Wang, D.S.; Wu, H.J.; Liu, A. Differential gene expression in the peripheral blood of chinese sanhe cattle exposed to severe cold stress. Genet. Mol. Res. 2017, 16. [Google Scholar] [CrossRef] [PubMed]
- Bruckmaier, R.M.; Gross, J.J. Lactational challenges in transition dairy cows. Anim. Prod. Sci. 2017, 57, 1471–1481. [Google Scholar] [CrossRef]
- Gross, J.J.; Bruckmaier, R.M. Review: Metabolic challenges in lactating dairy cows and their assessment via established and novel indicators in milk. Animal 2019, 13, s75–s81. [Google Scholar] [CrossRef] [PubMed]
- Bauman, D.E.; Mather, I.H.; Wall, R.J.; Lock, A.L. Major advances associated with the biosynthesis of milk. J. Dairy Sci. 2006, 89, 1235–1243. [Google Scholar] [CrossRef] [PubMed]
- McManaman, J.L.; Neville, M.C. Mammary physiology and milk secretion. Adv. Drug Deliv. Rev. 2003, 55, 629–641. [Google Scholar] [CrossRef]
- Yoshinaga, K.; Obi, J.; Tago, A.; Kato, Y.; Nagai, T.; Yoshida, A.; Gotoh, N. Analysis of hydroxy triacylglycerol as a lactone precursor in milk fat using liquid chromatography electrospray ionization tandem mass spectrometry. Food Chem. 2019, 274, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Cho, K.; Park, M.; Choi, T.; Kim, S.; Do, C. Genetic parameters of milk β-hydroxybutyric acid and acetone and their genetic association with milk production traits of holstein cattle. Asian-Australas. J. Anim. Sci. 2016, 29, 1530–1540. [Google Scholar] [CrossRef]
- Zhao, Y.; Guo, X.; Yan, S.; Shi, B.; Sheng, R. Acetate regulates milk fat synthesis through the mammalian target of rapamycin/eukaryotic initiation factor 4e signaling pathway in bovine mammary epithelial cells. J. Dairy Sci. 2021, 104, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Jiang, A.; Lv, X.; Zhou, C.; Tan, Z. Metabolic changes in serum and milk of holstein cows in their first to fourth parity revealed by biochemical analysis and untargeted metabolomics. Animals 2024, 14, 407. [Google Scholar] [CrossRef]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Kim, J.; Tran, T.T.H.; Hong, S.-P.; Jeong, J.-S. A reference measurement procedure for amino acids in blood using isotope dilution ultra-performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. B 2017, 1055–1056, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Bauman, C.A.; Barkema, H.W.; Dubuc, J.; Keefe, G.P.; Kelton, D.F. Canadian national dairy study: Herd-level milk quality. J. Dairy Sci. 2018, 101, 2679–2691. [Google Scholar] [CrossRef] [PubMed]
- Cabiddu, A.; Peratoner, G.; Valenti, B.; Monteils, V.; Martin, B.; Coppa, M. A quantitative review of on-farm feeding practices to enhance the quality of grassland-based ruminant dairy and meat products. Animal 2022, 16, 100375. [Google Scholar] [CrossRef] [PubMed]
- Orjales, I.; Lopez-Alonso, M.; Miranda, M.; Alaiz-Moretón, H.; Resch, C.; López, S. Dairy cow nutrition in organic farming systems. Comparison with the conventional system. Animal 2019, 13, 1084–1093. [Google Scholar] [CrossRef]
