Machine Learning Metabolomics Profiling of Dietary Interventions from a Six-Week Randomised Trial
Abstract
1. Introduction
2. Materials and Methods
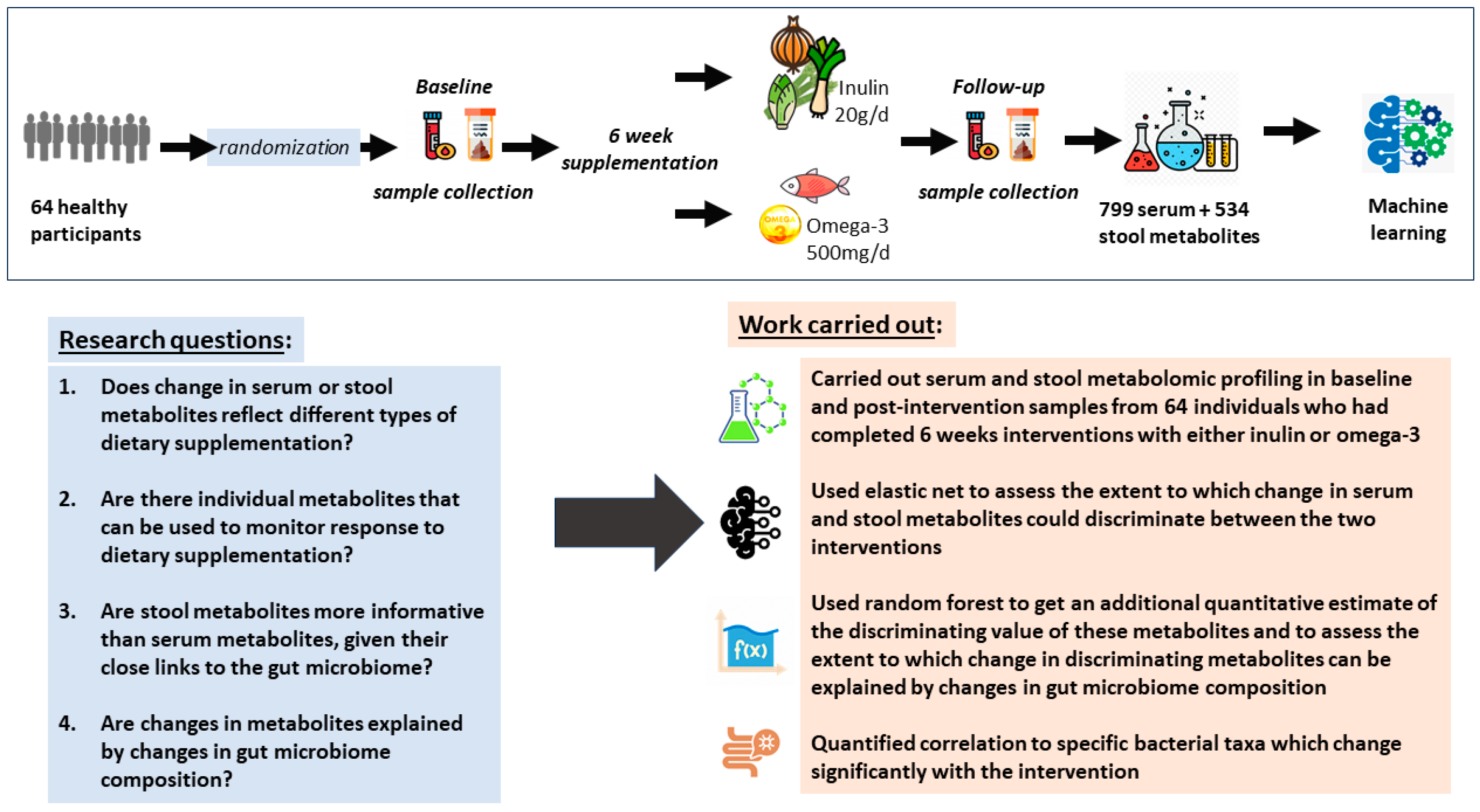
2.1. Study Aim and Design
2.2. Study Population
2.3. Sample Collection
2.4. Metabolite Profiling
2.5. Microbiota Analysis
2.6. Data Analysis
2.6.1. Metabolite QC
2.6.2. Elastic Net for Identifying Changes in Metabolites Reflecting Supplementation with Inulin or Omega-3
2.6.3. Sensitivity Analyses and Cross-Validation Using Random Forest and Logistic Regression
2.6.4. Associations of Differentiating Metabolites with SCFAs and Gut Microbiome Composition
3. Results
3.1. Descriptive Characteristics of the Participants
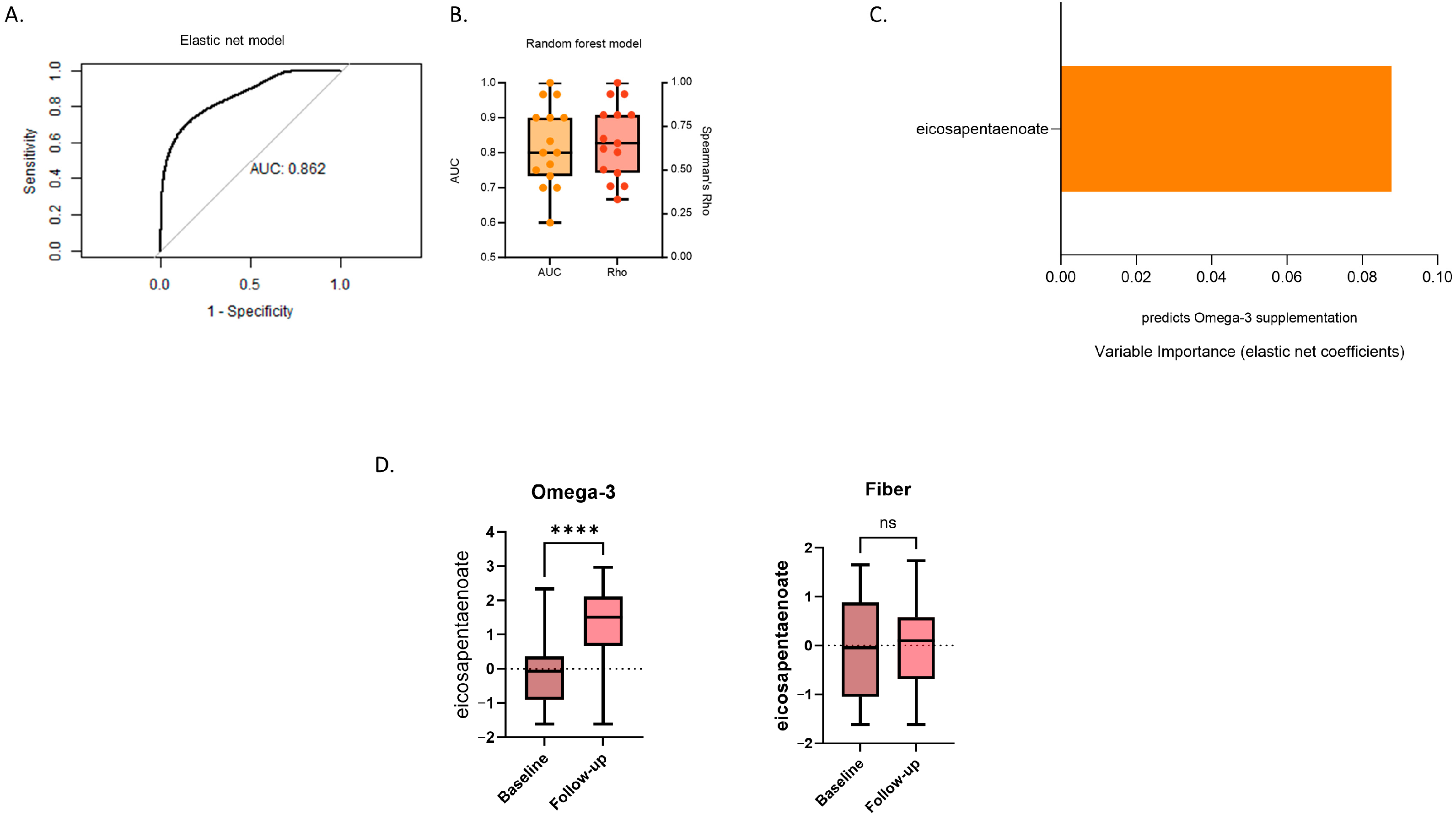
3.2. Changes in Serum Metabolites Differentiating between Inulin and Omega-3 Supplementation
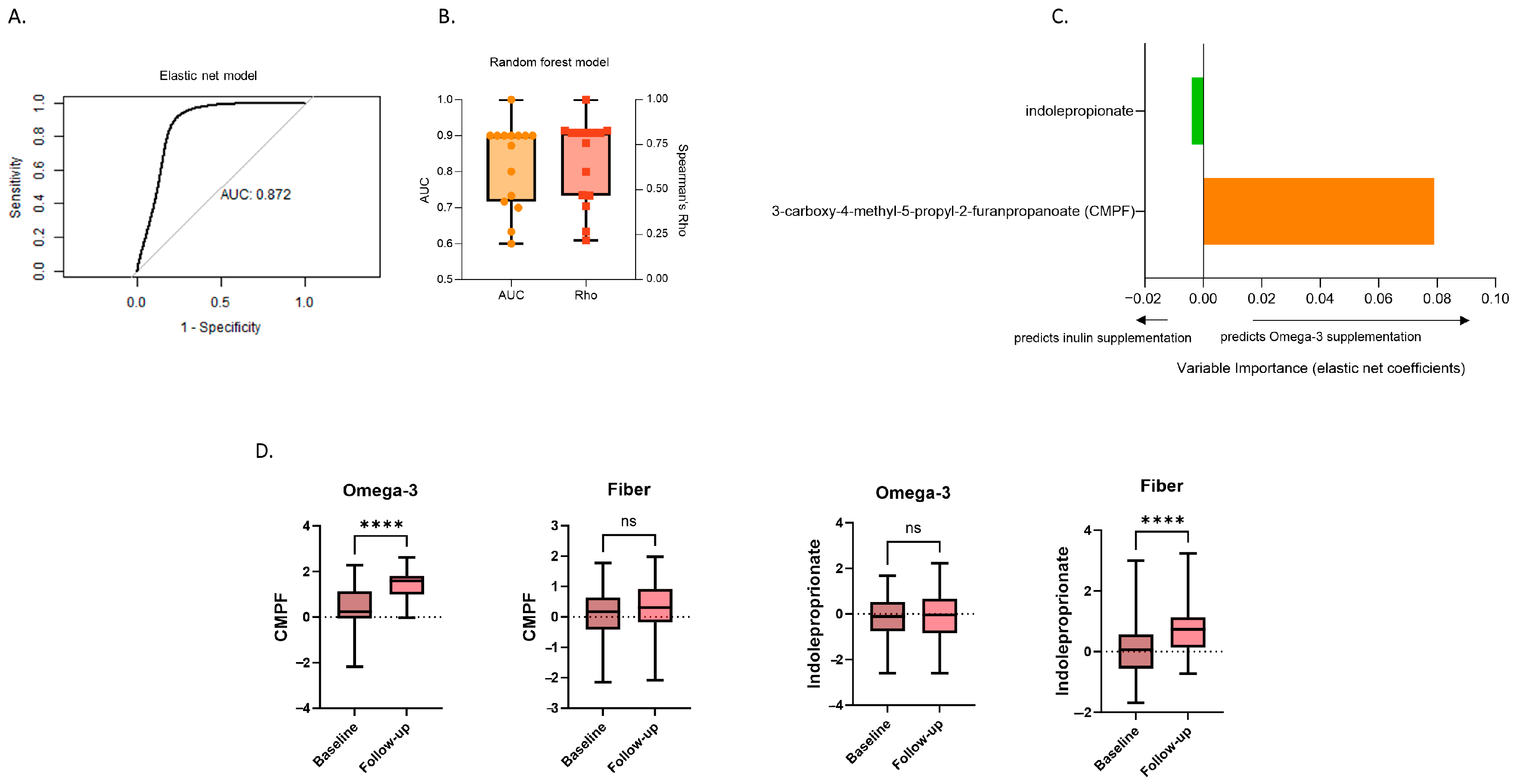
3.3. Changes in Stool Metabolites Differentiating between Inulin and Omega-3 Supplementation
3.4. Associations of Differentiating Metabolites with SCFAs and Gut Microbiome Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, F.F.; Barr, S.I.; McNulty, H.; Li, D.; Blumberg, J.B. Health effects of vitamin and mineral supplements. BMJ 2020, 369, m2511. [Google Scholar] [CrossRef] [PubMed]
- Stumpf, F.; Keller, B.; Gressies, C.; Schuetz, P. Inflammation and Nutrition: Friend or Foe? Nutrients 2023, 15, 1159. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Pathak, S.; Patil, K. Antioxidant Nutraceuticals: Preventive and Healthcare Applications; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Chaudhary, N.; Kumar, V.; Sangwan, P.; Pant, N.C.; Saxena, A.; Joshi, S.; Yadav, A.N. 3.36-Personalized Nutrition and -Omics. In Comprehensive Foodomics; Cifuentes, A., Ed.; Elsevier: Oxford, UK, 2021; pp. 495–507. [Google Scholar]
