Metabolite Predictors of Breast and Colorectal Cancer Risk in the Women’s Health Initiative
Abstract
:1. Introduction
2. Materials and Methods
2.1. Women’s Health Initiative
2.2. Case and Control Selection
2.3. Metabololite Profiling
2.3.1. Measurement of Serum Metabolites
2.3.2. Measurements of Urine Metabolites
2.4. Metabolite Quality Controls (QC)
2.5. Statistical Analysis
2.5.1. Participant Data
2.5.2. Imputing Missing Data
2.5.3. Algorithms Used for Selecting a Set of Metabolites
2.5.4. Assessing Prediction Performance
2.5.5. Final Selection of Metabolites
2.5.6. Post Hoc Sensitivity Analyses
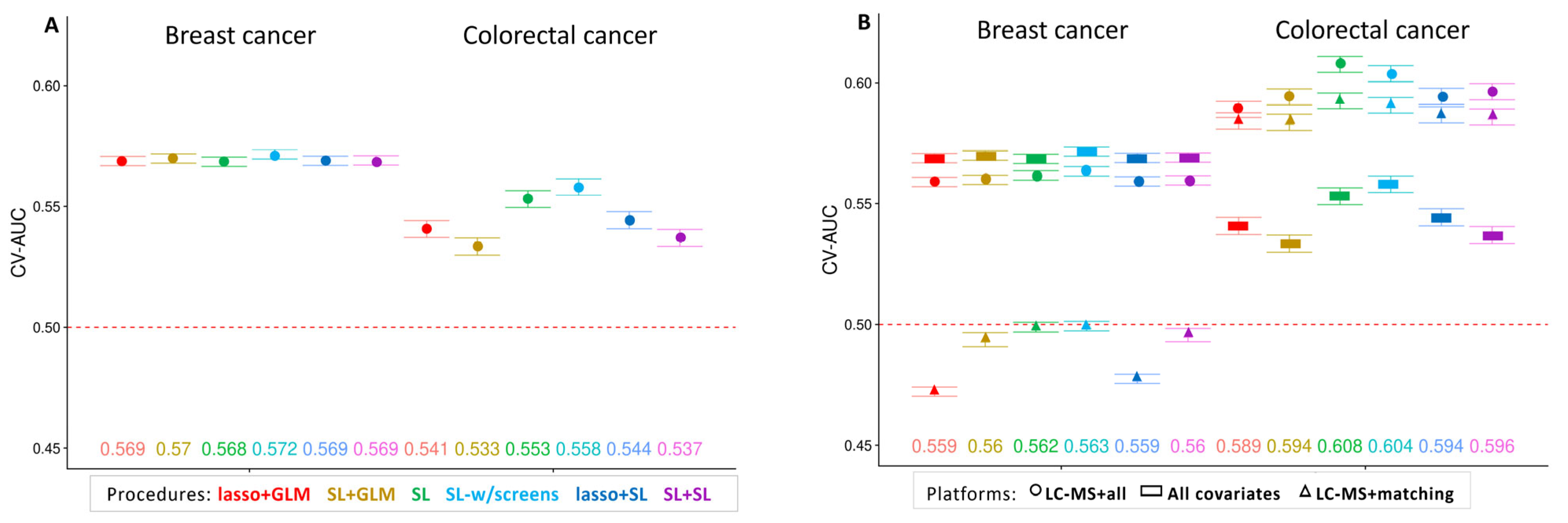
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef] [PubMed]
- American Institute for Cancer Research. Continuous Update Project Expert Report: Diet, Nutrition, Physical Activity and Cancer: A Global Perspective; World Cancer Research Fund: London, UK, 2018; Available online: www.dietandcancerreport.org (accessed on 2 April 2023).
- Yusof, A.S.; Isa, Z.M.; Shah, S.A. Dietary patterns and risk of colorectal cancer: A systematic review of cohort studies (2000–2011). Asian Pac. J. Cancer Prev. 2012, 13, 4713–4717. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Xia, J.; Li, L.; Ke, Y.; Cheng, J.; Xie, Y.; Chu, W.; Cheung, P.; Kim, J.H.; Colditz, G.A.; et al. Associations between dietary patterns and the risk of breast cancer: A systematic review and meta-analysis of observational studies. Breast Cancer Res. 2019, 21, 16. [Google Scholar] [CrossRef] [PubMed]
- Clinton, S.K.; Giovannucci, E.L.; Hursting, S.D. The World Cancer Research Fund/American Institute for Cancer Research Third Expert Report on diet, nutrition, physical activity, and cancer: Impact and future directions. J. Nutr. 2020, 150, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.O.; Berdzuli, N.; Ilbawi, A.; Kestel, D.; Kluge, H.P.; Krech, R.; Mikkelsen, B.; Neufeld, M.; Poznyak, V.; Rekve, D.; et al. Health and cancer risks associated with low levels of alcohol consumption. Lancet Public Health 2023, 8, e6–e7. [Google Scholar] [CrossRef] [PubMed]
- Key, T.J.; Bradbury, K.E.; Perez-Cornago, A.; Sinha, R.; Tsilidis, K.K.; Tsugane, S. Diet, nutrition, and cancer risk: What do we know and what is the way forward? BMJ 2020, 368, m511. [Google Scholar] [CrossRef] [PubMed]
- Cross, A.J.; Sinha, R. Meat-related mutagens/carcinogens in the etiology of colorectal cancer. Environ. Mol. Mutagen. 2004, 44, 44–55. [Google Scholar] [CrossRef]
- Mei, J.; Qian, M.; Hou, Y.; Liang, M.; Chen, Y.; Wang, C.; Zhang, J. Association of saturated fatty acids with cancer risk: A systematic review and meta-analysis. Lipids Health Dis. 2024, 23, 32. [Google Scholar] [CrossRef] [PubMed]
- Talukdar, J.R.; Steen, J.P.; Goldenberg, J.Z.; Zhang, Q.; Vernooij, R.W.M.; Ge, L.; Zeraatkar, D.; Bala, M.M.; Ball, G.D.C.; Thabane, L.; et al. Saturated fat, the estimated absolute risk and certainty of risk for mortality and major cancer and cardiometabolic outcomes: An overview of systematic reviews. Syst. Rev. 2023, 12, 179. [Google Scholar] [CrossRef]
- Epner, M.; Yang, P.; Wagner, R.W.; Cohen, L. Understanding the link between sugar and cancer: An examination of the preclinical and clinical evidence. Cancers 2022, 14, 6042. [Google Scholar] [CrossRef]
- Hariri, N.; Gougeon, R.; Thibault, L. A highly saturated fat-rich diet is more obesogenic than diets with lower saturated fat content. Nutr. Res. 2010, 30, 632–643. [Google Scholar] [CrossRef]