- Scarso, S.; McParland, S.; Visentin, G.; Berry, D.P.; McDermott, A.; De Marchi, M. Genetic and nongenetic factors associated with milk color in dairy cows. J. Dairy Sci. 2017, 100, 7345–7361. [Google Scholar] [CrossRef] [PubMed]
- Timlin, M.; Tobin, J.T.; Brodkorb, A.; Murphy, E.G.; Dillon, P.; Hennessy, D.; O’Donovan, M.; Pierce, K.M.; O’Callaghan, T.F. The impact of seasonality in pasture-based production systems on milk composition and functionality. Foods 2021, 10, 607. [Google Scholar] [CrossRef] [PubMed]
- Hristov, A.N.; Harper, M.; Oh, J.; Giallongo, F.; Lopes, J.C.; Cudoc, G.; Clay, J.; Ward, R.; Chase, L.E. Short communication: Variability in milk urea nitrogen and dairy total mixed ration composition in the northeastern united states. J. Dairy Sci. 2018, 101, 1579–1584. [Google Scholar] [CrossRef] [PubMed]
- Bastin, C.; Soyeurt, H.; Gengler, N. Genetic parameters of milk production traits and fatty acid contents in milk for holstein cows in parity 1–3. J. Anim. Breed. Genet. 2013, 130, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Coffey, M.P.; Hickey, J.; Brotherstone, S. Genetic aspects of growth of holstein-friesian dairy cows from birth to maturity. J. Dairy Sci. 2006, 89, 322–329. [Google Scholar] [CrossRef]
- Adrien, M.L.; Mattiauda, D.A.; Artegoitia, V.; Carriquiry, M.; Motta, G.; Bentancur, O.; Meikle, A. Nutritional regulation of body condition score at the initiation of the transition period in primiparous and multiparous dairy cows under grazing conditions: Milk production, resumption of post-partum ovarian cyclicity and metabolic parameters. Animal 2012, 6, 292–299. [Google Scholar] [CrossRef]
- Wathes, D.C.; Bourne, N.; Cheng, Z.; Mann, G.E.; Taylor, V.J.; Coffey, M.P. Multiple correlation analyses of metabolic and endocrine profiles with fertility in primiparous and multiparous cows. J. Dairy Sci. 2007, 90, 1310–1325. [Google Scholar] [CrossRef] [PubMed]
- Macmillan, K.; Gobikrushanth, M.; Helguera, I.L.; Behrouzi, A.; Colazo, M.G. Relationships between early postpartum nutritional and metabolic profiles and subsequent reproductive performance of lactating dairy cows. Theriogenology 2020, 151, 52–57. [Google Scholar] [CrossRef]
- Daneshvar, D.; Ghasemi, E.; Hashemzadeh, F.; Mahdavi, A.H.; Khorvash, M. Nutrient intake, digestibility, and serum metabolites in dairy cows fed diets differing in starch concentration with palmitic acid or stearic acid supplementation postpartum. Trop. Anim. Health Prod. 2022, 54, 284. [Google Scholar] [CrossRef] [PubMed]
- Neubert, E.; Senger-Sander, S.N.; Manzke, V.S.; Busse, J.; Polo, E.; Scheidmann, S.E.F.; Schon, M.P.; Kruss, S.; Erpenbeck, L. Serum and serum albumin inhibit in vitro formation of neutrophil extracellular traps (nets). Front. Immunol. 2019, 10, 12. [Google Scholar] [CrossRef]
- Gärtner, T.; Gernand, E.; Gottschalk, J.; Donat, K. Relationships between body condition, body condition loss, and serum metabolites during the transition period in primiparous and multiparous cows. J. Dairy Sci. 2019, 102, 9187–9199. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Mori, A.; Oda, H.; Murayama, I.; Kouno, M.; Sako, T. Comparison of cholesterol levels among lipoprotein fractions separated by anion-exchange high-performance liquid chromatography in periparturient holstein–friesian dairy cows. J. Vet. Med. Sci. 2021, 83, 260–266. [Google Scholar] [CrossRef]
