Comprehensive Metabolomics Profiling and Bioactivity Study of Lycium shawii (Awsaj) Extracts with Particular Emphasis on Potential Anti-Malarial Properties
Abstract
1. Introduction
2. Materials and Methods
2.1. Botanical Description and Collection
2.2. Extraction of L. shawii Samples
2.3. Metabolomic Profiling
2.3.1. NMR Spectroscopy Analyses of L. shawii Extracts
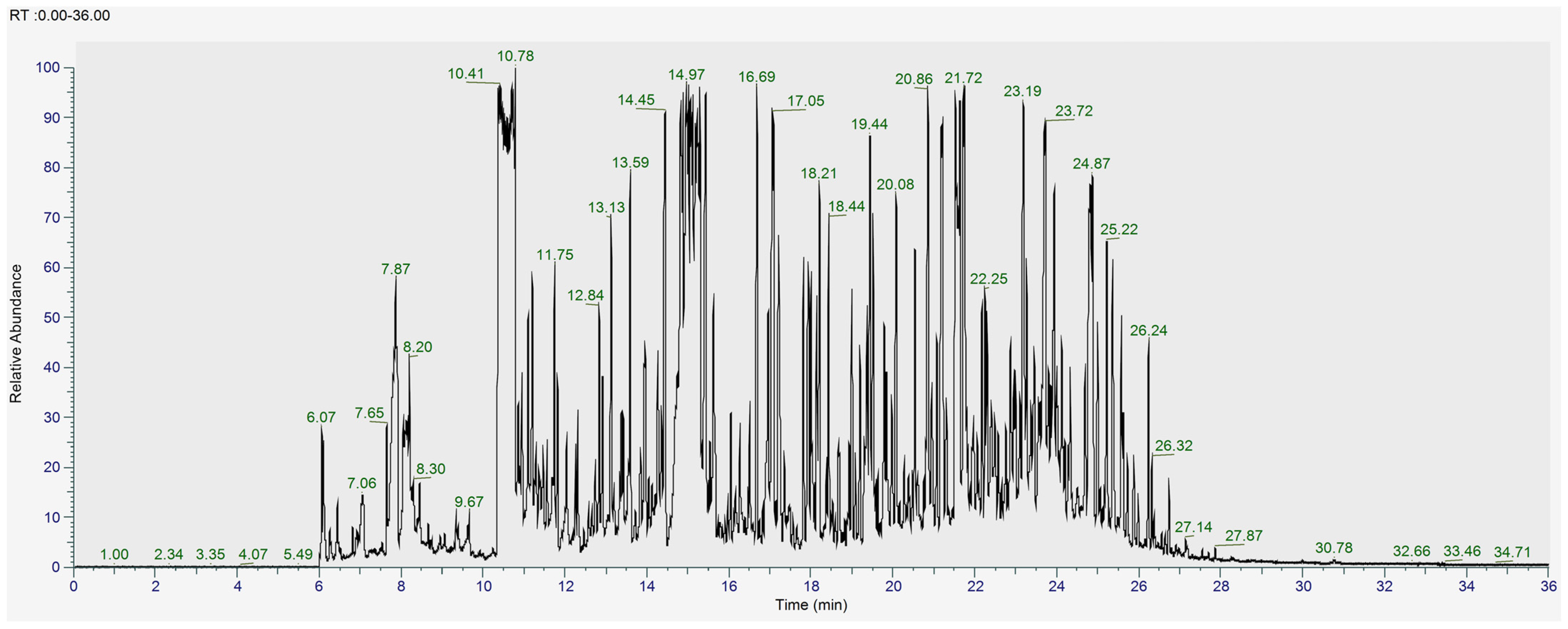
2.3.2. GC-MS Analysis of L. shawii Extracts
2.3.3. UHPLC-MS Analysis of L. shawii Extracts
2.4. Biological Activity Assays
2.4.1. Determination of Total Phenolic Content (TPC) of L. shawii Extracts
2.4.2. Cell Line Cultures
2.4.3. Viability Assay L. shawii Water Extract
2.4.4. In Vitro Semi-Quantitative Antimalarial Activity Assay of L. shawii Water Extract
2.4.5. Anti-Inflammatory Activity Assay of L. shawii Water Extract
2.4.6. Antibacterial Activity Assay of L. shawii Water Extract