- Chilton, F.H.; Dutta, R.; Reynolds, L.M.; Sergeant, S.; Mathias, R.A.; Seeds, M.C. Precision Nutrition and Omega-3 Polyunsaturated Fatty Acids: A Case for Personalized Supplementation Approaches for the Prevention and Management of Human Diseases. Nutrients 2017, 9, 1165. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Xie, B.; Zhang, H.; He, Q.; Guo, L.; Subramanieapillai, M.; Fan, B.; Lu, C.; McIntyre, R.S. Efficacy of omega-3 PUFAs in depression: A meta-analysis. Transl. Psychiatry 2019, 9, 190. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yu, X.; Shao, S. Effects of Omega-3 Fatty Acid Supplementation on Glucose Control and Lipid Levels in Type 2 Diabetes: A Meta-Analysis. PLoS ONE 2015, 10, e0139565. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N. Pro-resolving lipid mediators are leads for resolution physiology. Nature 2014, 510, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.L.; Alvarado, D.A.; Swanson, K.S.; Holscher, H.D. The Prebiotic Potential of Inulin-Type Fructans: A Systematic Review. Adv. Nutr. 2022, 13, 492–529. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.-Q.; Wang, L.-Y.; Yang, X.-Y.; Xu, Y.-J.; Fan, G.; Fan, Y.-G.; Ren, J.-N.; An, Q.; Li, X. Inulin: Properties and health benefits. Food Funct. 2023, 14, 2948–2968. [Google Scholar] [CrossRef]
- Ďásková, N.; Modos, I.; Krbcová, M.; Kuzma, M.; Pelantová, H.; Hradecký, J.; Heczková, M.; Bratová, M.; Videňská, P.; Šplíchalová, P.; et al. Multi-omics signatures in new-onset diabetes predict metabolic response to dietary inulin: Findings from an observational study followed by an interventional trial. Nutr. Diabetes 2023, 13, 7. [Google Scholar] [CrossRef]
- Cai, X.; Yu, H.; Liu, L.; Lu, T.; Li, J.; Ji, Y.; Le, Z.; Bao, L.; Ma, W.; Xiao, R.; et al. Milk Powder Co-Supplemented with Inulin and Resistant Dextrin Improves Glycemic Control and Insulin Resistance in Elderly Type 2 Diabetes Mellitus: A 12-Week Randomized, Double-Blind, Placebo-Controlled Trial. Mol. Nutr. Food Res. 2018, 62, e1800865. [Google Scholar] [CrossRef]
- Xiong, Q.; Li, L.; Xiao, Y.; He, S.; Zhao, J.; Lin, X.; He, Y.; Wang, J.; Guo, X.; Liang, W.; et al. The Effect of Inulin-Type Fructans on Plasma Trimethylamine N-Oxide Levels in Peritoneal Dialysis Patients: A Randomized Crossover Trial. Mol. Nutr. Food Res. 2023, 67, e2200531. [Google Scholar] [CrossRef] [PubMed]
