Excess Branched-Chain Amino Acids Suppress Mitochondrial Function and Biogenic Signaling but Not Mitochondrial Dynamics in a Myotube Model of Skeletal Muscle Insulin Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
2.3. Immunoblotting
2.4. Seahorse Metabolic Assays
2.5. Fluorescent Staining and Microscopy
2.6. Statistical Analyses
3. Results
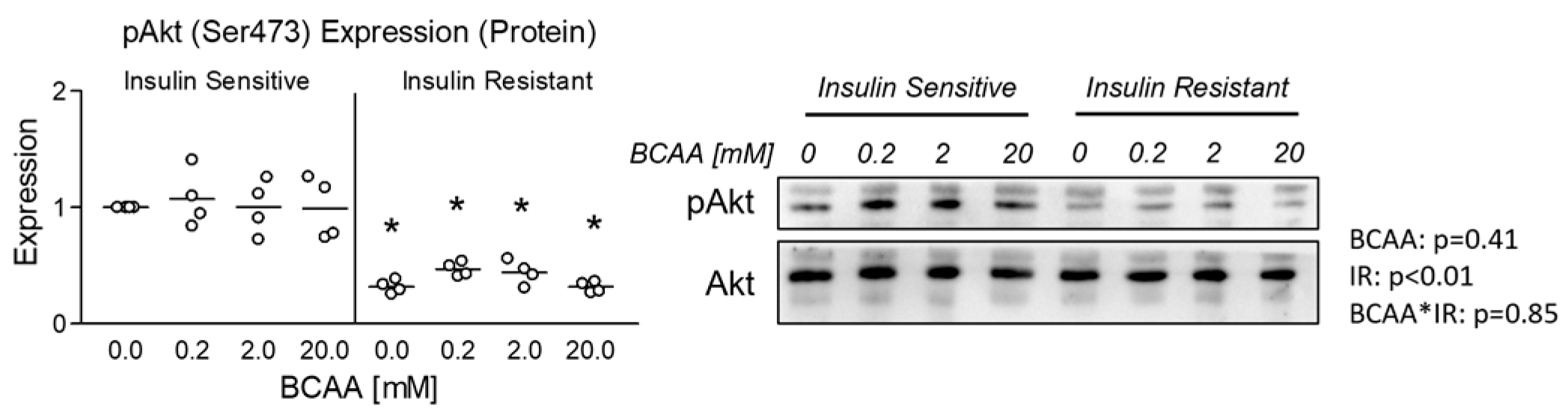
3.1. Effect of BCAA Treatment on Insulin Sensitivity
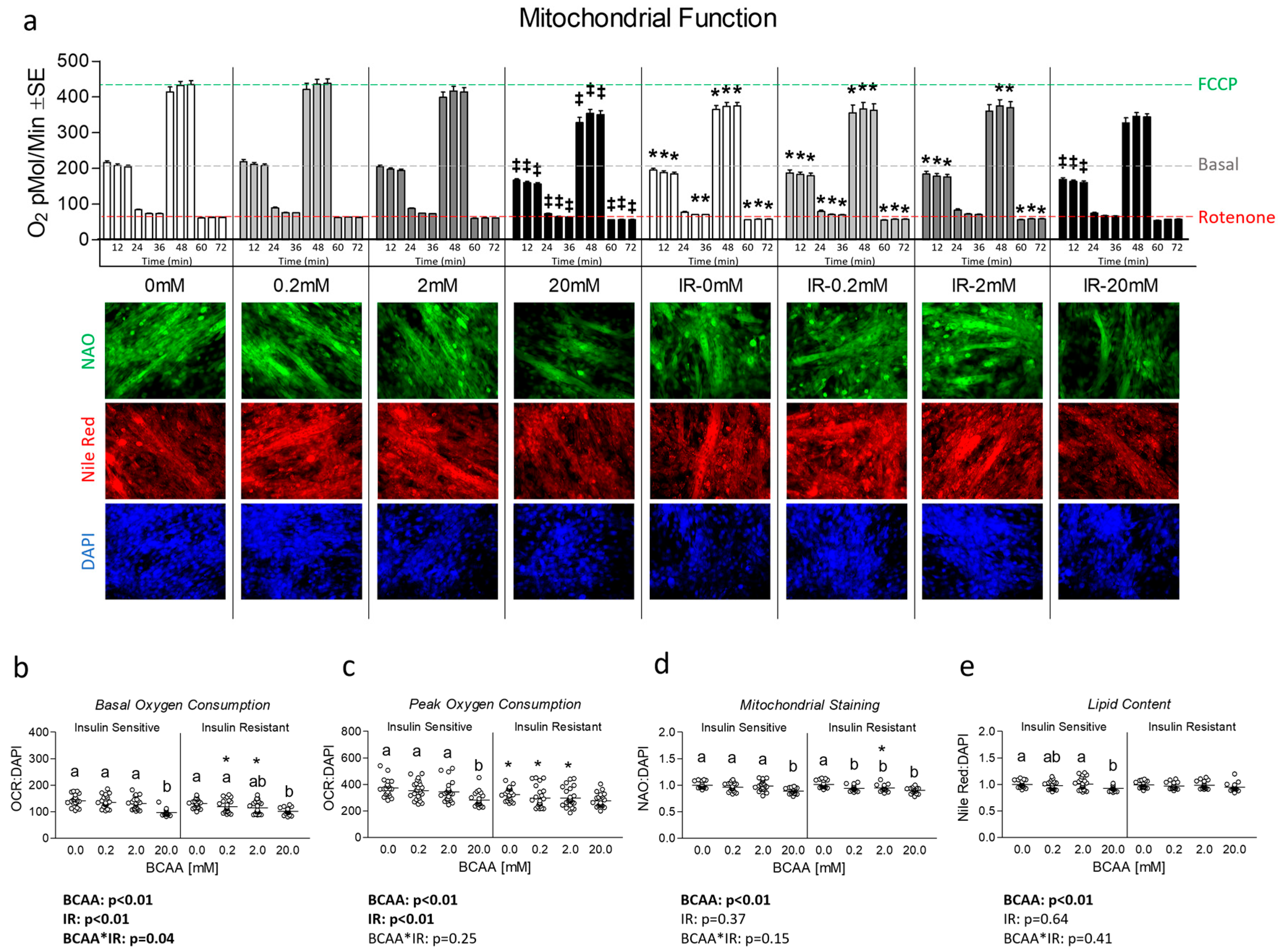
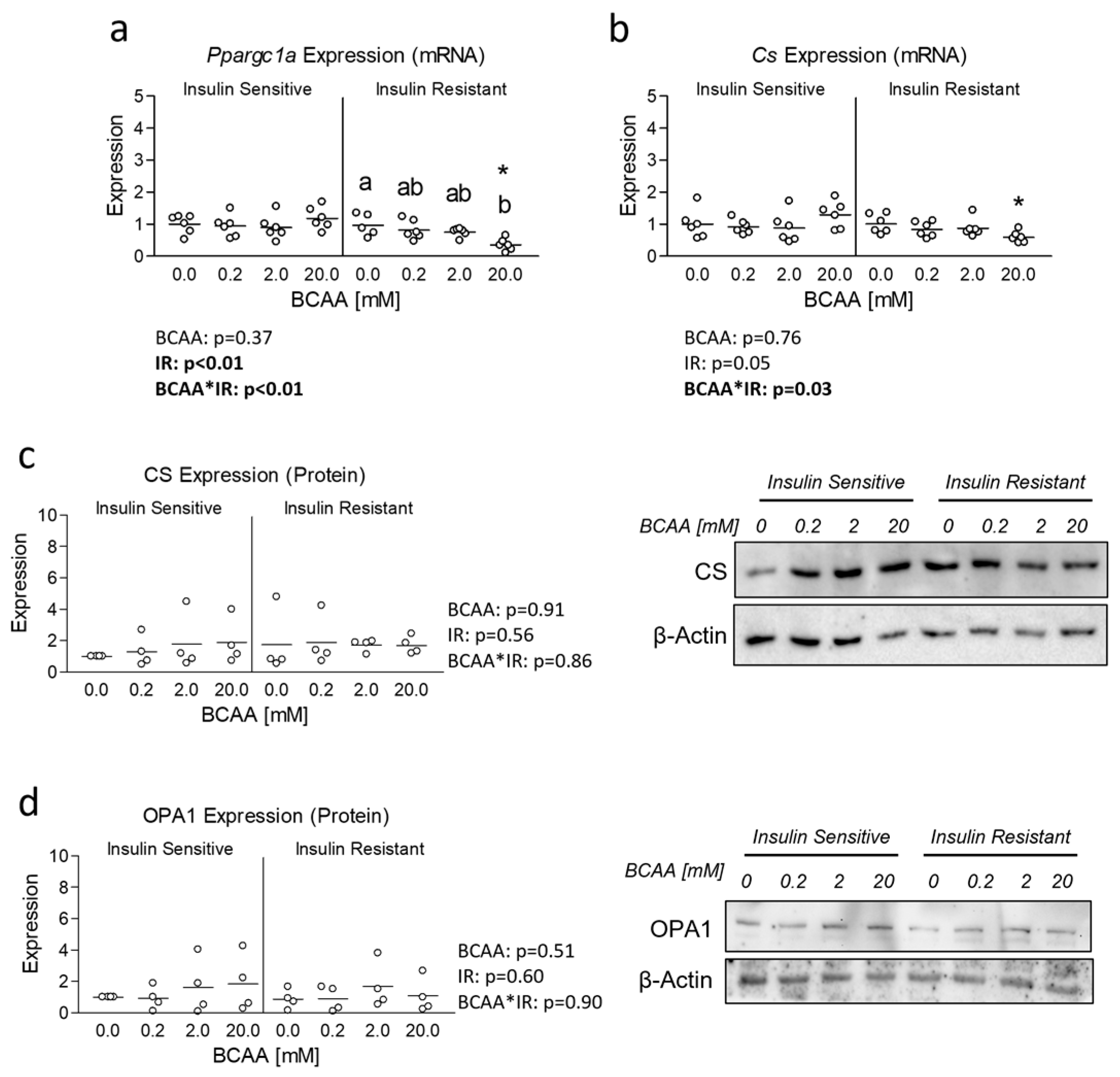
3.2. Effect of BCAA Treatment on Mitochondrial Function and Content
3.3. Effect of BCAA Treatment on Mitochondrial Dynamics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gannon, N.P.; Schnuck, J.K.; Vaughan, R.A. BCAA Metabolism and Insulin Sensitivity—Dysregulated by Metabolic Status? Mol. Nutr. Food Res. 2018, 62, 1700756. [Google Scholar] [CrossRef] [PubMed]
- Holeček, M. Why Are Branched-Chain Amino Acids Increased in Starvation and Diabetes? Nutrients 2020, 12, 3087. [Google Scholar] [CrossRef] [PubMed]
- Lynch, C.J.; Adams, S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 2014, 10, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Newgard, C.B. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012, 15, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Gannon, N.P.; Vaughan, R.A. Leucine-induced anabolic-catabolism: Two sides of the same coin. Amino Acids 2016, 48, 321–336. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, F.; Guo, Q.; Duan, Y.; Wang, W.; Zhong, Y.; Yang, Y.; Yin, Y. Leucine Supplementation: A Novel Strategy for Modulating Lipid Metabolism and Energy Homeostasis. Nutrients 2020, 12, 1299. [Google Scholar] [CrossRef] [PubMed]