2.4.7. Data Processing and Statistical Analysis
3. Results and Discussion
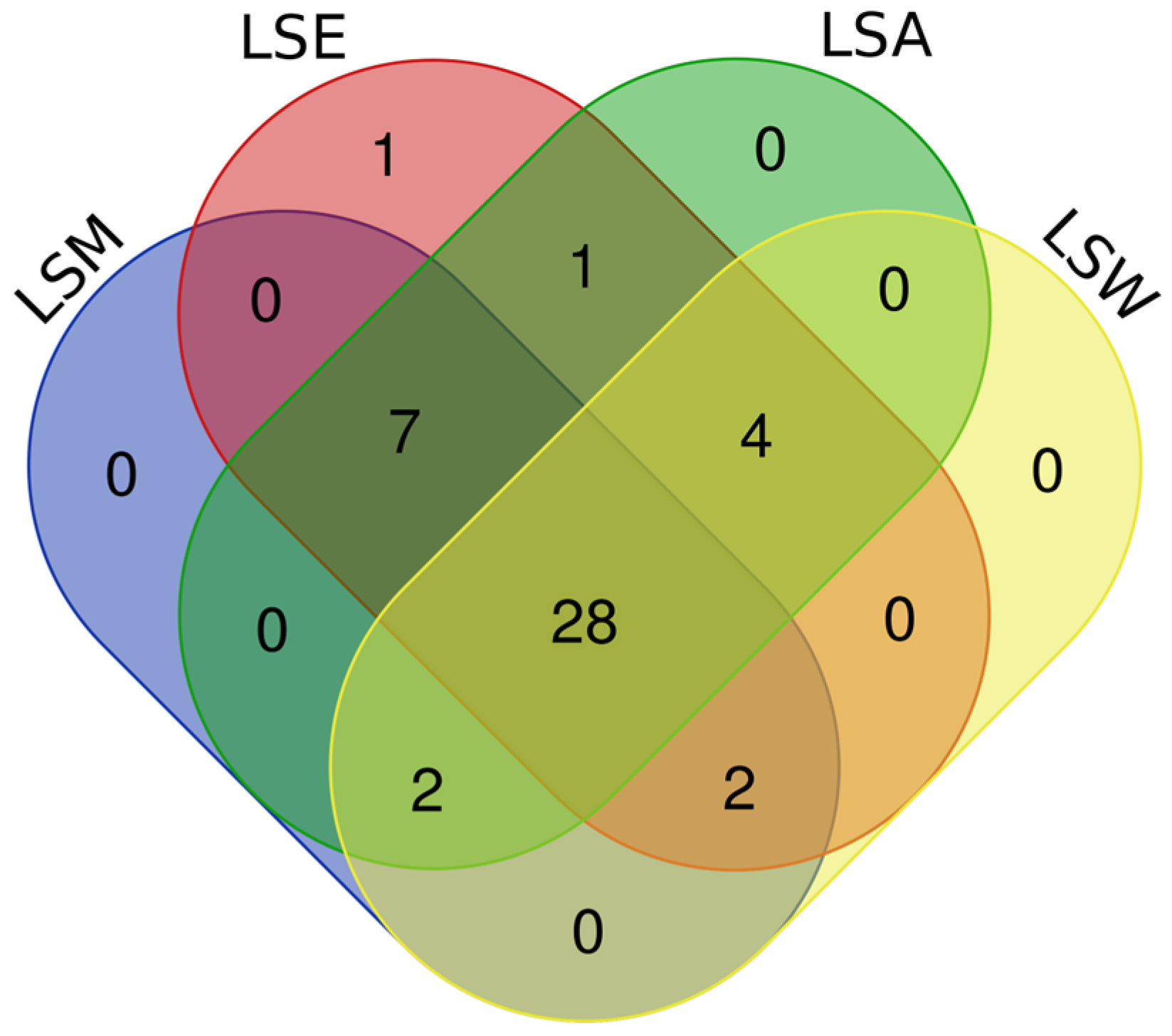
3.1. Metabolomic and Phytochemical Composition of L. shawii Extracts
3.2. Bioactivities of L. shawii Water Extract
3.2.1. Total Phenolic Content of L. shawii Extracts
3.2.2. Antibacterial Activity of L. shawii Water Extract
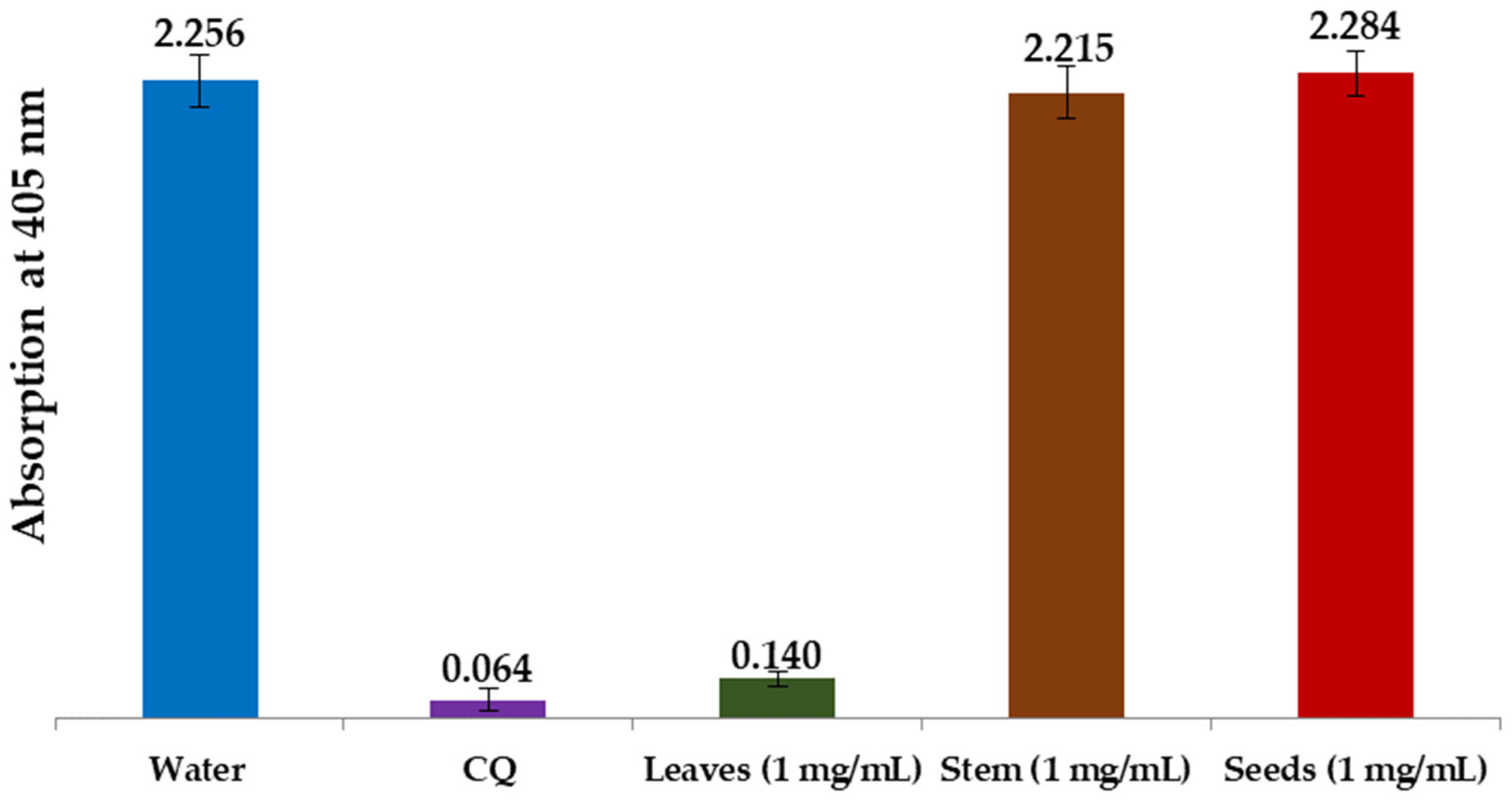
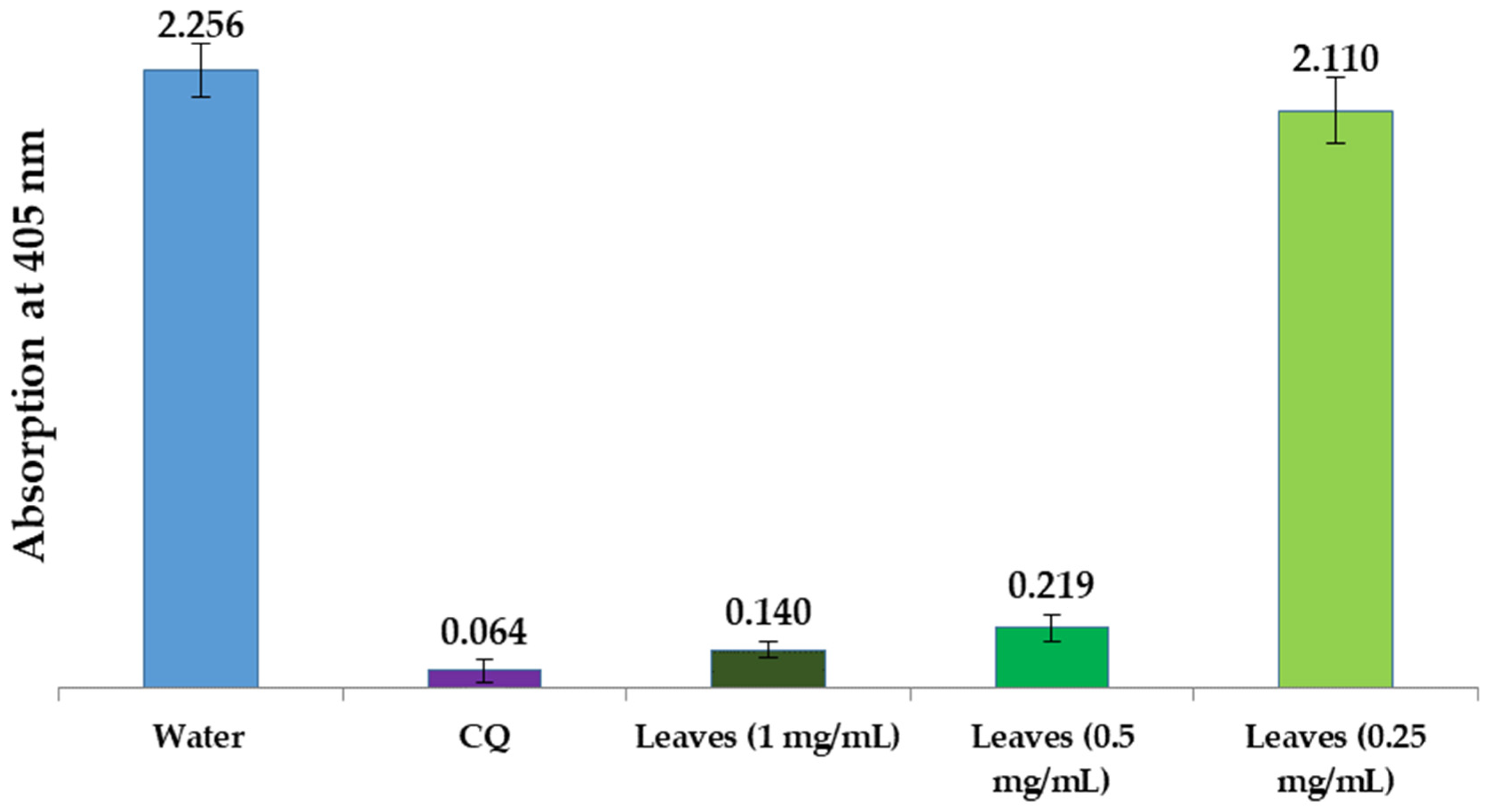
3.2.3. In Vitro Antimalarial Activity of L. shawii Water Extracts
3.2.4. Anti-Inflammatory Activity of L. shawii Water Extracts
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cox, F.E. History of the discovery of the malaria parasites and their vectors. Parasit. Vectors 2010, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- UNICEF. Malaria. Available online: https://data.unicef.org/topic/child-health/malaria/ (accessed on 27 July 2024).