- Zhang, P. Influence of foods and nutrition on the gut microbiome and complications for intestinal health. Int. J. Mol. Sci. 2022, 23, 9588. [Google Scholar] [CrossRef]
- Anderson, S.M.; Sears, C.L. The role of the gut microbiome in cancer: A review, with special focus on colorectal neoplasia and clostridioides difficile. Clin. Infect. Dis. 2023, 77 (Suppl. 6), S471–S478. [Google Scholar] [CrossRef] [PubMed]
- Putri, S.P.; Nakayama, Y.; Matsuda, F.; Uchikata, T.; Kobayashi, S.; Matsubara, A.; Fukusaki, E. Current metabolomics: Practical applications. J. Biosci. Bioeng. 2013, 115, 579–589. [Google Scholar] [CrossRef]
- Nannini, G.; Meoni, G.; Amedei, A.; Tenori, L. Metabolomics profile in gastrointestinal cancers: Update and future perspectives. World J. Gastroenterol. 2020, 26, 2514–2532. [Google Scholar] [CrossRef] [PubMed]
- Cheung, P.K.; Ma, M.H.; Tse, H.F.; Yeung, K.F.; Tsang, H.F.; Chu, M.K.M.; Kan, C.M.; Cho, W.C.S.; Ng, L.B.W.; Chan, L.W.C.; et al. The applications of metabolomics in the molecular diagnostics of cancer. Expert Rev. Mol. Diagn. 2019, 19, 785–793. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Y.; Cai, H.; Wang, S.; Shen, Y.; Ke, C. Application of metabolomics in the diagnosis of breast cancer: A systematic review. J. Cancer 2020, 11, 2540–2551. [Google Scholar] [CrossRef] [PubMed]
- His, M.; Gunter, M.J.; Keski-Rahkonen, P.; Rinaldi, S. Application of metabolomics to epidemiologic studies of breast cancer: New perspectives for etiology and prevention. J. Clin. Oncol. 2023, 42, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Orsini, A.; Diquigiovanni, C.; Bonora, E. Omics technologies improving breast cancer research and diagnostics. Int. J. Mol. Sci. 2023, 24, 12690. [Google Scholar] [CrossRef]
- The Women’s Health Initiative Study Group. Design of the Women’s Health Initiative clinical trial and observational study. Control. Clin. Trials 1998, 19, 61–109. [Google Scholar] [CrossRef]
- Bergstralh, E.J.; Kosanke, J.L. Computerized Matching of Cases to Controls. Technical Report #56. Department of Health Sciences Research; Mayo Clinic: Rochester, MN, USA, 1995. [Google Scholar]
- Kurup, K.; Matyi, S.; Giles, C.B.; Wren, J.D.; Jones, K.; Ericsson, A.; Raftery, D.; Wang, L.; Promislow, D.; Richardson, A.; et al. Calorie restriction prevents age-related changes in the intestinal microbiota. Aging 2021, 13, 6298–6329. [Google Scholar] [CrossRef] [PubMed]
- Hwangbo, N.; Zhang, X.; Raftery, D.; Gu, H.; Hu, S.C.; Montine, T.J.; Quinn, J.F.; Chung, K.A.; Hiller, A.L.; Wang, D.; et al. A Metabolomic Aging Clock Using Human Cerebrospinal Fluid. J. Gerontol. A Biol. Sci. Med. Sci. 2022, 77, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Hanson, A.J.; Banks, W.A.; Bettcher, L.F.; Pepin, R.; Raftery, D.; Craft, S. Cerebrospinal fluid lipidomics: Effects of an intravenous triglyceride infusion and apoE status. Metabolomics 2019, 16, 6. [Google Scholar] [CrossRef] [PubMed]
- Ghorasaini, M.; Mohammed, Y.; Adamski, J.; Bettcher, L.; Bowden, J.A.; Cabruja, M.; Contrepois, K.; Ellenberger, M.; Gajera, B.; Haid, M.; et al. Cross-Laboratory Standardization of Preclinical Lipidomics Using Differential Mobility Spectrometry and Multiple Reaction Monitoring. Anal. Chem. 2021, 93, 16369–16378. [Google Scholar] [CrossRef] [PubMed]