- Ruocco, C.; Segala, A.; Valerio, A.; Nisoli, E. Essential amino acid formulations to prevent mitochondrial dysfunction and oxidative stress. Curr. Opin. Clin. Nutr. Metab. Care 2021, 24, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Hinkle, J.S.; Rivera, C.N.; Vaughan, R.A. Branched-Chain Amino Acids and Mitochondrial Biogenesis: An Overview and Mechanistic Summary. Mol. Nutr. Food Res. 2022, 66, e2200109. [Google Scholar] [CrossRef] [PubMed]
- Rivera, M.E.; Rivera, C.N.; Vaughan, R.A. Excess branched-chain amino acids alter myotube metabolism and substrate preference which is worsened by concurrent insulin resistance. Endocrine 2021, 76, 18–28. [Google Scholar] [CrossRef]
- Van Huynh, T.; Rethi, L.; Chen, C.H.; Chen, Y.J.; Kao, Y.H. The Complex Interplay between Imbalanced Mitochondrial Dynamics and Metabolic Disorders in Type 2 Diabetes. Cells 2023, 12, 1223. [Google Scholar] [CrossRef]
- Axelrod, C.L.; Fealy, C.E.; Erickson, M.L.; Davuluri, G.; Fujioka, H.; Dantas, W.S.; Huang, E.; Pergola, K.; Mey, J.T.; King, W.T.; et al. Lipids activate skeletal muscle mitochondrial fission and quality control networks to induce insulin resistance in humans. Metabolism 2021, 121, 154803. [Google Scholar] [CrossRef] [PubMed]
- Kugler, B.A.; Deng, W.; Duguay, A.L.; Garcia, J.P.; Anderson, M.C.; Nguyen, P.D.; Houmard, J.A.; Zou, K. Pharmacological inhibition of dynamin-related protein 1 attenuates skeletal muscle insulin resistance in obesity. Physiol. Rep. 2021, 9, e14808. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.O.; Zeng, R.X.; Margerum, C.L.; Papadopoli, D.; Monnin, C.; Punter, K.B.; Chu, C.; Al-Rofaidi, M.; Al-Tannak, N.F.; Berardi, D.; et al. Mitochondrial hyperfusion via metabolic sensing of regulatory amino acids. Cell Rep. 2022, 40, 111198. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Dridi, S.; Huang, Y.; Baum, J.I. Leucine decreases intramyocellular lipid deposition in an mTORC1-independent manner in palmitate-treated C2C12 myotubes. Am. J. Physiol. Endocrinol. Metab. 2020, 318, E152–E163. [Google Scholar] [CrossRef] [PubMed]