- Elshafie, H.S.; Camele, I.; Mohamed, A.A. A Comprehensive review on the biological, agricultural and pharmaceutical properties of secondary metabolites based-plant origin. Int. J. Mol. Sci. 2023, 24, 3266. [Google Scholar] [CrossRef] [PubMed]
- Moyo, P.; Invernizzi, L.; Mianda, S.M.; Rudolph, W.; Andayi, A.W.; Wang, M.; Crouch, N.R.; Maharaj, V.J. Prioritised identification of structural classes of natural products from higher plants in the expedition of antimalarial drug discovery. Nat. Prod. Bioprospect. 2023, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Lou, J.-S.; Zhao, L.-P.; Huang, Z.-H.; Chen, X.-Y.; Xu, J.-T.; Tai, W.C.-S.; Tsim, K.W.K.; Chen, Y.-T.; Xie, T. Ginkgetin derived from Ginkgo biloba leaves enhances the therapeutic effect of cisplatin via ferroptosis-mediated disruption of the Nrf2/HO-1 axis in EGFR wild-type non-small-cell lung cancer. Phytomedicine 2021, 80, 153370. [Google Scholar] [CrossRef]
- Tu, Y. Artemisinin-A Gift from Traditional Chinese Medicine to the World (Nobel Lecture). Angew. Chem. Int. Ed. 2016, 55, 10210–10226. [Google Scholar] [CrossRef]
- Consortium, M. Artemisinin-Based Combination Therapy (ACT). Available online: https://www.malariaconsortium.org/pages/112.htm (accessed on 17 September 2024).
- Hawkes, J.G.; Lester, R.; Nee, M.; Estrada, N. Solanaceae III: Taxonomy, chemistry, evolution. R. Bot. Gard. 1991, 47, 785–788. [Google Scholar]
- Bosch, C.H. Lycium shawii Roem. & Schult. Record from PROTA4U. Available online: https://www.prota4u.org/database/ (accessed on 13 May 2024).
- Ali, S.S.; El-Zawawy, N.A.; Al-Tohamy, R.; El-Sapagh, S.; Mustafa, A.M.; Sun, J. Lycium shawii Roem. & Schult.: A new bioactive antimicrobial and antioxidant agent to combat multi-drug/pan-drug resistant pathogens of wound burn infections. J. Tradit. Complement. Med. 2020, 10, 13–25. [Google Scholar] [CrossRef]
- Mohammed, H.A.; Ali, H.M.; Qureshi, K.A.; Alsharidah, M.; Kandil, Y.I.; Said, R.; Mohammed, S.A.A.; Al-Omar, M.S.; Rugaie, O.A.; Abdellatif, A.A.H.; et al. Comparative Phytochemical Profile and Biological Activity of Four Major Medicinal Halophytes from Qassim Flora. Plants 2021, 10, 2208. [Google Scholar] [CrossRef]
- Sher, H.; Alyemeni, M.N. Evaluation of anti-diabetic activity and toxic potential of Lycium shawii in animal models. J. Med. Plants Res. 2011, 5, 3387–3395. [Google Scholar]
- Dahech, I.; Farah, W.; Trigui, M.; Hssouna, A.B.; Belghith, H.; Belghith, K.S.; Abdallah, F.B. Antioxidant and antimicrobial activities of Lycium shawii fruits extract. Int. J. Biol. Macromol. 2013, 60, 328–333. [Google Scholar] [CrossRef]
- Gaweesh, A.; Sengab, A.; Osman, H.; Abdou, A. Phytoconstituents, cytotoxic, antioxidant and hepatoprotective activities of the aerial parts of Lycium shawii R. growing in Egypt. Med. Aromat. Plants 2015, 4, 1–7. [Google Scholar]
- Jaafer, Z.A.; Sultan, M.S.; Nasif, Z.N. Bioanalytical applications of some Iraqi wild herbal Awsaj (Lycium shawii) extracts. Egypt. J. Chem. 2023, 66, 151–158. [Google Scholar]
- Abdel-Sattar, E.; Harraz, F.M.; Al-Ansari, S.M.; El-Mekkawy, S.; Ichino, C.; Kiyohara, H.; Otoguro, K.; Omura, S.; Yamada, H. Antiplasmodial and antitrypanosomal activity of plants from the Kingdom of Saudi Arabia. J. Nat. Med. 2009, 63, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Hassan, W.H.; Abdallah, H. UPLC-PDA-ESI-MS/MS analysis, isolation of chemical constituents, cytotoxic, antioxidant, antiviral and antimicrobial activities of the aerial parts of Lycium shawii roem. & schult. Eur. J. Pharm. Med. Res. 2017, 4, 18–29. [Google Scholar]
- Ur Rehman, N.; Halim, S.A.; Khan, M.; Hussain, H.; Yar Khan, H.; Khan, A.; Abbas, G.; Rafiq, K.; Al-Harrasi, A. Antiproliferative and carbonic anhydrase II inhibitory potential of chemical constituents from Lycium shawii and Aloe vera: Evidence from in silico target fishing and in vitro testing. Pharmaceuticals 2020, 13, 94. [Google Scholar] [CrossRef]
- Al-Eisawi, D.M. Field Guide to Wild Flowers of Jordan and Neighbouring Countries; Jordan Press Foundation: Amman, Jordan, 1998. [Google Scholar]
- Rehman, N.U.; Hussain, H.; Al-Riyami, S.A.; Green, I.; Al-Harrasi, A. Chemical constituents isolated from Lycium shawii and their chemotaxonomic significance. Rec. Nat. Prod. 2017, 12, 380–384. [Google Scholar] [CrossRef]
- Ahmed, F.A.; Abdallah, N.M.; Ezz, M.K.; El-Azab, M.M. Seasonal variations and identification of biologically active constituents of Lycium shawii plant roem. & shult.(family solanaceae). Int. J. Innov. Sci. Eng. Technol. 2017, 4, 2348–7968. [Google Scholar]
- Emwas, A.-H.M.; Salek, R.M.; Griffin, J.L.; Merzaban, J. NMR-based metabolomics in human disease diagnosis: Applications, limitations, and recommendations. Metabolomics 2013, 9, 1048–1072. [Google Scholar] [CrossRef]
- Pandohee, J.; Kyereh, E.; Kulshrestha, S.; Xu, B.; Mahomoodally, M.F. Review of the recent developments in metabolomics-based phytochemical research. Crit. Rev. Food Sci. Nutr. 2023, 63, 3734–3749. [Google Scholar] [CrossRef]
- Emwas, A.-H.M.; Al-Rifai, N.; Szczepski, K.; Alsuhaymi, S.; Rayyan, S.; Almahasheer, H.; Jaremko, M.; Brennan, L.; Lachowicz, J.I. You Are What You Eat: Application of Metabolomics Approaches to Advance Nutrition Research. Foods 2021, 10, 1249. [Google Scholar] [CrossRef]
- Emwas, A.H.; Szczepski, K.; Al-Younis, I.; Lachowicz, J.I.; Jaremko, M. Fluxomics–New Metabolomics Approaches to Monitor Metabolic Pathways. Front. Pharmacol. 2022, 13, 805782. [Google Scholar] [CrossRef] [PubMed]
- Gorrochategui, E.; Jaumot, J.; Lacorte, S.; Tauler, R. Data analysis strategies for targeted and untargeted LC-MS metabolomic studies: Overview and workflow. TrAC Trends Anal. Chem. 2016, 82, 425–442. [Google Scholar] [CrossRef]
- Troisi, J. Metabolomics Perspectives: From Theory to Practical Application; Academic Press: Cambridge, MA, USA, 2022. [Google Scholar]
- Emwas, A.-H.M. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Metabonomics Methods Protoc. 2015, 1277, 161–193. [Google Scholar]
- Scalbert, A.; Brennan, L.; Fiehn, O.; Hankemeier, T.; Kristal, B.S.; van Ommen, B.; Pujos-Guillot, E.; Verheij, E.; Wishart, D.; Wopereis, S. Mass-spectrometry-based metabolomics: Limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics 2009, 5, 435–458. [Google Scholar] [CrossRef]