- Chan, E.C.; Pasikanti, K.K.; Nicholson, J.K. Global urinary metabolic profiling procedures using gas chromatography-mass spectrometry. Nat. Protoc. 2011, 6, 1483–1499. [Google Scholar] [CrossRef] [PubMed]
- Prentice, R.L.; Pettinger, M.; Neuhouser, M.L.; Tinker, L.F.; Huang, Y.; Zheng, C.; Manson, J.E.; Mossavar-Rahmani, Y.; Anderson, G.L.; Lampe, J.W. Application of blood concentration biomarkers in nutritional epidemiology: Example of carotenoid and tocopherol intake in relation to chronic disease risk. Am. J. Clin. Nutr. 2019, 10, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Mansournia, M.A.; Jewell, N.P.; Greenland, S. Case-control matching: Effects, misconceptions, and recommendations. Eur. J. Epidemiol. 2018, 33, 5–14. [Google Scholar] [CrossRef] [PubMed]
- van Buuren, S.; Groothuis-Oudshoorn, K. Mice: Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef]
- Jaeger, B.C.; Cantor, R.; Sthanam, V.; Xie, R.; Kirklin, J.K.; Rudraraju, R. Improving outcome predictions for patients receiving mechanical circulatory support by optimizing imputation of missing values. Circ. Cardiovasc. Qual. Outcomes 2021, 14, e007071. [Google Scholar] [CrossRef]
- Zheng, C.; Gowda, G.A.N.; Raftery, D.; Neuhouser, M.L.; Tinker, L.F.; Prentice, R.L.; Beresford, S.A.A.; Zhang, Y.; Bettcher, L.; Pepin, R.; et al. Evaluation of potential metabolomic-based biomarkers of protein, carbohydrate and fat intakes using a controlled feeding study. Eur. J. Nutr. 2021, 60, 4207–4218. [Google Scholar] [CrossRef]
- Rubin, D.B. The use of matched samling and regression adjustment to remove bias in observational studies. Biometrics 1973, 29, 185–203. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- van der Laan, M.J.; Polley, E.C.; Hubbard, A.E. Super learner. Stat. Appl. Genet. Mol. Biol. 2007, 6. [Google Scholar] [CrossRef] [PubMed]
- Polley, E.; LeDell, E.; Kennedy, C.; van der Laan, M.J. SuperLearner: Super Learner Prediction. R Package, Version 2.0-26. 2023. Available online: https://github.com/ecpolley/SuperLearner (accessed on 18 February 2022).
- Wolpert, D.H. Stacked generalization. Neural Netw. 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Serial. B 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Leeb, H.; Potscher, B.M. Model selection and inference: Facts and fiction. Econom. Theory 2005, 21, 21–59. [Google Scholar] [CrossRef]
- Gunter, M.J.; Hoover, D.R.; Yu, H.; Wassertheil-Smoller, S.; Rohan, T.E.; Manson, J.E.; Li, J.; Ho, G.Y.; Xue, X.; Anderson, G.L.; et al. Insulin, insulin-like growth factor-I, and risk of breast cancer in postmenopausal women. J. Natl. Cancer Inst. 2009, 101, 48–60. [Google Scholar] [CrossRef]
- Gunter, M.J.; Wang, T.; Cushman, M.; Xue, X.; Wassertheil-Smoller, S.; Strickler, H.D.; Rohan, T.E.; Manson, J.E.; McTiernan, A.; Kaplan, R.C.; et al. Circulating adipokines and inflammatory markers and postmenopausal breast cancer risk. J. Natl. Cancer Inst. 2015, 107, djv169. [Google Scholar] [CrossRef]
- Kolb, R.; Zhang, W. Obesity and breast cancer: A case of inflamed adipose tissue. Cancers 2020, 12, 1686. [Google Scholar] [CrossRef]
- Pati, S.; Irfan, W.; Jameel, A.; Ahmed, S.; Shahid, R.K. Obesity and cancer: A current overview of epidemiology, pathogenesis, outcomes, and management. Cancers 2023, 15, 485. [Google Scholar] [CrossRef]
- Society, A.C. Does Body Weight Affect Cancer Risk? Available online: https://www.cancer.org/cancer/risk-prevention/diet-physical-activity/body-weight-and-cancer-risk/effects.html (accessed on 15 May 2024).