- Alves, P.K.N.; Schauer, A.; Augstein, A.; Prieto Jarabo, M.E.; Männel, A.; Barthel, P.; Vahle, B.; Moriscot, A.S.; Linke, A.; Adams, V. Leucine Supplementation Prevents the Development of Skeletal Muscle Dysfunction in a Rat Model of HFpEF. Cells 2024, 13, 502. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, Y.; Tamura, Y.; Takahashi, K.; Kitaoka, Y.; Takahashi, Y.; Hoshino, D.; Kadoguchi, T.; Hatta, H. Branched-chain amino acid supplementation suppresses the detraining-induced reduction of mitochondrial content in mouse skeletal muscle. FASEB J. 2022, 36, e22628. [Google Scholar] [CrossRef] [PubMed]
- Genders, A.J.; Holloway, G.P.; Bishop, D.J. Are Alterations in Skeletal Muscle Mitochondria a Cause or Consequence of Insulin Resistance? Int. J. Mol. Sci. 2020, 21, 6948. [Google Scholar] [CrossRef] [PubMed]
- Di Meo, S.; Iossa, S.; Venditti, P. Skeletal muscle insulin resistance: Role of mitochondria and other ROS sources. J. Endocrinol. 2017, 233, R15–R42. [Google Scholar] [CrossRef] [PubMed]
- Halling, J.F.; Pilegaard, H. PGC-1α-mediated regulation of mitochondrial function and physiological implications. Appl. Physiol. Nutr. Metab. 2020, 45, 927–936. [Google Scholar] [CrossRef]
- Adams, S.H. Emerging perspectives on essential amino acid metabolism in obesity and the insulin-resistant state. Adv. Nutr. 2011, 2, 445–456. [Google Scholar] [CrossRef]
- Lackey, D.E.; Lynch, C.J.; Olson, K.C.; Mostaedi, R.; Ali, M.; Smith, W.H.; Karpe, F.; Humphreys, S.; Bedinger, D.H.; Dunn, T.N.; et al. Regulation of adipose branched-chain amino acid catabolism enzyme expression and cross-adipose amino acid flux in human obesity. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E1175–E1187. [Google Scholar] [CrossRef] [PubMed]