- Vijay, A.; Astbury, S.; Le Roy, C.; Spector, T.D.; Valdes, A.M. The prebiotic effects of omega-3 fatty acid supplementation: A six-week randomised intervention trial. Gut Microbes 2021, 13, 1863133. [Google Scholar] [CrossRef] [PubMed]
- Moayyeri, A.; Hammond, C.J.; Valdes, A.M.; Spector, T.D. Cohort Profile: TwinsUK and healthy ageing twin study. Int. J. Epidemiol. 2013, 42, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.M.; DeHaven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef] [PubMed]
- Nogal, A.; Tettamanzi, F.; Dong, Q.; Louca, P.; Visconti, A.; Christiansen, C.; Breuninger, T.; Linseisen, J.; Grallert, H.; Wawro, N.; et al. A Fecal Metabolite Signature of Impaired Fasting Glucose: Results From Two Independent Population-Based Cohorts. Diabetes 2023, 72, 1870–1880. [Google Scholar] [CrossRef] [PubMed]
- Attaye, I.; Beynon-Cobb, B.; Louca, P.; Nogal, A.; Visconti, A.; Tettamanzi, F.; Wong, K.; Michellotti, G.; Spector, T.D.; Falchi, M.; et al. Cross-sectional analyses of metabolites across biological samples mediating dietary acid load and chronic kidney disease. iScience 2024, 27, 109132. [Google Scholar] [CrossRef] [PubMed]
- Louca, P.; Nogal, A.; Moskal, A.; Goulding, N.J.; Shipley, M.J.; Alkis, T.; Lindbohm, J.V.; Hu, J.; Kifer, D.; Wang, N.; et al. Cross-Sectional Blood Metabolite Markers of Hypertension: A Multicohort Analysis of 44,306 Individuals from the COnsortium of METabolomics Studies. Metabolites 2022, 12, 601. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Davenport, E.R.; Beaumont, M.; Jackson, M.A.; Knight, R.; Ober, C.; Spector, T.D.; Bell, J.T.; Clark, A.G.; Ley, R.E. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016, 19, 731–743. [Google Scholar] [CrossRef]
- Boers, S.A.; Hiltemann, S.D.; Stubbs, A.P.; Jansen, R.; Hays, J.P. Development and evaluation of a culture-free microbiota profiling platform (MYcrobiota) for clinical diagnostics. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1081–1089. [Google Scholar] [CrossRef]