- Rothwell, J.A.; Besevic, J.; Dimou, N.; Breeur, M.; Murphy, N.; Jenab, M.; Wedekind, R.; Viallon, V.; Ferrari, P.; Achaintre, D.; et al. Circulating amino acid levels and colorectal cancer risk in the European Prospective Investigation into Cancer and Nutrition and UK Biobank cohorts. BMC Med. 2023, 21, 80. [Google Scholar] [CrossRef] [PubMed]
- Caldovic, L.; Ah Mew, N.; Shi, D.; Morizono, H.; Yudkoff, M.; Tuchman, M. N-acetylglutamate synthase: Structure, function and defects. Mol. Genet. Metab. 2010, 100 (Suppl. 1), S13–S19. [Google Scholar] [CrossRef]
- Jalandra, R.; Dalal, N.; Yadav, A.K.; Verma, D.; Sharma, M.; Singh, R.; Khosla, A.; Kumar, A.; Solanki, P.R. Emerging role of trimethylamine-N-oxide (TMAO) in colorectal cancer. Appl. Microbiol. Biotechnol. 2021, 105, 7651–7660. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Ulrich, C.M.; Neuhouser, M.L.; Malysheva, O.; Bailey, L.B.; Xiao, L.R.; Brown, E.C.; Cushing-Haugen, K.L.; Zheng, Y.Y.; Cheng, T.Y.D.; et al. Plasma choline metabolites and colorectal cancer risk in the Women’s Health Initiative Observational Study. Cancer Res. 2014, 74, 7442–7452. [Google Scholar] [CrossRef]
- Byrd, D.A.; Zouiouich, S.; Karwa, S.; Li, X.S.; Wang, Z.; Sampson, J.N.; Loftfield, E.; Huang, W.Y.; Hazen, S.L.; Sinha, R. Associations of serum trimethylamine N-oxide and its precursors with colorectal cancer risk in the Prostate, Lung, Colorectal, Ovarian Cancer Screening Trial Cohort. Cancer 2024, 130, 1982–1990. [Google Scholar] [CrossRef]
- Chan, D.S.; Lau, R.; Aune, D.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Red and processed meat and colorectal cancer incidence: Meta-analysis of prospective studies. PLoS ONE 2011, 6, e20456. [Google Scholar] [CrossRef]
- Aune, D.; Chan, D.S.; Vieira, A.R.; Navarro Rosenblatt, D.A.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Red and processed meat intake and risk of colorectal adenomas: A systematic review and meta-analysis of epidemiological studies. Cancer Causes Control 2013, 24, 611–627. [Google Scholar] [CrossRef] [PubMed]
- Playdon, M.C.; Ziegler, R.G.; Sampson, J.N.; Stolzenberg-Solomon, R.; Thompson, H.J.; Irwin, M.L.; Mayne, S.T.; Hoover, R.N.; Moore, S.C. Nutritional metabolomics and breast cancer risk in a prospective study. Am. J. Clin. Nutr. 2017, 106, 637–649. [Google Scholar] [CrossRef]
- Moore, S.C.; Mazzilli, K.M.; Sampson, J.N.; Matthews, C.E.; Carter, B.D.; Playdon, M.C.; Wang, Y.; Stevens, V.L. A metabolomics analysis of postmenopausal breast cancer risk in the cancer prevention study II. Metabolites 2021, 11, 95. [Google Scholar] [CrossRef]
- His, M.; Viallon, V.; Dossus, L.; Schmidt, J.A.; Travis, R.C.; Gunter, M.J.; Overvad, K.; Kyro, C.; Tjonneland, A.; Lecuyer, L.; et al. Lifestyle correlates of eight breast cancer-related metabolites: A cross-sectional study within the EPIC cohort. BMC Med. 2021, 19, 312. [Google Scholar] [CrossRef]
- Moore, S.C.; Playdon, M.C.; Sampson, J.N.; Hoover, R.N.; Trabert, B.; Matthews, C.E.; Ziegler, R.G. A metabolomics analysis of body mass index and postmenopausal breast cancer risk. J. Natl. Cancer Inst. 2018, 110, 588–597. [Google Scholar] [CrossRef]
- His, M.; Viallon, V.; Dossus, L.; Gicquiau, A.; Achaintre, D.; Scalbert, A.; Ferrari, P.; Romieu, I.; Onland-Moret, N.C.; Weiderpass, E.; et al. Prospective analysis of circulating metabolites and breast cancer in EPIC. BMC Med. 2019, 17, 178. [Google Scholar] [CrossRef] [PubMed]
- Lecuyer, L.; Dalle, C.; Lefevre-Arbogast, S.; Micheau, P.; Lyan, B.; Rossary, A.; Demidem, A.; Petera, M.; Lagree, M.; Centeno, D.; et al. Diet-related metabolomic signature oflLong-termbBreast cancer risk using penalized regression: An exploratory study in the SU.VI.MAX cohort. Cancer Epidemiol. Biomark. Prev. 2020, 29, 396–405. [Google Scholar] [CrossRef]
- Stevens, V.L.; Carter, B.D.; Jacobs, E.J.; McCullough, M.L.; Teras, L.R.; Wang, Y. A prospective case-cohort analysis of plasma metabolites and breast cancer risk. Breast Cancer Res. 2023, 25, 5. [Google Scholar] [CrossRef] [PubMed]