- White, P.J.; McGarrah, R.W.; Herman, M.A.; Bain, J.R.; Shah, S.H.; Newgard, C.B. Insulin action, type 2 diabetes, and branched-chain amino acids: A two-way street. Mol. Metab. 2021, 52, 101261. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.A.; Gannon, N.P.; Schnuck, J.K.; Lyon, E.S.; Sunderland, K.L.; Vaughan, R.A. Leucine, Palmitate, or Leucine/Palmitate Cotreatment Enhances Myotube Lipid Content and Oxidative Preference. Lipids 2018, 53, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Rivera, M.E.; Lyon, E.S.; Johnson, M.A.; Vaughan, R.A. Leucine increases mitochondrial metabolism and lipid content without altering insulin signaling in myotubes. Biochimie 2020, 168, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Schnuck, J.K.; Sunderland, K.L.; Gannon, N.P.; Kuennen, M.R.; Vaughan, R.A. Leucine stimulates PPARbeta/delta-dependent mitochondrial biogenesis and oxidative metabolism with enhanced GLUT4 content and glucose uptake in myotubes. Biochimie 2016, 128–129, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Curry, B.J.; Brown, P.L.; Zemel, M.B. Leucine Modulates Mitochondrial Biogenesis and SIRT1-AMPK Signaling in C2C12 Myotubes. J. Nutr. Metab. 2014, 2014, 239750. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Zeng, L.; Deng, J.; Duan, Y.; Li, F. β-hydroxy-β-methylbutyrate (HMB) improves mitochondrial function in myocytes through pathways involving PPARβ/δ and CDK4. Nutrition 2018, 60, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Stancliffe, R.A. Role of Beta-Hydroxy-Beta-Methylbutyrate (HMB) in Leucine Stimulation of Mitochondrial Biogenesis and Fatty Acid Oxidation. Master’s Thesis, University of Tennessee, Knoxville, TN, USA, 2012. [Google Scholar]
- Sato, Y.; Obeng, K.A.; Yoshizawa, F. Acute oral administration of L-leucine upregulates slow-fiber- and mitochondria-related genes in skeletal muscle of rats. Nutr. Res. 2018, 57, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xiang, L.; Jia, G.; Liu, G.; Zhao, H.; Huang, Z. Leucine regulates slow-twitch muscle fibers expression and mitochondrial function by Sirt1/AMPK signaling in porcine skeletal muscle satellite cells. Anim. Sci. J. 2019, 90, 255–263. [Google Scholar] [CrossRef]