- Master, P.B.Z.; Macedo, R.C.O. Effects of dietary supplementation in sport and exercise: A review of evidence on milk proteins and amino acids. Crit. Rev. Food Sci. Nutr. 2021, 61, 1225–1239. [Google Scholar] [CrossRef]
- Moyes, K.M.; Larsen, T.; Sørensen, P.; Ingvartsen, K.L. Changes in various metabolic parameters in blood and milk during experimental escherichia coli mastitis for primiparous holstein dairy cows during early lactation. J. Anim. Sci. Biotechnol. 2014, 5, 47. [Google Scholar] [CrossRef]
- Cant, J.P.; Reyes, G.C.; Seymour, D.J. Review: Influence of postabsorptive metabolism on essential amino acid partitioning in lactating dairy cows. Animal 2022, 16, 100573. [Google Scholar] [CrossRef]
- Church, D.D.; Hirsch, K.R.; Park, S.; Kim, I.-Y.; Gwin, J.A.; Pasiakos, S.M.; Wolfe, R.R.; Ferrando, A.A. Essential amino acids and protein synthesis: Insights into maximizing the muscle and whole-body response to feeding. Nutrients 2020, 12, 3717. [Google Scholar] [CrossRef]
- Park, S.; Church, D.D.; Azhar, G.; Schutzler, S.E.; Ferrando, A.A.; Wolfe, R.R. Anabolic response to essential amino acid plus whey protein composition is greater than whey protein alone in young healthy adults. J. Int. Soc. Sports Nutr. 2020, 17, 9. [Google Scholar] [CrossRef]
- Swaisgood, H.E. Protein and amino acid composition of bovine milk. In Handbook of Milk Composition; Jensen, R.G., Ed.; Academic Press: San Diego, CA, USA, 1995; pp. 464–468. [Google Scholar]
- Bochniarz, M.; Piech, T.; Kocki, T.; Iskra, M.; Krukowski, H.; Jagielski, T. Tryptophan, kynurenine and kynurenic acid concentrations in milk and serum of dairy cows with prototheca mastitis. Animals 2021, 11, 3608. [Google Scholar] [CrossRef]
- Smith, S.L.; Denholm, S.J.; Coffey, M.P.; Wall, E. Energy profiling of dairy cows from routine milk mid-infrared analysis. J. Dairy Sci. 2019, 102, 11169–11179. [Google Scholar] [CrossRef] [PubMed]
- Sadovnikova, A.; Garcia, S.C.; Hovey, R.C. A comparative review of the extrinsic and intrinsic factors regulating lactose synthesis. J. Mammary Gland Biol. Neoplasia 2021, 26, 197–215. [Google Scholar] [CrossRef]
- Rigout, S.; Lemosquet, S.; van Eys, J.E.; Blum, J.W.; Rulquin, H. Duodenal glucose increases glucose fluxes and lactose synthesis in grass silage-fed dairy cows. J. Dairy Sci. 2002, 85, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Baumgard, L.H.; Collier, R.J.; Bauman, D.E. A 100-year review: Regulation of nutrient partitioning to support lactation. J. Dairy Sci. 2017, 100, 10353–10366. [Google Scholar] [CrossRef]
- Takeuchi, N.; Higashida, K.; Li, X.; Nakai, N. Glucose enhances catecholamine-stimulated lipolysis via increased glycerol-3-phosphate synthesis in 3t3-l1 adipocytes and rat adipose tissue. Mol. Biol. Rep. 2021, 48, 6269–6276. [Google Scholar] [CrossRef]