- Emwas, A.-H.M.; Al-Talla, Z.A.; Yang, Y.; Kharbatia, N.M. Gas Chromatography–Mass Spectrometry of Biofluids and Extracts. In Metabonomics: Methods and Protocols; Bjerrum, J.T., Ed.; Springer: New York, NY, USA, 2015; pp. 91–112. [Google Scholar]
- Johnson, J.; Mani, J.; Naiker, M. Development and Validation of a 96-Well Microplate Assay for the Measurement of Total Phenolic Content in Ginger Extracts. Food Anal. Methods 2021, 15, 413–420. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Deharo, E.; Garcıa, R.; Oporto, P.; Gimenez, A.; Sauvain, M.; Jullian, V.; Ginsburg, H. A non-radiolabelled ferriprotoporphyrin IX biomineralisation inhibition test for the high throughput screening of antimalarial compounds. Exp. Parasitol. 2002, 100, 252–256. [Google Scholar] [CrossRef]
- Park, J.-W.; Kwon, O.-K.; Yuniato, P.; Marwoto, B.; Lee, J.; Oh, S.-R.; Kim, J.-H.; Ahn, K.-S. Amelioration of an LPS-induced inflammatory response using a methanolic extract of Lagerstroemia ovalifolia to suppress the activation of NF-κB in RAW264. 7 macrophages. Int. J. Mol. Med. 2016, 38, 482–490. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute: Wayne, MI, USA, 2008. [Google Scholar]
- Al-Nemi, R.; Makki, A.A.; Sawalha, K.; Hajjar, D.; Jaremko, M. Untargeted Metabolomic Profiling and Antioxidant Capacities of Different Solvent Crude Extracts of Ephedra foeminea. Metabolites 2022, 12, 451. [Google Scholar] [CrossRef]
- Wolfender, J.-L.; Marti, G.; Thomas, A.; Bertrand, S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr. A 2015, 1382, 136–164. [Google Scholar] [CrossRef]
- Shukri, S.Z.M.; Daud, N.M.A.N.; Bakar, A.R.A.; Arsad, S.S.; Zainudin, M.A.M. Profiling of Bioactive Compounds and Bioactivity of the Kenaf (Hibiscus cannabinus L.) Leaf Extract. In Proceedings of the Emerging Technologies for Future Sustainability, Singapore, 1 September 2023; pp. 415–427. [Google Scholar]
- Kaur, N.; Kumar, R.; Alhan, S.; Sharma, H.; Singh, N.; Yogi, R.; Chhokar, V.; Beniwal, V.; Kumar Ghosh, M.; Kumar Chandraker, S.; et al. Lycium shawii mediated green synthesis of silver nanoparticles, characterization and assessments of their phytochemical, antioxidant, antimicrobial properties. Inorg. Chem. Commun. 2024, 159, 111735. [Google Scholar] [CrossRef]
- Antony, A.; Farid, M. Effect of Temperatures on Polyphenols during Extraction. Appl. Sci. 2022, 12, 2107. [Google Scholar] [CrossRef]
- Gamage, D.; Abeysinghe, D.; Wijesekara, R.; Prathapasinghe, G.; Dharmadasa, R.; Someya, T. Assessment of Phytochemical Contents and Total Antioxidant Capacity of Five Medicinal Plants with Cosmetic Potential under Three Different Drying Methods. World J. Agric. Res. 2021, 9, 24–28. [Google Scholar] [CrossRef]
- Zhang, C.; Suen, C.L.-C.; Yang, C.; Quek, S.Y. Antioxidant capacity and major polyphenol composition of teas as affected by geographical location, plantation elevation and leaf grade. Food Chem. 2018, 244, 109–119. [Google Scholar] [CrossRef]
- Mamede, L.; Ledoux, A.; Jansen, O.; Frédérich, M. Natural phenolic compounds and derivatives as potential antimalarial agents. Planta Medica 2020, 86, 585–618. [Google Scholar] [CrossRef]
- Freije García, F.E.; Bravo, S.C.; García Liñares, G. Chapter 6–Promising role of phenolic acids as antimalarial and antiviral drugs. In Advancement of Phenolic Acids in Drug Discovery; Kumar, N., Goel, N., Gandara, J.S., Eds.; Academic Press: Cambridge, MA, USA, 2024; pp. 211–254. [Google Scholar]
- El-Amier, Y.A.; Alghanem, S.M.; Wafa’a, A.-T.A.; Elsayed, A.; Tokmina-Lukaszewska, M. Lycium shawii Methanolic Extract: Chemical Characterization, and its Biological Implications Supported by Computational Studies. SSRN 2024. preprint. [Google Scholar] [CrossRef]
- Tayel, A.A.; Shaban, S.M.; Moussa, S.H.; Elguindy, N.M.; Diab, A.M.; Mazrou, K.E.; Ghanem, R.A.; El-Sabbagh, S.M. Bioactivity and application of plant seeds’ extracts to fight resistant strains of Staphylococcus aureus. Ann. Agric. Sci. 2018, 63, 47–53. [Google Scholar] [CrossRef]
- Kumar, S.; Guha, M.; Choubey, V.; Maity, P.; Bandyopadhyay, U. Antimalarial drugs inhibiting hemozoin (β-hematin) formation: A mechanistic update. Life Sci. 2007, 80, 813–828. [Google Scholar] [CrossRef]
- Goldberg, D.E.; Slater, A.; Cerami, A.; Henderson, G.B. Hemoglobin degradation in the malaria parasite Plasmodium falciparum: An ordered process in a unique organelle. Proc. Natl. Acad. Sci. USA 1990, 87, 2931–2935. [Google Scholar] [CrossRef]
- Egan, T.J.; Ncokazi, K.K. Quinoline antimalarials decrease the rate of β-hematin formation. J. Inorg. Biochem. 2005, 99, 1532–1539. [Google Scholar] [CrossRef]
- Egan, T.J.; Ross, D.C.; Adams, P.A. Quinoline anti-malarial drugs inhibit spontaneous formation of β-haematin (malaria pigment). FEBS Lett. 1994, 352, 54–57. [Google Scholar] [CrossRef] [PubMed]
- Usman, M.A.; Ibrahim, F.B.; Mohammed, H.-O.; Awogbamila, S.O.; Idris, U.A.; Suleiman, M.A. Antiplasmodial Activity of β-Ionone and the Effect of the Compound on Amelioration of Anaemia and Oxidative Organ Damage in Mice Infected with Plasmodium berghei. Acta Parasitol. 2024, 69, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Chikambwe, V.M.; Mubambe, P.; Maseka, K.; Banda, L. Therapeutic Potential of Combretum Mossambicense Extracts Against P. Falciparum Parasite. Res. Sq. 2024. preprint. [Google Scholar] [CrossRef]
- Yusuf, A.J.; Abdullahi, M.I.; Nasir, I.; Yunusa, A.; Alebiosu, C.O.; Muhammad, A.A. Isolation and Characterization of Prophylactic Antimalarial Agents from Ochna kibbiensis Leaves. Drugs Drug Candidates 2023, 2, 37–51. [Google Scholar] [CrossRef]
- Abbas, F.A.; Al-massarany, S.M.; Khan, S.; Al-howiriny, T.A.; Mossa, J.S.; Abourashed, E.A. Phytochemical and biological studies on Saudi Commiphora opobalsamum L. Natural Product. Res. 2007, 21, 383–391. [Google Scholar] [CrossRef]
- Olaosebikan, O.J.; Faboro, E.O.; Fadare, O.A.; Ikotun, A.A. In silico Investigation of the Antimalarial Activity of some Selected Alkaloids and Terpenoids Present in the Aerial Parts of Andrographis paniculata. Trop. J. Nat. Product. Res. 2023, 7, 3787–3799. [Google Scholar]