- Xiang, L.; Huang, Z.; Chen, X.; Jia, G.; Liu, G.; Zhao, H. Leucine regulates porcine muscle fiber type transformation via adiponectin signaling pathway. Anim. Biotechnol. 2021, 33, 330–338. [Google Scholar] [CrossRef]
- Yin, Y.; Liu, Y.; Duan, G.; Han, M.; Gong, S.; Yang, Z.; Duan, Y.; Guo, Q.; Chen, Q.; Li, F. The Effect of Dietary Leucine Supplementation on Antioxidant Capacity and Meat Quality of Finishing Pigs under Heat Stress. Antioxidants 2022, 11, 1373. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiong, Z.; Yan, W.; Gao, E.; Cheng, H.; Wu, G.; Liu, Y.; Zhang, L.; Li, C.; Wang, S.; et al. Branched chain amino acids exacerbate myocardial ischemia/reperfusion vulnerability via enhancing GCN2/ATF6/PPAR-α pathway-dependent fatty acid oxidation. Theranostics 2020, 10, 5623–5640. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Sato, Y.; Obeng, K.A.; Suzuki, D.; Komiya, Y.; Adachi, S.I.; Yoshizawa, F. Energy metabolism profile of the effects of amino acid treatment on skeletal muscle cells: Leucine inhibits glycolysis of myotubes. Nutrition 2020, 77, 110794. [Google Scholar] [CrossRef] [PubMed]
- Rivera, M.E.; Lyon, E.S.; Johnson, M.A.; Sunderland, K.L.; Vaughan, R.A. Effect of valine on myotube insulin sensitivity and metabolism with and without insulin resistance. Mol. Cell. Biochem. 2020, 468, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Zhang, X.; Azhar, G.; Patyal, P.; Verma, A.; Kc, G.; Wei, J.Y. Valine improves mitochondrial function and protects against oxidative stress. Biosci. Biotechnol. Biochem. 2024, 88, 168–176. [Google Scholar] [CrossRef] [PubMed]