- Miyazaki, R.; Kato, S.; Otoki, Y.; Rahmania, H.; Sakaino, M.; Takeuchi, S.; Sato, T.; Imagi, J.; Nakagawa, K. Elucidation of decomposition pathways of linoleic acid hydroperoxide isomers by gc-ms and lc-ms/ms. Biosci. Biotechnol. Biochem. 2023, 87, 179–190. [Google Scholar] [CrossRef]
- Wigger, D.; Gulbins, E.; Kleuser, B.; Schumacher, F. Monitoring the sphingolipid de novo synthesis by stable-isotope labeling and liquid chromatography-mass spectrometry. Front. Cell Dev. Biol. 2019, 7, 210. [Google Scholar] [CrossRef]
- Jung, S.; Kim, M.; Ryu, H.J.; Chae, J.S.; Lee, S.-H.; Lee, J.H. Age-related increase in ldl-cholesterol is associated with enhanced oxidative stress and disturbed sphingolipid metabolism. Metabolomics 2015, 11, 40–49. [Google Scholar] [CrossRef]

| Item 1 | Group 2 | SEM 3 | p-Value | |||
|---|---|---|---|---|---|---|
| S1 | S2 | S3 | S4 | |||
| BW (kg) | 569 | 572 | 591 | 575 | 10.26 | 0.889 |
| DMI (kg/d) | 21.88 | 21.28 | 21.79 | 21.86 | 0.30 | 0.430 |
| Milk yield (kg/d) | 29.43 | 30.56 | 31.68 | 30.85 | 2.64 | 0.937 |
| Somatic cell count (×1000 cells/mL) | 29.10 | 33.78 | 31.40 | 40.10 | 8.51 | 0.793 |
| Milk fat content (%) | 4.30 | 4.06 | 4.05 | 4.38 | 0.32 | 0.828 |
| Milk protein content (%) | 3.14 b | 3.49 a | 3.52 a | 3.13 b | 0.09 | 0.001 |
| Lactose content (%) | 5.24 a | 4.93 b | 4.99 b | 4.97 b | 0.07 | 0.009 |
| Solid not fat (%) | 9.41 a | 9.31 a | 9.28 a | 8.97 b | 0.09 | 0.011 |
| Urea nitrogen content (%) | 18.99 | 17.38 | 18.53 | 19.22 | 0.78 | 0.458 |
| Total solids (%) | 11.71 | 11.56 | 11.50 | 10.98 | 0.35 | 0.342 |
| Free amino acid content (g/100 mL) | ||||||
| EAA | 1.41 | 1.62 | 1.64 | 1.39 | 0.074 | 0.076 |
| Arginine | 0.10 | 0.09 | 0.10 | 0.08 | 0.006 | 0.135 |
| Threonine | 0.13 | 0.14 | 0.13 | 0.11 | 0.006 | 0.273 |
| Valine | 0.19 | 0.24 | 0.23 | 0.20 | 0.011 | 0.203 |
| Methionine | 0.07 a | <0.01 b | 0.05 ab | 0.04 ab | 0.009 | 0.009 |
| Isoleucine | 0.17 | 0.20 | 0.20 | 0.17 | 0.008 | 0.182 |
| Leucine | 0.29 | 0.35 | 0.34 | 0.29 | 0.015 | 0.166 |
| Phenylalanine | 0.15 | 0.18 | 0.18 | 0.15 | 0.010 | 0.087 |
| Lysine | 0.23 | 0.33 | 0.30 | 0.26 | 0.016 | 0.093 |
| Histidine | 0.09 | 0.10 | 0.10 | 0.08 | 0.004 | 0.101 |
| NEAA | 1.65 | 1.59 | 1.61 | 1.35 | 0.091 | 0.077 |
| Aspartate | 0.22 | 0.25 | 0.25 | 0.21 | 0.012 | 0.144 |
| Serine | 0.14 | 0.15 | 0.13 | 0.11 | 0.009 | 0.428 |
| Glutamate | 0.51 | 0.65 | 0.63 | 0.53 | 0.037 | 0.265 |
| Glycine | 0.06 | 0.06 | 0.06 | 0.06 | 0.003 | 0.088 |
| Alanine | 0.10 | 0.12 | 0.11 | 0.10 | 0.005 | 0.126 |
| Tyrosine | 0.09 a | 0.03 b | 0.08 a | 0.06 ab | 0.013 | 0.003 |
| Proline | 0.54 a | 0.34 b | 0.34 b | 0.29 b | 0.044 | 0.001 |
| TAA | 3.06 | 3.21 | 3.25 | 2.74 | 0.158 | 0.085 |
| EAA/TAA | 0.46 b | 0.51 a | 0.51 a | 0.51 a | 0.008 | 0.019 |
| Item 1 | Group 2 | SEM 3 | p-Value | |||
|---|---|---|---|---|---|---|
| S1 | S2 | S3 | S4 | |||
| Total protein (g/L) | 76.94 b | 83.24 a | 84.45 a | 81.15 ab | 1.79 | 0.017 |
| Albumin (g/L) | 42.11 | 42.87 | 41.56 | 40.24 | 1.06 | 0.337 |
| Glucose (mM) | 3.61 b | 3.88 a | 3.88 a | 3.84 a | 0.07 | 0.017 |