- Jobard, E.; Dossus, L.; Baglietto, L.; Fornili, M.; Lecuyer, L.; Mancini, F.R.; Gunter, M.J.; Tredan, O.; Boutron-Ruault, M.C.; Elena-Herrmann, B.; et al. Investigation of circulating metabolites associated with breast cancer risk by untargeted metabolomics: A case-control study nested within the French E3N cohort. Br. J. Cancer 2021, 124, 1734–1743. [Google Scholar] [CrossRef]
- Kuhn, T.; Floegel, A.; Sookthai, D.; Johnson, T.; Rolle-Kampczyk, U.; Otto, W.; von Bergen, M.; Boeing, H.; Kaaks, R. Higher plasma levels of lysophosphatidylcholine 18:0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med. 2016, 14, 13. [Google Scholar] [CrossRef]
- Brantley, K.D.; Zeleznik, O.A.; Dickerman, B.A.; Balasubramanian, R.; Clish, C.B.; Avila-Pacheco, J.; Rosner, B.; Tamimi, R.M.; Eliassen, A.H. A metabolomic analysis of adiposity measures and pre- and postmenopausal breast cancer risk in the Nurses’ Health Studies. Br. J. Cancer 2022, 127, 1076–1085. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Xiang, Y.B.; Rothman, N.; Yu, D.; Li, H.L.; Yang, G.; Cai, H.; Ma, X.; Lan, Q.; Gao, Y.T.; et al. Prospective study of blood metabolites associated with colorectal cancer risk. Int. J. Cancer 2018, 143, 527–534. [Google Scholar] [CrossRef]
- Cross, A.J.; Moore, S.C.; Boca, S.; Huang, W.Y.; Xiong, X.; Stolzenberg-Solomon, R.; Sinha, R.; Sampson, J.N. A prospective study of serum metabolites and colorectal cancer risk. Cancer 2014, 120, 3049–3057. [Google Scholar] [CrossRef] [PubMed]
- Harewood, R.; Rothwell, J.A.; Besevic, J.; Viallon, V.; Achaintre, D.; Gicquiau, A.; Rinaldi, S.; Wedekind, R.; Prehn, C.; Adamski, J.; et al. Association between pre-diagnostic circulating lipid metabolites and colorectal cancer risk: A nested case-control study in the European Prospective Investigation into Cancer and Nutrition (EPIC). EBioMedicine 2024, 101, 105024. [Google Scholar] [CrossRef]
- Perttula, K.; Schiffman, C.; Edmands, W.M.B.; Petrick, L.; Grigoryan, H.; Cai, X.; Gunter, M.J.; Naccarati, A.; Polidoro, S.; Dudoit, S.; et al. Untargeted lipidomic features associated with colorectal cancer in a prospective cohort. BMC Cancer 2018, 18, 996. [Google Scholar] [CrossRef]
- Kliemann, N.; Viallon, V.; Murphy, N.; Beeken, R.J.; Rothwell, J.A.; Rinaldi, S.; Assi, N.; van Roekel, E.H.; Schmidt, J.A.; Borch, K.B.; et al. Metabolic signatures of greater body size and their associations with risk of colorectal and endometrial cancers in the European Prospective Investigation into Cancer and Nutrition. BMC Med. 2021, 19, 101. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Ismond, K.; Liu, Z.; Constable, J.; Wang, H.; Alatise, O.I.; Weiser, M.R.; Kingham, T.P.; Chang, D. Urinary Metabolomics to identify a unique biomarker panel for detecting colorectal cancer: A multicenter study. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1283–1291. [Google Scholar] [CrossRef]
- McCullough, M.L.; Hodge, R.A.; Campbell, P.T.; Stevens, V.L.; Wang, Y. Pre-Diagnostic circulating metabolites and colorectal cancer risk in the Cancer Prevention Study-II Nutrition Cohort. Metabolites 2021, 11, 156. [Google Scholar] [CrossRef] [PubMed]
- Vidman, L.; Zheng, R.; Boden, S.; Ribbenstedt, A.; Gunter, M.J.; Palmqvist, R.; Harlid, S.; Brunius, C.; Van Guelpen, B. Untargeted plasma metabolomics and risk of colorectal cancer—An analysis nested within a large-scale prospective cohort. Cancer Metab. 2023, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Dai, R.; Huang, Y.; Neuhouser, M.L.; Lampe, J.W.; Raftery, D.; Tabung, F.K.; Zheng, C. Variable selection with FDR control for noisy data—An application to screening metabolites that are associated with breast and colorectal cancer. arXiv 2023, arXiv:2310.06696. [Google Scholar]
- Foygel Barber, R.; Candes, E.J. Controlling the false discovery rate via knockoffs. Ann. Stat. 2015, 45, 2055–2085. [Google Scholar]
