Characteristics and Expression Analyses of Trehalose-6-Phosphate Synthase Family in Prunus mume Reveal Genes Involved in Trehalose Biosynthesis and Drought Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Experimental Design
2.2. Relative Water Content Measurement
2.3. Trehalose Content Detection in P. mume
2.4. Genomic Data Collection of P. mume and 11 Other Plants
2.5. Identification of TPS Family Members in P. mume and 11 Other Plants
2.6. Multiple Sequence Alignment and Phylogenetic Tree Construction of the TPS Family
2.7. Gene Features Analyses and Protein Subcellular Localization of TPS Family in P. mume
2.8. TPS Genes Structure and Cis-Acting Elements Analyses in P. mume
2.9. Protein Motif and Conserved Domains Analysis
2.10. Gene Duplication Analyses of TPS Family in P. mume
2.11. Selection Force Estimation
2.12. Functional Divergence in TPS Family Protein Sequence Evolution
2.13. Expression Patterns of PmTPSs in Leaf, Stem, and Root
2.14. Quantitative Real-Time PCR
3. Results
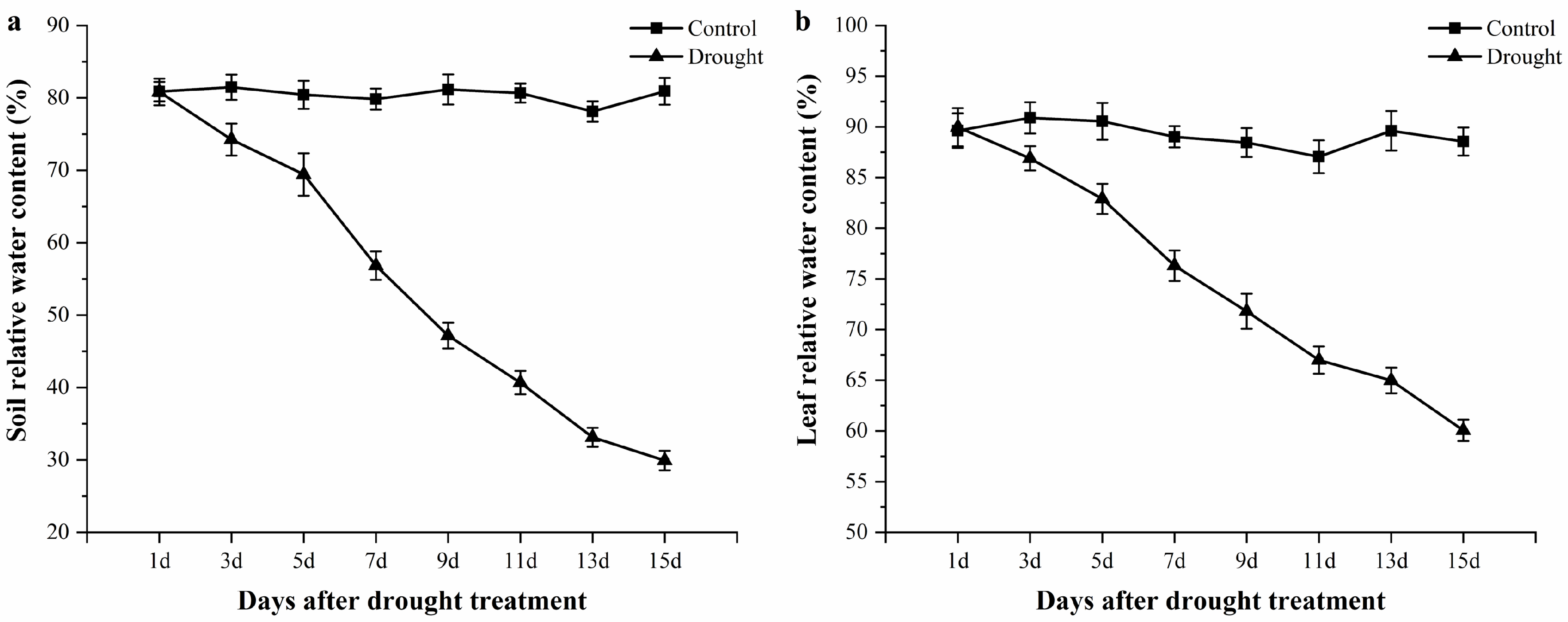