- Banzouzi, J.T.; Soh, P.N.; Ramos, S.; Toto, P.; Cavé, A.; Hemez, J.; Benoit-Vical, F. Samvisterin, a new natural antiplasmodial betulin derivative from Uapaca paludosa (Euphorbiaceae). J. Ethnopharmacol. 2015, 173, 100–104. [Google Scholar] [CrossRef]
- Ja’afar, N.S.A.; Zin, N.N.I.N.M.Z.; Mohamad, F.S.; Abu-Bakar, N. A Polyphenol, Pyrogallol Changes the Acidic Ph of The Digestive Vacuole of Plasmodium Falciparum. Life Sci. Med. Biomed. 2021, 5, 82. [Google Scholar] [CrossRef]
- Chaniad, P.; Techarang, T.; Phuwajaroanpong, A.; Plirat, W.; Viriyavejakul, P.; Septama, A.W.; Punsawad, C. Antimalarial efficacy and toxicological assessment of medicinal plant ingredients of Prabchompoothaweep remedy as a candidate for antimalarial drug development. BMC Complement. Med. Ther. 2023, 23, 12. [Google Scholar] [CrossRef]
- Oliveira, C.B.S.; Meurer, Y.S.R.; Oliveira, M.G.; Medeiros, W.M.T.Q.; Silva, F.O.N.; Brito, A.C.F.; Pontes, D.D.L.; Andrade-Neto, V.F. Comparative Study on the Antioxidant and Anti-Toxoplasma Activities of Vanillin and Its Resorcinarene Derivative. Molecules 2014, 19, 5898–5912. [Google Scholar] [CrossRef]
- Fordjour, P.A.; Adjimani, J.P.; Asare, B.; Duah-Quashie, N.O.; Quashie, N.B. Anti-malarial activity of phenolic acids is structurally related. Res. Sq. 2020. preprint. [Google Scholar] [CrossRef]
- Narayanaswamy, R.; Wai, L.K.; Ismail, I.S. Natural compounds as inhibitors of Plasmodium falciparum enoyl-acyl carrier protein reductase (PfENR): An in silico study. J. Chosun Nat. Sci. 2017, 10, 1–6. [Google Scholar] [CrossRef][Green Version]
- Nagarajan, K.; Ankita, S.; Unnati, B.; Meghna, T.; Parul, G.; Roma, G.; Sanjeev, C.; Garima, K. Bioassay Guided Fractionation And Identification of Boundary Fractions of Arnebia nobilis: Search for Antimalarial Potency in Vitro. Pharmacol. Online 2020, 2, 129–145. [Google Scholar]
- Memariani, H.; Memariani, M.; Ghasemian, A. Quercetin as a Promising Antiprotozoan Phytochemical: Current Knowledge and Future Research Avenues. BioMed Res. Int. 2024, 2024, 7632408. [Google Scholar] [CrossRef]
- Ganesh, D.; Fuehrer, H.-P.; Starzengrüber, P.; Swoboda, P.; Khan, W.A.; Reismann, J.A.B.; Mueller, M.S.K.; Chiba, P.; Noedl, H. Antiplasmodial activity of flavonol quercetin and its analogues in Plasmodium falciparum: Evidence from clinical isolates in Bangladesh and standardized parasite clones. Parasitol. Res. 2012, 110, 2289–2295. [Google Scholar] [CrossRef]
- Abu-Lafi, S.; Akkawi, M.; Al-Rimawi, F.; Abu-Remeleh, Q.; Lutgen, P. Morin, quercetin, catechin and quercitrin as novel natural antimalarial candidates. Pharm. Pharmacol. Int. J. 2020, 8, 184–190. [Google Scholar] [CrossRef]
- Bhatt, D.; Kumar, S.; Kumar, P.; Bisht, S.; Kumar, A.; Maurya, A.K.; Pal, A.; Bawankule, D.U. Rutin ameliorates malaria pathogenesis by modulating inflammatory mechanism: An in vitro and in vivo study. Inflammopharmacology 2022, 30, 159–171. [Google Scholar] [CrossRef]
- Al-Huqail, A.A.; Bekhit, A.A.; Ullah, H.; Ayaz, M.; Mostafa, N.M. Antimalarial and Antileishmanial Flavonoids from Calendula officinalis Flowers. Agronomy 2023, 13, 2765. [Google Scholar] [CrossRef]
- Somsak, V.; Damkaew, A.; Onrak, P. Antimalarial Activity of Kaempferol and Its Combination with Chloroquine in Plasmodium berghei Infection in Mice. J. Pathog. 2018, 2018, 3912090. [Google Scholar] [CrossRef]
- Maulana Yusuf, A.; Nia, Y.; Afiat, B.; Anas, S. Antimalaria Activities of Several Active Compounds from Medicinal Plants. Pharmacogn. J. 2022, 14, 245–252. [Google Scholar]
- Marín, C.; Boutaleb-Charki, S.; Díaz, J.G.; Huertas, O.; Rosales, M.J.; Pérez-Cordon, G.; Guitierrez-Sánchez, R.; Sánchez-Moreno, M. Antileishmaniasis Activity of Flavonoids from Consolida oliveriana. J. Nat. Prod. 2009, 72, 1069–1074. [Google Scholar] [CrossRef] [PubMed]
- Makati, A.C.; Ananda, A.N.; Putri, J.A.; Amellia, S.F.; Setiawan, B. Molecular docking of ethanol extracts of katuk leaf (Sauropus androgynus) on functional proteins of severe acute respiratory syndrome coronavirus 2. S. Afr. J. Bot. 2022, 149, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Vargas, S.; Ndjoko Ioset, K.; Hay, A.E.; Ioset, J.R.; Wittlin, S.; Hostettmann, K. Screening medicinal plants for the detection of novel antimalarial products applying the inhibition of β-hematin formation. J. Pharm. Biomed. Anal. 2011, 56, 880–886. [Google Scholar] [CrossRef] [PubMed]
- Akkawi, M.; Aburemeleh, Q.; Jaber, S.; Qutob, M.; Lutgen, P. The effect of Artemisia sieberi extracts on the Formation of β-Hematin. Br. J. Pharmacol. Toxicol. 2014, 5, 49–54. [Google Scholar] [CrossRef]
- Ndjonka, D.; Bergmann, B.; Agyare, C.; Zimbres, F.M.; Lüersen, K.; Hensel, A.; Wrenger, C.; Liebau, E. In vitro activity of extracts and isolated polyphenols from West African medicinal plants against Plasmodium falciparum. Parasitol. Res. 2012, 111, 827–834. [Google Scholar] [CrossRef]
- Yuan, L.; Zhang, F.; Shen, M.; Jia, S.; Xie, J. Phytosterols Suppress Phagocytosis and Inhibit Inflammatory Mediators via ERK Pathway on LPS-Triggered Inflammatory Responses in RAW264.7 Macrophages and the Correlation with Their Structure. Foods 2019, 8, 582. [Google Scholar] [CrossRef]
- Hunthayung, K.; Bhawamai, S. Polyphenol compounds of freeze-dried Moringa oleifera Lam pods and their anti-inflammatory effects on RAW 264.7 macrophages stimulated with lipopolysaccharide. Bioact. Compd. Health Dis. 2024, 7, 185–198. [Google Scholar] [CrossRef]
- Fernando, I.P.S.; Sanjeewa, K.K.A.; Samarakoon, K.W.; Lee, W.W.; Kim, H.S.; Jeon, Y.J. Squalene isolated from marine macroalgae Caulerpa racemosa and its potent antioxidant and anti-inflammatory activities. J. Food Biochem. 2018, 42, e12628. [Google Scholar] [CrossRef]
- Kim, H.N.; Kim, J.D.; Park, S.B.; Son, H.-J.; Park, G.H.; Eo, H.J.; Kim, H.-S.; Jeong, J.B. Anti-inflammatory activity of the extracts from Rodgersia podophylla leaves through activation of Nrf2/HO-1 pathway, and inhibition of NF-κB and MAPKs pathway in mouse macrophage cells. Inflamm. Res. 2020, 69, 233–244. [Google Scholar] [CrossRef]
- Kwon, K.W.; Jang, W.Y.; Kim, J.W.; Noh, J.K.; Yi, D.-K.; Cho, J.Y. Anti-Inflammatory Effect of Meriania hexamera Sprague by Targeting Syk Kinase in NF-κB Signaling. Plants 2023, 12, 3044. [Google Scholar] [CrossRef]