| Blood urea nitrogen (mM) | 6.54 | 6.01 | 6.15 | 6.74 | 0.38 | 0.477 |
| Triglyceride (mM) | 0.13 b | 0.16 a | 0.16 a | 0.13 b | 0.01 | 0.010 |
| Cholesterol (mM) | 5.95 | 5.71 | 5.77 | 5.02 | 0.31 | 0.154 |
| Low-density lipoprotein (mM) | 3.00 | 2.96 | 2.96 | 2.49 | 0.35 | 0.399 |
| High-density lipoprotein (mM) | 4.38 a | 4.26 a | 4.16 ab | 3.58 b | 0.22 | 0.047 |
| Serum ammonia (μM) | 319.17 a | 250.62 b | 268.43 b | 242.08 b | 13.30 | 0.002 |
| Lipase (U/L) | 17.85 | 19.31 | 19.75 | 19.59 | 0.98 | 0.608 |
| Free amino acid content (μg/mL) | ||||||
| EAA | 151.74 | 157.67 | 148.90 | 143.14 | 0.15 | 0.324 |
| Threonine | 34.82 | 32.49 | 30.78 | 30.33 | 2.46 | 0.534 |
| Valine | 26.31 | 28.71 | 27.77 | 25.54 | 0.43 | 0.343 |
| Methionine | 5.13 a | 4.72 ab | 4.45 ab | 3.45 b | 1.22 | 0.024 |
| Isoleucine | 13.13 | 14.06 | 12.55 | 12.38 | 1.01 | 0.180 |
| Leucine | 16.40 | 18.75 | 16.58 | 15.65 | 1.08 | 0.126 |
| Phenylalanine | 8.57 | 8.66 | 7.40 | 7.96 | 0.32 | 0.115 |
| Lysine | 14.44 | 15.28 | 14.81 | 14.43 | 1.23 | 0.844 |
| Histidine | 9.24 | 10.40 | 10.18 | 9.27 | 0.39 | 0.120 |
| NEAA | 106.75 | 99.94 | 107.15 | 93.76 | 0.60 | 0.053 |
| Aspartate | 2.20 a | 1.56 b | 1.93 ab | 1.74 ab | 0.90 | 0.034 |
| Serine | 9.27 | 9.08 | 9.33 | 8.86 | 0.53 | 0.887 |
| Glutamate | 26.17 | 24.03 | 25.66 | 23.40 | 0.38 | 0.482 |
| Glycine | 27.87 a | 24.04 ab | 25.94 b | 22.81 ab | 0.78 | 0.006 |
| Alanine | 23.05 a | 18.98 ab | 21.41 ab | 18.56 b | 0.42 | 0.034 |
| Cysteine | 2.95 | 3.35 | 3.27 | 2.07 | 0.95 | 0.081 |
| Tyrosine | 8.79 | 8.42 | 7.60 | 7.58 | 1.10 | 0.249 |
| Arginine | 23.71 | 24.59 | 24.37 | 24.13 | 8.22 | 0.872 |
| Proline | 6.46 b | 10.48 ab | 12.01 a | 8.74 ab | 5.66 | 0.014 |
| TAA | 258.50 | 257.61 | 256.05 | 236.90 | 3.64 | 0.372 |
| EAA/TAA | 0.59 | 0.61 | 0.58 | 0.60 | 0.01 | 0.057 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Jiang, A.; Lv, X.; Fan, D.; Chen, Q.; Wu, Y.; Zhou, C.; Tan, Z. Combined Metabolomics and Biochemical Analyses of Serum and Milk Revealed Parity-Related Metabolic Differences in Sanhe Dairy Cattle. Metabolites 2024, 14, 227. https://doi.org/10.3390/metabo14040227
Liu Z, Jiang A, Lv X, Fan D, Chen Q, Wu Y, Zhou C, Tan Z. Combined Metabolomics and Biochemical Analyses of Serum and Milk Revealed Parity-Related Metabolic Differences in Sanhe Dairy Cattle. Metabolites. 2024; 14(4):227. https://doi.org/10.3390/metabo14040227
Chicago/Turabian StyleLiu, Zixin, Aoyu Jiang, Xiaokang Lv, Dingkun Fan, Qingqing Chen, Yicheng Wu, Chuanshe Zhou, and Zhiliang Tan. 2024. "Combined Metabolomics and Biochemical Analyses of Serum and Milk Revealed Parity-Related Metabolic Differences in Sanhe Dairy Cattle" Metabolites 14, no. 4: 227. https://doi.org/10.3390/metabo14040227
APA StyleLiu, Z., Jiang, A., Lv, X., Fan, D., Chen, Q., Wu, Y., Zhou, C., & Tan, Z. (2024). Combined Metabolomics and Biochemical Analyses of Serum and Milk Revealed Parity-Related Metabolic Differences in Sanhe Dairy Cattle. Metabolites, 14(4), 227. https://doi.org/10.3390/metabo14040227