3.1. Relative Water Content Changes of Soil and Leaf Tissues during the Drought Period
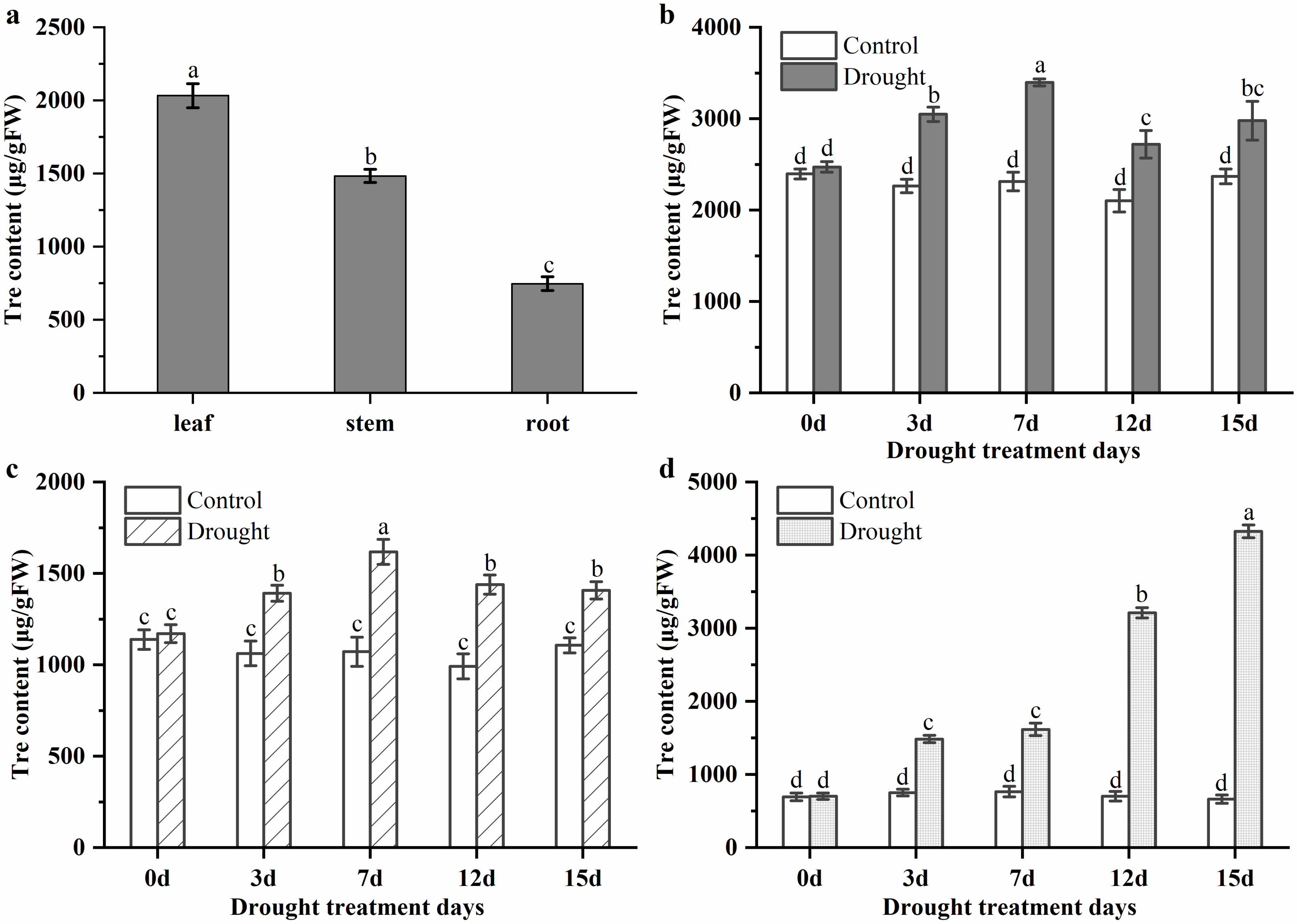
3.2. Trehalose Levels in Leaf, Stem, and Root of P. mume under Normal Conditions and Drought Stress
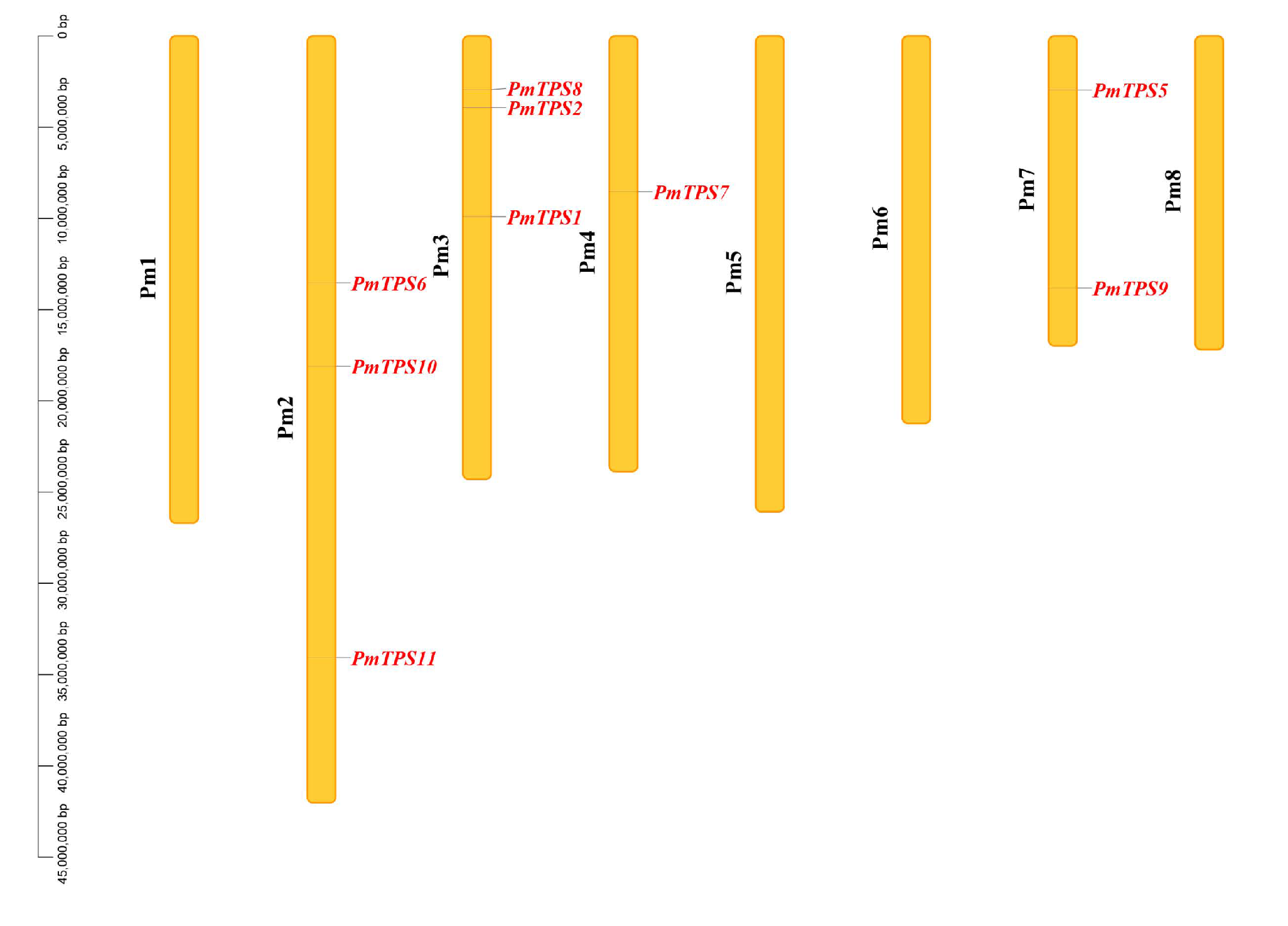
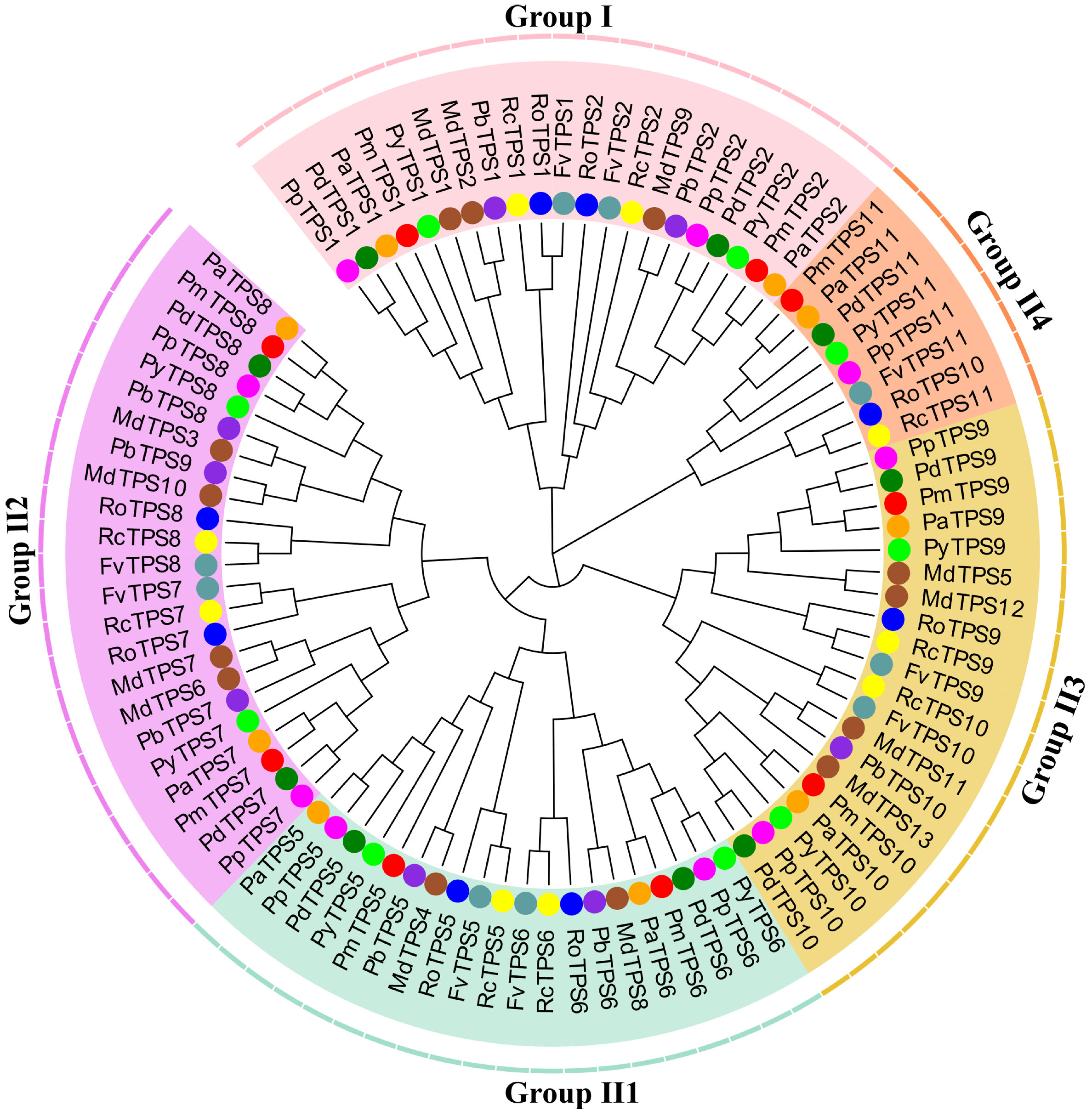
3.3. Identification, Characteristics, and Phylogenetic Analyses of TPS Family in P. mume
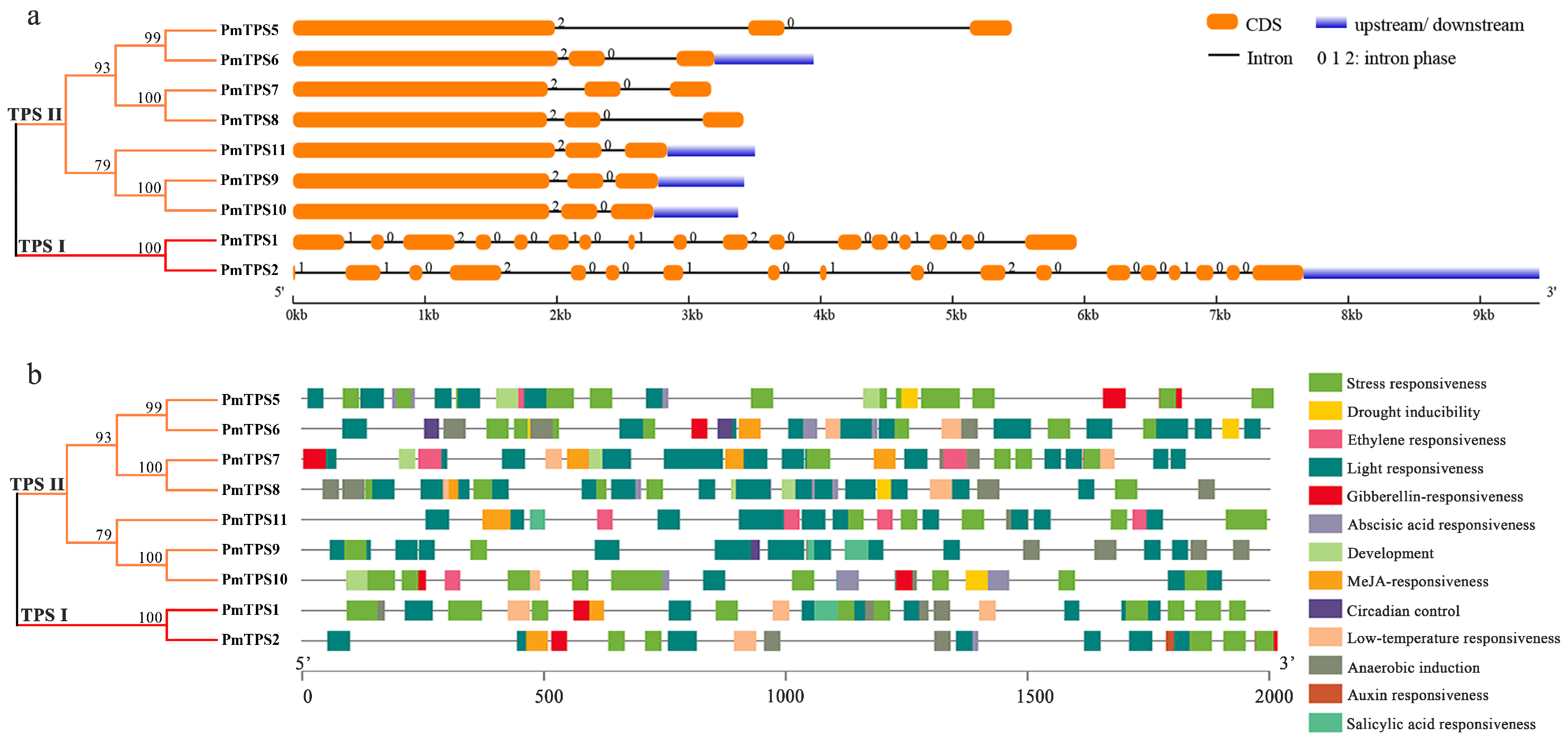
3.4. Gene Structure and Promoter Cis-Acting Elements Analyses of PmTPSs