- Deng, K.; Xu, J.-J.; Shen, L.; Zhao, H.; Gou, W.; Xu, F.; Fu, Y.; Jiang, Z.; Shuai, M.; Li, B.-Y.; et al. Comparison of fecal and blood metabolome reveals inconsistent associations of the gut microbiota with cardiometabolic diseases. Nat. Commun. 2023, 14, 571. [Google Scholar] [CrossRef] [PubMed]
- Visconti, A.; Le Roy, C.I.; Rosa, F.; Rossi, N.; Martin, T.C.; Mohney, R.P.; Li, W.; de Rinaldis, E.; Bell, J.T.; Venter, J.C.; et al. Interplay between the human gut microbiome and host metabolism. Nat. Commun. 2019, 10, 4505. [Google Scholar] [CrossRef]
- Zierer, J.; Jackson, M.A.; Kastenmüller, G.; Mangino, M.; Long, T.; Telenti, A.; Mohney, R.P.; Small, K.S.; Bell, J.T.; Steves, C.J.; et al. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 2018, 50, 790–795. [Google Scholar] [CrossRef]
- de Mello, V.D.; Paananen, J.; Lindström, J.; Lankinen, M.A.; Shi, L.; Kuusisto, J.; Pihlajamäki, J.; Auriola, S.; Lehtonen, M.; Rolandsson, O.; et al. Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish Diabetes Prevention Study. Sci. Rep. 2017, 7, 46337. [Google Scholar] [CrossRef]
- Menni, C.; Hernandez, M.M.; Vital, M.; Mohney, R.P.; Spector, T.D.; Valdes, A.M. Circulating levels of the anti-oxidant indoleproprionic acid are associated with higher gut microbiome diversity. Gut Microbes 2019, 10, 688–695. [Google Scholar] [CrossRef]
- Tuomainen, M.; Lindström, J.; Lehtonen, M.; Auriola, S.; Pihlajamäki, J.; Peltonen, M.; Tuomilehto, J.; Uusitupa, M.; de Mello, V.D.; Hanhineva, K. Associations of serum indolepropionic acid, a gut microbiota metabolite, with type 2 diabetes and low-grade inflammation in high-risk individuals. Nutr. Diabetes 2018, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Qi, Q.; Li, J.; Yu, B.; Moon, J.-Y.; Chai, J.C.; Merino, J.; Hu, J.; Ruiz-Canela, M.; Rebholz, C.; Wang, Z.; et al. Host and gut microbial tryptophan metabolism and type 2 diabetes: An integrative analysis of host genetics, diet, gut microbiome and circulating metabolites in cohort studies. Gut 2022, 71, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Pan, T.; Xiao, Y.; Wang, H.; Zhu, J.; Tian, F.; Lu, W.; Chen, W. The therapeutic potential of dietary intervention: Based on the mechanism of a tryptophan derivative-indole propionic acid on metabolic disorders. Crit. Rev. Food Sci. Nutr. 2024, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, R.; Gupta, H.; Jeong, J.