- Gunter, M.J.; Hoover, D.R.; Yu, H.; Wassertheil-Smoller, S.; Rohan, T.E.; Manson, J.E.; Howard, B.V.; Wylie-Rosett, J.; Anderson, G.L.; Ho, G.Y.; et al. Insulin, insulin-like growth factor-I, endogenous estradiol, and risk of colorectal cancer in postmenopausal women. Cancer Res. 2008, 68, 329–337. [Google Scholar] [CrossRef]
| Characteristics | BC Cases (n = 577) | CRC Cases (n = 181) | Controls (n = 758) |
|---|---|---|---|
| Demographic factors | |||
| Age (yrs) | 62; [56, 68] | 64; [58, 69] | 63; [57, 68] |
| Body Mass Index (kg/m)2 | 27.73; [24.45, 32.45] * | 28.05; [24.9, 31.98] * | 27.16; [24.19, 31.18] |
| Waist circumference (cm) | 86; [77, 96] * | 87; [78, 99] * | 84; [76, 93] * |
| Self-reported race or ethnicity | |||
| Alaska Native or American Indian | <10 * | <10 * | <10 * |
| Asian or Pacific Islander | <10 * | <10 * | <10 * |
| Hispanic or Latina | 25 (4%) | 10 (6%) | 35 (5%) |
| Non-Hispanic Black or African American | 67 (12%) | 26 (14%) | 93 (12%) |
| White | 477 (83) | 139 (77%) | 616 (81) |
| Unknown | <10 | <10 | <10 |
| Education | |||
| Less than high school | 37 (6%) | 15 (8%) | 41 (5%) |
| High school or GED | 120 (21%) | 43 (24%) | 166 (22%) |
| School after high school | 209 (36%) | 74 (41%) | 284 (38%) |
| College degree or higher | 207 (36%) | 48 (27%) | 267 (35%) |
| Unknown | <10 | <10 | 0 |
| Income | |||
| <$20,000 | 104 (18%) | 56 (31%) | 165 (22%) |
| $20,000–$35,000 | 171 (30%) | 47 (26%) | 232 (31%) |
| $35,000–$50,000 | 100 (17%) | 37 (20%) | 132 (17%) |
| $50,000–$75,000 | 85 (15%) | 21 (12%) | 134 (18%) |
| >$75,000 | 81 (14%) | 13 (7%) | 95 (13%) |
| Unknown | 36 (6%) | <10 | 0 |
| Lifestyle factors | |||
| Alcohol intake (svgs/wk) | 0.42; [0, 1.81] * | 0.21; [0, 2.73] * | 0.21; [0, 1.37] |
| Total calcium (mg/d) | 1024.1; [647.5, 1557.2] * | 1029.3; [711.8, 1532.0] * | 973.8; [649.7, 1530.9] * |
| Total folate (mcg/d) | 635.47; [420.94, 881.23] * | 619.21; [448.25, 861.41] * | 593.64; [419.06, 838.82] * |
| Red or processed meat (svgs/d) | 1.87; [1.04, 2.95] * | 1.85; [1.1, 3.31] * | 1.83; [1.07, 3] * |
| Energy expenditure (MET-hours/wk) | 7.08; [1.5, 16.75] * | 7.25; [1.38, 16.42] * | 7; [1.5, 15.5] * |
| History of smoking (current yes/no) | 45 (7.84%) * | 16 (8.94%)* | 47 (6.2%) |
| Any supplement use | 257 (44.54%) * | 71 (39.23%) | 328 (43.27%) |
| Uses anti-diabetes medication | 24 (4.16%) | 16 (8.84%) | 35 (4.62%) |
| Uses anti-hypertensive medication | 185 (32.06%) | 68 (37.57%) | 231 (30.47%) |
| Uses anti-lipid medication | 42 (7.28%) | 16 (8.84%) | 50 (6.6%) |
| Uses NSAIDs | 196 (33.97%) | 58 (32.04%) | 262 (34.56%) |
| Clinical risk factors | |||
| Gail 5-year risk score | 1.9; [1.16, 2.22] | 1.7; [1.1, 1.92] | 1.68; [1.15, 1.95] |
| Family history of cancer | |||
| Yes | 80 (14%) | 30 (17%) | 124 (16%) |
| No | 453 (79%) | 139 (77%) | 617 (8%) |
| Unknown | 44 (8%) | 12 (7%) | 17 (2%) |
| Personal history of cancer | |||
| Yes | 27 (5%) | 13 (7%) | 39 (5.2%) |
| No | 547 (95%) | 168 (93%) | 719 (95%) |
| Unknown | <10 | 0 | 0 |
| History of colonoscopy | |||
| Yes | 253 (44%) | 71 (39%) | 355 (47%) |
| No | 249 (43%) | 90 (50%) | 401 (53%) |
| Unknown | 75 (13%) | 20 (11%) | <10 |
| History of colon polyp removal | |||
| Yes | 39 (7%) | 11 (6%) | 73 (10%) |
| No | 454 (79%) | 145 (80%) | 677 (89%) |
| Unknown | 84 (15%) | 25 (14%) | <10 |
| History of treated diabetes | |||
| Yes | 27 (5%) | 19 (11%) | 40 (5%) |
| No | 549 (95%) | 162 (90%) | 717 (95%) |
| Unknown | <10 | 0 | <10 |
| History of treated hypertension | |||
| Yes | 144 (25%) | 55 (30%) | 186 (25%) |
| No | 356 (62%) | 104 (57%) | 568 (75%) |
| Unknown | 77 (13%) | 22 (12%) | <10 |
| Had at least one term pregnancy | 510 (88%) | 158 (87%) | 676 (89%) |
| Post-menopausal hormone therapy use | |||
| Never | 286 (50%) | 97 (54%) | 331 (44%) |
| Past | 75 (13%) | 30 (17%) | 125 (16%) |
| Current estrogen alone | 117 (20%) | 36 (20%) | 189 (25%) |
| Current estrogen and progestin | 98 (17%) | 18 (10%) | 113 (15%) |
| Study variables | |||
| WHI enrollment date | |||
| Baseline | 108 (19%) | 44 (24%) | 152 (20%) |
| Year 1 | 220 (38%) | 65 (36%) | 285 (38%) |