3.5. Protein Domain Analyses and Multiple Sequences Alignment of TPS Family in P. mume
3.6. Expansion and Evolution Analyses of TPS Family Members in P. mume
3.7. Evolution and Function Divergence Analyses of Rosaceae Plants
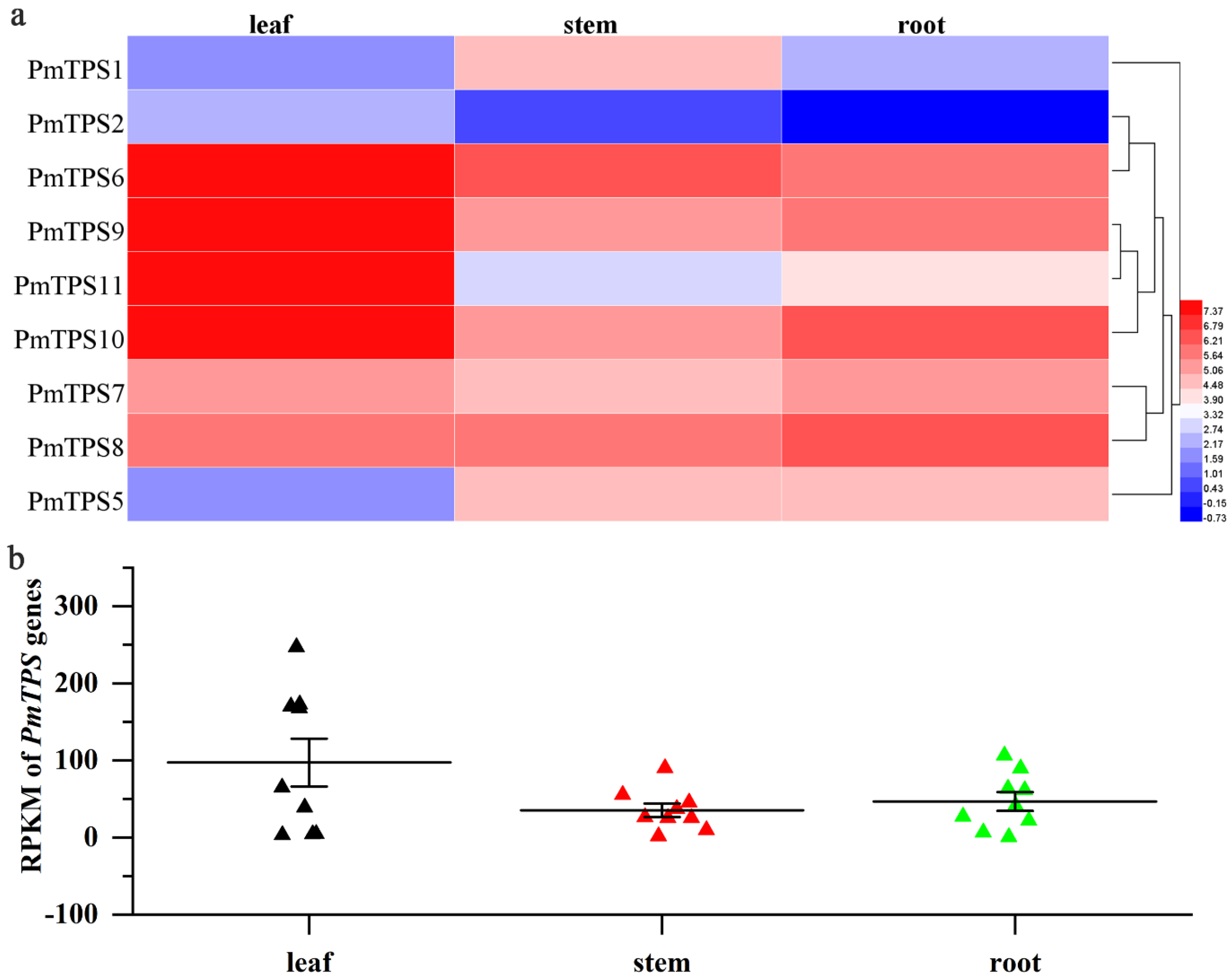
3.8. Expression Profile Analysis of TPS Genes in Leaf, Stem, and Root Tissues of P. mume
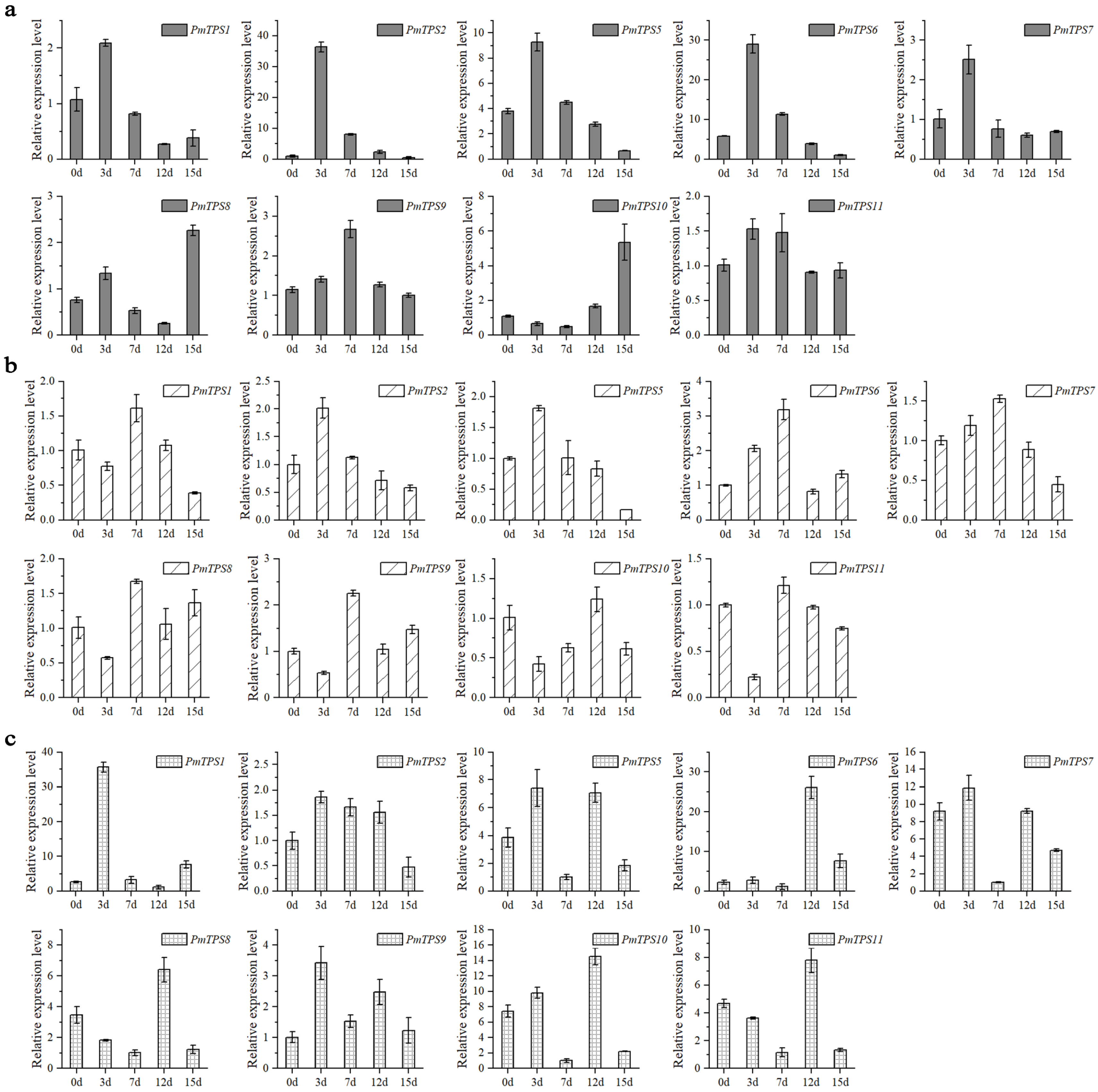
3.9. Expression Pattern Analysis of PmTPSs under Drought Stress
4. Discussion
4.1. Trehalose Function in P. mume Response to Drought
4.2. Characteristics of TPS Family in P. mume