-J.; Sharma, S.P.; Won, S.-M.; Oh, K.-K.; Yoon, S.J.; Han, S.H.; Yang, Y.J.; Baik, G.H.; et al. Characteristics of microbiome-derived metabolomics according to the progression of alcoholic liver disease. Hepatol. Int. 2023, 18, 486–499. [Google Scholar] [CrossRef]
- Cason, C.A.; Dolan, K.T.; Sharma, G.; Tao, M.; Kulkarni, R.; Helenowski, I.B.; Doane, B.M.; Avram, M.J.; McDermott, M.M.; Chang, E.B.; et al. Plasma microbiome-modulated indole- and phenyl-derived metabolites associate with advanced atherosclerosis and postoperative outcomes. J. Vasc. Surg. 2018, 68, 1552–1562.e7. [Google Scholar] [CrossRef]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Prentice, K.J.; Wendell, S.G.; Liu, Y.; Eversley, J.A.; Salvatore, S.R.; Mohan, H.; Brandt, S.L.; Adams, A.C.; Wang, X.S.; Wei, D.; et al. CMPF, a Metabolite Formed Upon Prescription Omega-3-Acid Ethyl Ester Supplementation, Prevents and Reverses Steatosis. EBioMedicine 2018, 27, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.-S.; Lin, M.; Imamura, F.; Cai, W.; Wang, L.; Feng, J.-P.; Ruan, Y.; Tang, J.; Wang, F.; Yang, H.; et al. Serum metabolomics profiles in response to n-3 fatty acids in Chinese patients with type 2 diabetes: A double-blind randomised controlled trial. Sci. Rep. 2016, 6, 29522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, P.; Jin, H.; Yi, J.; Xie, X.; Yang, M.; Gao, T.; Yang, L.; Hu, C.; Zhang, X.; et al. Circulating 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid (CMPF) levels are associated with hyperglycemia and β cell dysfunction in a Chinese population. Sci. Rep. 2017, 7, 3114. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Xiong, Q.; Huang, D.; Wu, Z.; Chen, Z. Causal relationship between blood metabolites and risk of five infections: A Mendelian randomization study. BMC Infect. Dis. 2023, 23, 663. [Google Scholar] [CrossRef] [PubMed]