| Year 3 | 243 (42%) | 68 (38%) | 311 (41%) |
| Year 6 | <10 | <10 | <10 |
| Year 9 | 0 | <10 | <10 |
| Calcium / Vitamin D (CaD) trial arm | |||
| Not randomized to CaD | 462 (80%) | 147 (81%) | 560 (74%) |
| Control arm | 54 (9%) | 19 (11%) | 104 (14%) |
| Intervention arm | 61 (11%) | 15 (8%) | 94 (12%) |
| Dietary Modification (DM) trial arm | |||
| Not randomized to DM | 384 (67%) | 122 (67%) | 488 (64%) |
| Control arm | 118 (20%) | 27 (15%) | 163 (22%) |
| Intervention arm | 75 (13%) | 32 (18%) | 107 (14%) |
| Hormone therapy (HT) trial arm | |||
| Not randomized to HT | 495 (86%) | 144 (80%) | 641 (85%) |
| Estrogen-only control arm | 19 (3%) | 13 (7%) | 27 (4%) |
| Estrogen-only intervention arm | 15 (3%) | 11 (6%) | 34 (4%) |
| Estrogen and progestin control arm | 16 (3%) | <10 | 30 (4%) |
| Estrogen and progestin intervention arm | 32 (6%) | <10 | 26 (3%) |
| Metabolites Selected | Proportion of Explained Variation 2 | Direction of Coefficient for Metabolites 3 |
|---|---|---|
| Breast Cancer | All covariates + metabolites: 0.27 | |
| Serum: | ||
| LC-MS | ||
| Azelaic acid | 0.23 | − |
| Choline | 0.23 | + |
| Cysteinyl glycine | 0.23 | − |
| Ethanolamine | 0.23 | + |
| Gamma tocopherol | 0.23 | + |
| Hippuric acid | 0.23 | − |
| Isovaleryl carnitine | 0.23 | + |
| N-isovaleryl glycine | 0.23 | − |
| Sucrose | 0.23 | − |
| Trimethylamine-N-oxide | 0.23 | + |
| Valine | 0.23 | + |
| Xylose | 0.23 | − |
| Lipidyzer 4 | ||
| Cholesteryl ester (CE 12:0) | 0.23 | − |
| Cholesteryl ester (CE 20:0) | 0.23 | − |
| Diacylglycerol (DAG 14:1) | 0.24 | + |
| Free fatty Acid (FFA 18:4) | 0.23 | − |
| Free fatty Acid (FFA 20:2) | 0.23 | + |
| Hexosylceramide (HCER 22:0) | 0.23 | + |
| Hexosylceramide (HCER 22:0) | 0.23 | − |
| Phosphatidylcholine (PC 18:1) | 0.23 | + |
| Phosphatidylcholine (PC 18:2) | 0.23 | + |
| Phosphatidylcholine (PC 16:0/18:2) | 0.24 | − |
| Phosphatidylethanolamine (PE 18:2) | 0.23 | + |
| Triacylglycerol (TAG 12:0) | 0.23 | − |
| Triacylglycerol (TAG 16:0) | 0.23 | − |
| Triacylglycerol (TAG 18:0) | 0.23 | − |
| Triacylglycerol (TAG 47:0/15:0) | 0.23 | − |
| Triacylglycerol (TAG 48:4/18:2) | 0.23 | − |
| Triacylglycerol (TAG 50:0/16:0) | 0.23 | + |
| Triacylglycerol (TAG 50:2/18:2) | 0.23 | − |
| Triacylglycerol (TAG 50:5/18:3) | 0.24 | − |
| Triacylglycerol (TAG 52:2/18:2) | 0.24 | + |
| Triacylglycerol (TAG 55:4/18:1) | 0.23 | − |
| Urine | ||
| NMR | ||
| Dimethylamine | 0.23 | − |
| Propanediol | 0.23 | − |
| Formate | 0.23 | + |
| Sucrose | 0.23 | − |
| Taurine | 0.23 | + |
| Uracil | 0.23 | − |
| Trimethylamine-N-oxide | 0.23 | − |
| 2-Hydroxyisobutyrate | 0.23 | + |
| 2-Oxoglutarate | 0.23 | − |
| GC-MS | ||
| Unknown 73.012.10 5 | 0.23 | − |
| Unknown 73.014.49 5 | 0.23 | + |
| Unknown 73.016.52 5 | 0.23 | + |
| Colorectal Cancer | All covariates + metabolites: 0.31 | |
| Serum | ||
| LC-MS | ||
| Adenosine | 0.23 | − |
| Leucic Acid | 0.21 | + |
| Glycerate | 0.25 | + |
| Myo-inositol | 0.22 | + |
| N-Acetyl-glutamate | 0.22 | − |
| N-Acetyl-glycine | 0.23 | + |
| N-Acetylneuraminate | 0.22 | + |
| 2-Hydroxyglutarate | 0.22 | + |
| Hydroxyproline | 0.21 | + |
| 7-Methylguanine | 0.22 | + |
| Lipidyzer4 | ||
| Lysophosphatidylcholine (LPC 20:3) | 0.22 | − |
| Urine | ||
| NMR | ||
| Acetate | 0.21 | + |
| Allantoin | 0.21 | − |
| Histidine | 0.22 | − |
| Isoleucine | 0.21 | + |
| Taurine | 0.22 | + |
| Threonine | 0.21 | + |
| Trimethylamine-N-oxide | 0.21 | + |
| Uracil | 0.22 | − |
| GC-MS | ||
| Unknown 103 17.03 5 | 0.21 | − |
| Unknown 285 22.41 5 | 0.22 | + |
| Unknown 57 9.58 5 | 0.22 | + |
| Unknown 73 10.76 5 | 0.21 | − |
| Unknown 73 17.66 5 | 0.21 | + |
| Metabolite | Class | Function/Relevance |
|---|---|---|
| 2-Hydroxyglutarate | Hydroxy acid | TCA intermediate; inhibitor of alpha-keto dehydrogenases, including histone demethylases; considered an oncometabolite |
| N-Acetyl-glycine | Alpha amino acid | Lipid signaling mediator |
| Taurine | Sulfur-containing amino acid | Metabolism of fats, bile acids |