- Kim, M.E.; Na, J.Y.; Park, Y.-D.; Lee, J.S. Anti-Neuroinflammatory Effects of Vanillin Through the Regulation of Inflammatory Factors and NF-κB Signaling in LPS-Stimulated Microglia. Appl. Biochem. Biotechnol. 2019, 187, 884–893. [Google Scholar] [CrossRef] [PubMed]
- Costantini, E.; Sinjari, B.; Falasca, K.; Reale, M.; Caputi, S.; Jagarlapodii, S.; Murmura, G. Assessment of the Vanillin Anti-Inflammatory and Regenerative Potentials in Inflamed Primary Human Gingival Fibroblast. Mediat. Inflamm. 2021, 2021, 5562340. [Google Scholar] [CrossRef] [PubMed]
- Winter, A.N.; Brenner, M.C.; Punessen, N.; Snodgrass, M.; Byars, C.; Arora, Y.; Linseman, D.A. Comparison of the Neuroprotective and Anti-Inflammatory Effects of the Anthocyanin Metabolites, Protocatechuic Acid and 4-Hydroxybenzoic Acid. Oxidative Med. Cell. Longev. 2017, 2017, 6297080. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Dai, Y.; Xu, M.; Zhang, C.; Ma, Y.; Gao, P.; Teng, M.; Jiao, K.; Huang, G.; Zhang, J.; et al. The Attenuation of 14-3-3 is Involved in the Caffeic Acid-Blocked Lipopolysaccharide-Stimulated Inflammatory Response in RAW264.7 Macrophages. Inflammation 2017, 40, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Búfalo, M.C.; Ferreira, I.; Costa, G.; Francisco, V.; Liberal, J.; Cruz, M.T.; Lopes, M.C.; Batista, M.T.; Sforcin, J.M. Propolis and its constituent caffeic acid suppress LPS-stimulated pro-inflammatory response by blocking NF-κB and MAPK activation in macrophages. J. Ethnopharmacol. 2013, 149, 84–92. [Google Scholar] [CrossRef]
- Lampiasi, N.; Montana, G. The molecular events behind ferulic acid mediated modulation of IL-6 expression in LPS-activated Raw 264.7 cells. Immunobiology 2016, 221, 486–493. [Google Scholar] [CrossRef]
- Kim, N.-H.; Park, H.-J.; Lee, E.-H.; Cho, E.-B.; Kang, I.-K.; Cho, Y.-J. The inflammatory activity of purified-ferulic acid from Tetragonia tetragonioides. J. Appl. Biol. Chem. 2019, 62, 239–246. [Google Scholar] [CrossRef]
- Park, C.M.; Yoon, H.-S. Chlorogenic Acid as a Positive Regulator in LPS-PG-Induced Inflammation via TLR4/MyD88-Mediated NF-κB and PI3K/MAPK Signaling Cascades in Human Gingival Fibroblasts. Mediat. Inflamm. 2022, 2022, 2127642. [Google Scholar] [CrossRef]
- Hwang, S.J.; Kim, Y.-W.; Park, Y.; Lee, H.-J.; Kim, K.-W. Anti-inflammatory effects of chlorogenic acid in lipopolysaccharide-stimulated RAW 264.7 cells. Inflamm. Res. 2014, 63, 81–90. [Google Scholar] [CrossRef]
- Lv, P.; Han, P.; Cui, Y.; Chen, Q.; Cao, W. Quercetin attenuates inflammation in LPS-induced lung epithelial cells via the Nrf2 signaling pathway. Immun. Inflamm. Dis. 2024, 12, e1185. [Google Scholar] [CrossRef]
- Tang, J.; Diao, P.; Shu, X.; Li, L.; Xiong, L. Quercetin and Quercitrin Attenuates the Inflammatory Response and Oxidative Stress in LPS-Induced RAW264.7 Cells: In Vitro Assessment and a Theoretical Model. Bio. Med. Res. Int. 2019, 2019, 7039802. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Sahu, K.; Kapil, L.; Singh, C.; Singh, A. Quercetin ameliorates lipopolysaccharide-induced neuroinflammation and oxidative stress in adult zebrafish. Mol. Biol. Rep. 2022, 49, 3247–3258. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Guo, Y.; Chang, Y.; Zhao, J.; Cui, C.; Liu, M. Dose-effect relationship on anti-inflammatory activity on LPS induced RAW 264.7 cells and antioxidant activity of rutin in vitro. Acta Pol. Pharm. Drug Res. 2019, 76, 511–522. [Google Scholar] [CrossRef]
- Lee, M.-H.; Jeong, J.-H.; Jeong, M.-S.; Chang, S.-H.; Her, E. Anti-inflammatory function of the sophora japonica extract rutin: The inhibitory effect of rutin of korean sophora japonica on the productions of NO and TNF-alpha from mouse peritoneal macrophages. Korean J. Med. Crop Sci. 2010, 18, 105–112. [Google Scholar]
- Choi, S.Y.; Choi, J.Y.; Lee, J.M.; Lee, S.; Cho, E.J. Tartary buckwheat on nitric oxide-induced inflammation in RAW264. 7 macrophage cells. Food Funct. 2015, 6, 2664–2670. [Google Scholar] [CrossRef]
- Tian, C.; Shao, Y.; Jin, Z.; Liang, Y.; Li, C.; Qu, C.; Sun, S.; Cui, C.; Liu, M. The protective effect of rutin against lipopolysaccharide induced acute lung injury in mice based on the pharmacokinetic and pharmacodynamic combination model. J. Pharm. Biomed. Anal. 2022, 209, 114480. [Google Scholar] [CrossRef]
- Huang, G.-J.; Deng, J.-S.; Huang, S.-S.; Wang, S.-Y.; Chang, Y.-S.; Kuo, Y.-H. Bioassay Guided Isolation and Identification of Anti-inflammatory Active Compounds from the Root of Ficus formosana. J. Agric. Food Chem. 2013, 61, 11008–11015. [Google Scholar] [CrossRef]
- Hämäläinen, M.; Nieminen, R.; Vuorela, P.; Heinonen, M.; Moilanen, E. Anti-inflammatory effects of flavonoids: Genistein, kaempferol, quercetin, and daidzein inhibit STAT-1 and NF-kappaB activations, whereas flavone, isorhamnetin, naringenin, and pelargonidin inhibit only NF-kappaB activation along with their inhibitory effect on iNOS expression and NO production in activated macrophages. Mediat. Inflamm. 2007, 2007, 45673. [Google Scholar] [CrossRef]
- Qi, F.; Sun, J.-h.; Yan, J.-q.; Li, C.-m.; Lv, X.-c. Anti-inflammatory effects of isorhamnetin on LPS-stimulated human gingival fibroblasts by activating Nrf2 signaling pathway. Microb. Pathog. 2018, 120, 37–41. [Google Scholar] [CrossRef]
- Hwang, Y.-J.; Lee, S.J.; Park, J.-Y.; Chun, W.; Nam, S.-J.; Park, J.M.; Park, S.C.; Choi, D.H.; Kang, C.D. Apocynin Suppresses Lipopolysaccharide-Induced Inflammatory Responses Through the Inhibition of MAP Kinase Signaling Pathway in RAW264.7 Cells. Drug Dev. Res. 2016, 77, 271–277. [Google Scholar] [CrossRef]
- Boshtam, M.; Kouhpayeh, S.; Amini, F.; Azizi, Y.; Najaflu, M.; Shariati, L.; Khanahmad, H. Anti-inflammatory effects of apocynin: A narrative review of the evidence. All Life 2021, 14, 997–1010. [Google Scholar] [CrossRef]
- Kang, M.J.; Choi, W.; Yoo, S.H.; Nam, S.W.; Shin, P.G.; Kim, K.K.; Kim, G.D. Modulation of Inflammatory Pathways and Adipogenesis by the Action of Gentisic Acid in RAW 264.7 and 3T3-L1 Cell Lines. J. Microbiol. Biotechnol. 2021, 31, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lee, H.-S.; Kim, S.-H.; Moon, B.; Lee, C. Antioxidant and Anti-inflammatory Activities of Methanol Extracts of Tremella fuciformis and Its Major Phenolic Acids. J. Food Sci. 2014, 79, C460–C468. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lin, J.; Ding, X.; Xuan, J.; Hu, Z.; Wu, D.; Zhu, X.; Feng, Z.; Ni, W.; Wu, A. The protective effect of sinapic acid in osteoarthritis: In vitro and in vivo studies. J. Cell. Mol. Med. 2019, 23, 1940–1950. [Google Scholar] [CrossRef]