- James, D.E.; Stöckli, J.; Birnbaum, M.J. The aetiology and molecular landscape of insulin resistance. Nat. Rev. Mol. Cell Biol. 2021, 22, 751–771. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Dey, C.S. PTEN and SHIP2 regulates PI3K/Akt pathway through focal adhesion kinase. Mol. Cell Endocrinol. 2009, 309, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Janssen, J.A.M.J.L. Hyperinsulinemia and Its Pivotal Role in Aging, Obesity, Type 2 Diabetes, Cardiovascular Disease and Cancer. Int. J. Mol. Sci. 2021, 22, 7797. [Google Scholar] [CrossRef]
- Vaidya, R.A.; Desai, S.; Moitra, P.; Salis, S.; Agashe, S.; Battalwar, R.; Mehta, A.; Madan, J.; Kalita, S.; Udipi, S.A.; et al. Hyperinsulinemia: An early biomarker of metabolic dysfunction. Front. Clin. Diabetes Healthc. 2023, 4, 1159664. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
VanDerStad, L.R.; Wyatt, E.C.; Vaughan, R.A. Excess Branched-Chain Amino Acids Suppress Mitochondrial Function and Biogenic Signaling but Not Mitochondrial Dynamics in a Myotube Model of Skeletal Muscle Insulin Resistance. Metabolites 2024, 14, 389. https://doi.org/10.3390/metabo14070389
VanDerStad LR, Wyatt EC, Vaughan RA. Excess Branched-Chain Amino Acids Suppress Mitochondrial Function and Biogenic Signaling but Not Mitochondrial Dynamics in a Myotube Model of Skeletal Muscle Insulin Resistance. Metabolites. 2024; 14(7):389. https://doi.org/10.3390/metabo14070389
Chicago/Turabian StyleVanDerStad, Lindsey R., Emily C. Wyatt, and Roger A. Vaughan. 2024. "Excess Branched-Chain Amino Acids Suppress Mitochondrial Function and Biogenic Signaling but Not Mitochondrial Dynamics in a Myotube Model of Skeletal Muscle Insulin Resistance" Metabolites 14, no. 7: 389. https://doi.org/10.3390/metabo14070389
APA StyleVanDerStad, L. R., Wyatt, E. C., & Vaughan, R. A. (2024). Excess Branched-Chain Amino Acids Suppress Mitochondrial Function and Biogenic Signaling but Not Mitochondrial Dynamics in a Myotube Model of Skeletal Muscle Insulin Resistance. Metabolites, 14(7), 389. https://doi.org/10.3390/metabo14070389