| Threonine | Amino acid | Metabolism of fats |
| TAG (53:2/FA18:1) | Triglyceride | Fat storage; energy |
| LPC (FA20:3) | Lysophosphatidyl choline | Cholesterol metabolism |
| CE (FA20) | Cholesteryl ester | Cholesterol metabolism |
| Acetate | Short chain fatty acid | Microbial metabolite; acetylation reactions, energy metabolism |
| Glycerate | Sugar acid | Generation of ATP |
| Adenosine | Nucleoside | Energy transfer, component of RNA/DNA |
| Hypoxanthine | Purine | Metabolism of adenosine |
| Uracil | Nucleic acid | Component of RNA |
| 7-Methylguanine | Purine | Component of RNA/DNA; potential biomarker of chicken |
| Histidine | Amino acid | Protein synthesis, histamine and carnosine biosynthesis, scavenger of ROS |
| Leucic acid | Hydroxy fatty acid | Leucine (branched-chain amino acid) metabolite; accelerates lipid peroxidation, oxidative stress |
| Isoleucine | Branched-chain amino acid | Protein metabolism, hemoglobin production, glucose control, immunity |
| N-Acetyl-glutamate | Alpha amino acid | Involved in the urea cycle |
| Allantoin | Imidazoles | Microbial metabolite; uric acid metabolite; found in dairy |
| N-Acetyl-neuraminate | Amino sugar | Component of glycoproteins and mucins involved in immunity |
| Hydroxyproline | Amino acid | Component of collagen; biomarker of meat |
| Myo-inositol | Sugar alcohol | Component of phosphatidylinositol; increases insulin sensitivity; biomarker of whole grains |
| Trimethylamine-N-oxide | Amine oxide | Microbial metabolite formed from choline, betaine, and carnitine; associated with cardiovascular disease; biomarker of fish and red meat |
| Metabolites Selected | Proportion of Explained Variation 2 | Direction of Coefficient for Metabolites 3 |
|---|---|---|
| Breast Cancer | All covariates + metabolites: 0.27 | |
| Serum | ||
| LC-MS | ||
| Cystenyl-glycine | 0.22 | − |
| Ethanolamine | 0.21 | + |
| Sucrose | 0.22 | − |
| Lipidyzer 4 | ||
| Free fatty acid (FFA 20:2) | 0.22 | + |
| Phosphatidylcholine (PC 16:0/18:2) | 0.23 | + |
| Triacylglyceride (TAG 48:4/18:2) | 0.22 | − |
| Triacylglyceride (TAG 50:5/18:3) | 0.22 | − |
| Triacylglyceride (TAG 52:2/18:2) | 0.22 | + |
| Urine | ||
| NMR | ||
| Uracil | 0.22 | − |
| 2-Hydroxyisobutyrate | 0.22 | + |
| GC-MS | ||
| Unknown 73.0 14.49 5 | 0.21 | + |
| Unknown 73.0 12.10 5 | 0.22 | − |
| Colorectal Cancer | All covariates + metabolites: 0.33 | |
| Serum | ||
| LC-MS | ||
| Adenosine | 0.20 | − |
| Leucic acid | 0.18 | + |
| Glycerate | 0.23 | + |
| Hypoxanthine | 0.18 | + |
| Myoinositol | 0.19 | + |
| N-Acetylneuraminate | 0.19 | − |
| 2-Hydroxyglutarate | 0.19 | + |
| 7-Methylguanine | 0.19 | + |
| Lipidyzer | ||
| CE (FA20) | 0.17 | − |
| TAG (53:2/18:1) | 0.18 | + |
| Urine | ||
| NMR | ||
| Histidine | 0.18 | − |
| Taurine | 0.19 | + |
| Threonine | 0.17 | + |
| GC-MS | ||
| Unknown 103 17.03 5 | 0.18 | − |
| Unknown 285 22.41 5 | 0.18 | + |
| Unknown 57 9.58 5 | 0.18 | + |
| Unknown 73 10.76 5 | 0.18 | − |
| Unknown 73 17.27 5 | 0.17 | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navarro, S.L.; Williamson, B.D.; Huang, Y.; Nagana Gowda, G.A.; Raftery, D.; Tinker, L.F.; Zheng, C.; Beresford, S.A.A.; Purcell, H.; Djukovic, D.; et al. Metabolite Predictors of Breast and Colorectal Cancer Risk in the Women’s Health Initiative. Metabolites 2024, 14, 463. https://doi.org/10.3390/metabo14080463
Navarro SL, Williamson BD, Huang Y, Nagana Gowda GA, Raftery D, Tinker LF, Zheng C, Beresford SAA, Purcell H, Djukovic D, et al. Metabolite Predictors of Breast and Colorectal Cancer Risk in the Women’s Health Initiative. Metabolites. 2024; 14(8):463. https://doi.org/10.3390/metabo14080463
Chicago/Turabian StyleNavarro, Sandi L., Brian D. Williamson, Ying Huang, G. A. Nagana Gowda, Daniel Raftery, Lesley F. Tinker, Cheng Zheng, Shirley A. A. Beresford, Hayley Purcell, Danijel Djukovic, and et al. 2024. "Metabolite Predictors of Breast and Colorectal Cancer Risk in the Women’s Health Initiative" Metabolites 14, no. 8: 463. https://doi.org/10.3390/metabo14080463