- Huang, X.; Pan, Q.; Mao, Z.; Zhang, R.; Ma, X.; Xi, Y.; You, H. Sinapic Acid Inhibits the IL-1β-Induced Inflammation via MAPK Downregulation in Rat Chondrocytes. Inflammation 2018, 41, 562–568. [Google Scholar] [CrossRef]
- Kong, Y.-H.; Lee, Y.-C.; Choi, S.-Y. Neuroprotective and anti-inflammatory effects of phenolic compounds in Panax ginseng CA Meyer. J. Ginseng Res. 2009, 33, 111–114. [Google Scholar]
- D’Andrea, G.; D’Arrigo, A.; Facchinetti, F.; Del Giudice, E.; Colavito, D.; Bernardini, D.; Leon, A. Octopamine, unlike other trace amines, inhibits responses of astroglia-enriched cultures to lipopolysaccharide via a β-adrenoreceptor-mediated mechanism. Neurosci. Lett. 2012, 517, 36–40. [Google Scholar] [CrossRef]
- Steiner, L.; Gold, M.; Mengel, D.; Dodel, R.; Bach, J.-P. The endogenous α7 nicotinic acetylcholine receptor antagonist kynurenic acid modulates amyloid-β-induced inflammation in BV-2 microglial cells. J. Neurol. Sci. 2014, 344, 94–99. [Google Scholar] [CrossRef]
- Kongpuckdee, S.; Phadoongsombut, N.; Ardhanwanich, S.; Mahattanadul, S. Role of Scopoletin Containing in an Aqueous Morinda citrifolia Fruit Extract on the Gastric Mucosal Integrity and Gastric Ulcer Healing Process. Open Access J. 2024, 13, 160. [Google Scholar]
- Kang, T.-H.; Pae, H.-O.; Jeong, S.-J.; Yoo, J.-C.; Choi, B.-M.; Jun, C.-D.; Chung, H.-T.; Miyamoto, T.; Higuchi, R.; Kim, Y.-C. Scopoletin: An inducible nitric oxide synthesis inhibitory active constituent from Artemisia feddei. Planta Medica 1999, 65, 400–403. [Google Scholar] [CrossRef]
- Mukherjee, S.; Pal, M. Quinolines: A new hope against inflammation. Drug Discov. Today 2013, 18, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Ding, H.; Zhong, L.; Xin, W.; Yi, X.; Fang, L. Spectrum-Effect Relationship-Based Strategy Combined with Molecular Docking to Explore Bioactive Flavonoids from Sceptridium ternatum. Molecules 2022, 27, 5698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yang, L.; Gong, Q.; Liu, X.; Deng, F. Metabolic profiling of different parts of Physalis alkekengi L. var. franchetii (Mast.) Makino based on UPLC-Q-Orbitrap-HRMS coupled with bioactivity assays. J. Pharm. Biomed. Anal. 2024, 249, 116388. [Google Scholar] [CrossRef]
- Alkuwari, A.; Al-Naemi, M.; Vito, P.; Stilo, R.; Ahmed, T.A.; Al Naemi, H. Biological activities of Lycium shawii leaves extract. Int. J. Pharm. Biol. Arch 2010, 3, 697–700. [Google Scholar]
- Popa, G.L.; Popa, M.I. Recent Advances in Understanding the Inflammatory Response in Malaria: A Review of the Dual Role of Cytokines. J. Immunol. Res. 2021, 2021, 7785180. [Google Scholar] [CrossRef]









| Botanical Name | Lycium shawii Roem & Schult |
|---|---|
| Local name | Awsaj |
| Family | Solanaceae |
| Habitat | Semi-desert, extreme desert, and arid environment |
| Distribution | Africa, Middle East, Indian subcontinent |
| Medicinal uses | To treat mouth ulcers, eye ailments, cough, backache, jaundice, constipation, and stomach ache; to act as a hypotensive in humans; and to cure tick fever in livestock |
| References | [8,9,19] |
| Tentative Phytochemicals | Method | Extract | ||
|---|---|---|---|---|
| Isoprenoids | (a) C30 isoprenoids | Squalene | GC-MS | A, E, M |
| Phenols | (a) Methoxyphenols | 4-Hydroxy-3-methoxyphenylglycol | GC-MS | A, E, M, W |
| (b) Benzenetriols | Pyrogallol | GC-MS | A, E, W | |
| Phenylpropanoids | (a) Cinnamic acids | 4-Hydroxyphenylacetic acid | GC-MS | A, E, M, W |
| Caffeic acid | GC-MS | A, E, M, W | ||
| Benzeneacetic acid | GC-MS | A, E, M, W | ||
| Chlorogenic acid | GC-MS, UHPLC-ESI-MS (−) | A, E, M, W | ||
| Ferulic acid | GC-MS, UHPLC-ESI-MS (+) | A, E, M, W | ||
| Cinnamic acid | GC-MS | A, E, M | ||
| Sinapic acid | GC-MS | A, E, M | ||
| p-Coumaric acid | GC-MS | E, M, W | ||
| Phenyllactic acid | UHPLC-ESI-MS (−) | A, E, M, W | ||
| Vanillylmandelic acid | GC-MS | A, E, W | ||
| Flavonoids | (a) Flavonols | Quercetin | GC-MS, UHPLC-ESI-MS (+) | A, E, M, W |
| Quercetin 3-O-rhamnoside-7-O-glucoside | UHPLC-ESI-MS (+) | A, E, M, W | ||
| Quercetin-3β-D-glucoside | UHPLC-ESI-MS (+) | A, E, M, W | ||
| Quercitrin | UHPLC-ESI-MS (+) | A, E, M, W | ||
| Kaempferol | UHPLC-ESI-MS (+) | A, E, M, W | ||
| Isorhamnetin | UHPLC-ESI-MS (+) | A, E, M, W | ||
| Rutin | UHPLC-ESI-MS (−) | A, E, M, W | ||
| Trifolin | UHPLC-ESI-MS (+) | A, E, M, W | ||
| (b) Coumarins | 7-hydroxy-6-methoxy-2H-chromen-2-one | UHPLC-ESI-MS (+) | A, E | |
| Alkaloids | (a) Tryptophan alkaloids | Tryptamine | GC-MS | A, E, M, W |
| Indole-3-acrylic acid | UHPLC-ESI-MS (+) | A, E, M, W | ||
| Indole-3-carboxylic acid | GC-MS | A, E, M | ||
| (b) Anthranilic acid alkaloids | Kynurenic acid | GC-MS, UHPLC-ESI-MS (+) | A, E, M, W | |
| (c) Tyrosine alkaloids | Octopamine | GC-MS | A, E, M, W | |
| Norepinephrine | UHPLC-ESI-MS (+) | A, E, M, W | ||
| (d) Pyridine alkaloids | Nicotinic acid | GC-MS | A, E, M, W | |
| Nicotinamide | UHPLC-ESI-MS (+) | A, E | ||
| Picolinic acid | UHPLC-ESI-MS (+) | A, E, M, W | ||
| (e) Quinazoline alkaloids | Quinoline | UHPLC-ESI-MS (+) | A, M, W | |
| Benezenoids | (a) Hydroxybenzoic acids | 4-Hydroxybenzoic acid | GC-MS | A, E, M, W |
| Benzoic acid | GC-MS | A, E, M, W | ||
| Vanillic acid | GC-MS | A, E, M, W | ||
| Benzyl salicylate | GC-MS | E, M, W | ||
| Protocatechuic acid | GC-MS | A, E, M | ||
| Gentisic acid | UHPLC-ESI-MS (−) | A, E, M, W | ||
| Vanillin | GC-MS | A, E, W | ||
| Apocynin | UHPLC-ESI-MS (+) | A, E, M, W | ||
| Quinic acid | UHPLC-ESI-MS (−) | A, M, W | ||
| Syringic acid | GC-MS | A, E, W | ||
| (b) Acylaminobenzoic acids | 4-Acetamidobenzoic acid | UHPLC-ESI-MS (−) | A, E, M | |
| Quinones | (a) Quinones | alpha-Tocopherol | GC-MS | A, E, M |
| Sterols | (a) Stigmasterols | beta-Sitosterol | GC-MS | E |
| Sample | MIC (μg/mL) | |||
|---|---|---|---|---|
| P. aeruginosa | MRSA | A. baumannii | K. pneumoniae | |
| LSW extract | 500 | 500 | 500 | 500 |