- Brustad, N.; Olarini, A.; Kim, M.; Chen, L.; Ali, M.; Wang, T.; Cohen, A.S.; Ernst, M.; Hougaard, D.; Schoos, A.-M.; et al. Diet-associated vertically transferred metabolites and risk of asthma, allergy, eczema, and infections in early childhood. Pediatr. Allergy Immunol. 2023, 34, e13917. [Google Scholar] [CrossRef] [PubMed]
- Dyall, S.C. Long-chain omega-3 fatty acids and the brain: A review of the independent and shared effects of EPA, DPA and DHA. Front. Aging Neurosci. 2015, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Koudoufio, M.; Desjardins, Y.; Feldman, F.; Spahis, S.; Delvin, E.; Levy, E. Insight into Polyphenol and Gut Microbiota Crosstalk: Are Their Metabolites the Key to Understand Protective Effects against Metabolic Disorders? Antioxidants 2020, 9, 982. [Google Scholar] [CrossRef] [PubMed]
- Nogal, A.; Valdes, A.M.; Menni, C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 2021, 13, 1897212. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kouraki, A.; Nogal, A.; Nocun, W.; Louca, P.; Vijay, A.; Wong, K.; Michelotti, G.A.; Menni, C.; Valdes, A.M. Machine Learning Metabolomics Profiling of Dietary Interventions from a Six-Week Randomised Trial. Metabolites 2024, 14, 311. https://doi.org/10.3390/metabo14060311
Kouraki A, Nogal A, Nocun W, Louca P, Vijay A, Wong K, Michelotti GA, Menni C, Valdes AM. Machine Learning Metabolomics Profiling of Dietary Interventions from a Six-Week Randomised Trial. Metabolites. 2024; 14(6):311. https://doi.org/10.3390/metabo14060311
Chicago/Turabian StyleKouraki, Afroditi, Ana Nogal, Weronika Nocun, Panayiotis Louca, Amrita Vijay, Kari Wong, Gregory A. Michelotti, Cristina Menni, and Ana M. Valdes. 2024. "Machine Learning Metabolomics Profiling of Dietary Interventions from a Six-Week Randomised Trial" Metabolites 14, no. 6: 311. https://doi.org/10.3390/metabo14060311
APA StyleKouraki, A., Nogal, A., Nocun, W., Louca, P., Vijay, A., Wong, K., Michelotti, G. A., Menni, C., & Valdes, A. M. (2024). Machine Learning Metabolomics Profiling of Dietary Interventions from a Six-Week Randomised Trial. Metabolites, 14(6), 311. https://doi.org/10.3390/metabo14060311