| Vancomycin | 62.5 | 1.95 | 125 | 62.5 |
| GC-MS | UHPLC-ESI-MS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | Phytochemical Name | Extract | RT (min) | Calc. RI | Delta RI | RT (min) | Annot. DeltaMass (ppm) | m/z | Adduct | Antimalarial Activity |
| 1 | beta-Sitosterol | E | 26.24 | 3344 | 20 | – | – | – | – | [51] |
| 2 | Syringic acid | A, E, W | 15.22 | 1889 | 6 | – | – | – | – | [52,53] |
| 3 | Squalene | A, E, M | 22.845 | 2811 | 0 | – | – | – | – | [54] |
| 4 | Pyrogallol | A, E, W | 12.035 | 1596 | 78 | – | – | – | – | [55,56] |
| 5 | Vanillin | A, E, W | 12.607 | 1647 | 14 | [57,58] | ||||
| 6 | Protocatechuic acid | A, E, M | 14.422 | 1813 | 16 | – | – | – | – | [59] |
| 7 | Caffeic acid | A, E, M, W | 17.5 | 2133 | 17 | – | – | – | – | [60] |
| 8 | Ferulic acid | A, E, M, W | 17.139 | 2092 | 160 | – | – | – | – | [60] |
| 9 | Chlorogenic acid | A, E, M, W | 25.112 | 3106 | 3 | 4.625 | −0.93 | 353.08751 | [M−H] −1 | [60,61] |
| 10 | Quercetin | A, E, M, W | 25.252 | 3177 | 43 | 5.509 | 0.32 | 303.05004 | [M+H]+1 | [62,63,64,65] |
| 11 | Quercitrin | A, E, M, W | – | – | – | 5.745 | 0.34 | 449.10799 | [M+H]+1 | [65] |
| 12 | Rutin | A, E, M, W | – | – | – | 5.307 | 1.54 | 609.14705 | [M−H]−1 | [64,66,67] |
| 13 | Kaempferol | A, E, M, W | – | – | – | 5.988 | 0.455 | 287.05514 | [M+H]+1 | [68,69] |
| 14 | Trifolin | A, E, M, W | – | – | – | 5.978 | 0.45 | 449.10804 | [M+H]+1 | [70,71] |
| 15 | Isorhamentin | A, E, M, W | – | – | – | 6.018 | 0.43 | 317.06572 | [M+H]+1 | [72] |
| 16 | 7-hydroxy-6-methoxy-2H-chromen-2-one isomer 1 | A, E | – | – | – | 4.813 | −0.04 | 193.04953 | [M+H]+1 | [73] |
| 17 | 7-hydroxy-6-methoxy-2H-chromen-2-one isomer 2 | A, E | – | – | – | 5.935 | −0.13 | 193.04951 | [M+H]+1 | [73] |
| 18 | Quinoline | A, M, W | – | – | – | 6.168 | 0.46 | 130.06518 | [M+H]+1 | [49] |
| 19 | Gentisic acid | A, E, M, W | – | – | – | 8.671 | 0.56 | 153.01942 | [M−H] −1 | [74] |
| GC-MS | UHPLC-ESI-MS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | Phytochemical Name | Extract | RT (min) | Calc. RI | Delta RI | RT (min) | Annot. DeltaMass (ppm) | m/z | Adduct | Anti-Inflammatory Activity |
| 1 | beta-Sitosterol | E | 26.24 | 3344 | 20 | – | – | – | – | [75] |
| 2 | Syringic acid | A, E, W | 15.22 | 1889 | 6 | – | – | – | – | [76] |
| 3 | Squalene | A, E, M | 22.845 | 2811 | 0 | – | – | – | – | [77] |
| 4 | Pyrogallol | A, E, W | 12.035 | 1596 | 78 | – | – | – | – | [78,79] |
| 5 | Vanillin | A, E, W | 12.607 | 1647 | 14 | – | – | – | – | [80,81] |
| 6 | Protocatechuic acid | A, E, M | 14.422 | 1813 | 16 | – | – | – | – | [82] |
| 7 | Caffeic acid | A, E, M, W | 17.5 | 2133 | 17 | – | – | – | – | [83,84] |
| 8 | Ferulic acid | A, E, M, W | 17.139 | 2092 | 160 | – | – | – | – | [85,86] |
| 9 | Chlorogenic acid | A, E, M, W | 25.112 | 3106 | 3 | 4.625 | −0.93 | 353.08751 | [M−H] −1 | [87,88] |
| 10 | Quercetin | A, E, M, W | 25.252 | 3177 | 43 | 5.509 | 0.32 | 303.05004 | [M+H]+1 | [89,90,91] |
| 11 | Quercitrin | A, E, M, W | – | – | – | 5.745 | 0.34 | 449.10799 | [M+H]+1 | [92] |
| 12 | Rutin | A, E, M, W | – | – | – | 5.307 | 1.54 | 609.14705 | [M−H] −1 | [92,93,94,95] |
| 13 | Kaempferol | A, E, M, W | – | – | – | 5.988 | 0.455 | 287.05514 | [M+H]+1 | [96,97] |
| 15 | Isorhamentin | A, E, M, W | – | – | – | 6.018 | 0.43 | 317.06572 | [M+H]+1 | [97,98] |
| 16 | Apocynin | A, E, M, W | 0.99 | −0.04 | 167.07026 | [M+H]+1 | [99,100] | |||
| 17 | Gentisic acid | A, M, W | – | – | – | 6.168 | 0.46 | 130.06518 | [M+H]+1 | [101] |
| 18 | p-Coumaric acid | E, M, W | 15.6935 | 1933 | 13 | – | – | – | – | [102] |
| 19 | Sinapic acid | A, E, M | 18.443 | 2242 | 33 | – | – | – | – | [103,104] |
| 20 | Cinnamic acid | A, E, M | 11.49 | 1550 | 3 | – | – | – | – | [105] |
| 21 | Octopamine | A, E, M, W | 15.55 | 1923 | 186 | – | – | – | – | [106] |
| 22 | Kynurenic acid | A, E, M, W | 16.872 | 2064 | 22 | 6.102 | −0.06 | 190.04986 | [M+H]+1 | [107] |
| 23 | 7-hydroxy-6-methoxy-2H-chromen-2-one isomer 1 | A, E | – | – | – | 4.813 | −0.04 | 193.04953 | [M+H]+1 | [108,109] |
| 24 | 7-hydroxy-6-methoxy-2H-chromen-2-one isomer 2 | A, E | – | – | – | 5.935 | −0.13 | 193.04951 | [M+H]+1 | [108,109] |
| 25 | Quinoline | A, M, W | – | – | – | 6.168 | 0.46 | 130.06518 | [M+H]+1 | [110] |
| 25 | Quercetin 3-O-rhamnoside-7-O-glucoside | A, E, M, W | – | – | – | 6.191 | 0.37 | 611.16089 | [M+H]+1 | [111] |
| 25 | Quercetin-3β-D-glucoside isomer 1 | A, E, M, W | – | – | – | 5.75 | 0.11 | 465.1028 | [M+H]+1 | [112] |
| 25 | Quercetin-3β-D-glucoside isomer 2 | A, E, M, W | – | – | – | 6.189 | 0.38 | 465.10293 | [M+H]+1 | [112] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Nemi, R.; Akkawi, M.; Sawalha, K.; Kusumastuti, S.A.; Nuralih; Kusumaningrum, S.; Okselni, T.; Situmorang, V.C.; Septama, A.W.; Jaremko, M.; et al. Comprehensive Metabolomics Profiling and Bioactivity Study of Lycium shawii (Awsaj) Extracts with Particular Emphasis on Potential Anti-Malarial Properties. Metabolites 2025, 15, 84. https://doi.org/10.3390/metabo15020084
Al-Nemi R, Akkawi M, Sawalha K, Kusumastuti SA, Nuralih, Kusumaningrum S, Okselni T, Situmorang VC, Septama AW, Jaremko M, et al. Comprehensive Metabolomics Profiling and Bioactivity Study of Lycium shawii (Awsaj) Extracts with Particular Emphasis on Potential Anti-Malarial Properties. Metabolites. 2025; 15(2):84. https://doi.org/10.3390/metabo15020084
Chicago/Turabian StyleAl-Nemi, Ruba, Mutaz Akkawi, Khalid Sawalha, Siska Andrina Kusumastuti, Nuralih, Susi Kusumaningrum, Tia Okselni, Vania Chlarisa Situmorang, Abdi Wira Septama, Mariusz Jaremko, and et al. 2025. "Comprehensive Metabolomics Profiling and Bioactivity Study of Lycium shawii (Awsaj) Extracts with Particular Emphasis on Potential Anti-Malarial Properties" Metabolites 15, no. 2: 84. https://doi.org/10.3390/metabo15020084
APA StyleAl-Nemi, R., Akkawi, M., Sawalha, K., Kusumastuti, S. A., Nuralih, Kusumaningrum, S., Okselni, T., Situmorang, V. C., Septama, A. W., Jaremko, M., & Emwas, A.-H. (2025). Comprehensive Metabolomics Profiling and Bioactivity Study of Lycium shawii (Awsaj) Extracts with Particular Emphasis on Potential Anti-Malarial Properties. Metabolites, 15(2), 84. https://doi.org/10.3390/metabo15020084