4.3. Functional Evolution Analyses of TPS Family in Rosaceae
4.4. Roles of PmTPSs in Trehalose Biosynthesis in P. mume
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Elbein, A.D.; Pan, Y.T.; Pastuszak, I.; Carroll, D. New insights on trehalose: A multifunctional molecule. Glycobiology 2003, 13, 17R–27R. [Google Scholar] [CrossRef]
- Garg, A.K.; Kim, J.K.; Owens, T.G.; Ranwala, A.P.; Choi, Y.D.; Kochian, L.V.; Wu, R.J. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. USA 2002, 99, 15898–15903. [Google Scholar] [CrossRef]
- Bell, W.; Sun, W.; Hohmann, S.; Wera, S.; Reinders, A.; De Virgilio, C.; Wiemken, A.; Thevelein, J.M. Composition and functional analysis of the Saccharomyces cerevisiae trehalose synthase complex. J. Biol. Chem. 1998, 273, 33311–33319. [Google Scholar] [CrossRef] [PubMed]
- Zentella, R.; Mascorro-Gallardo, J.O.; Van Dijck, P.; Folch-Mallol, J.; Bonini, B.; Van Vaeck, C.; Gaxiola, R.; Covarrubias, A.A.; Nieto-Sotelo, J.; Thevelein, J.M.; et al. A Selaginella lepidophylla trehalose-6-phosphate synthase complements growth and stress-tolerance defects in a yeast tps1 mutant. Plant Physiol. 1999, 119, 1473–1482. [Google Scholar] [CrossRef] [PubMed]
- El-Bashiti, T.; Hamamcı, H.; Öktem, H.A.; Yücel, M. Biochemical analysis of trehalose and its metabolizing enzymes in wheat under abiotic stress conditions. Plant Sci. 2005, 169, 47–54. [Google Scholar] [CrossRef]
- Cortina, C.; Culiáñez-Macià, F.A. Tomato abiotic stress enhanced tolerance by trehalose biosynthesis. Plant Sci. 2005, 169, 75–82. [Google Scholar] [CrossRef]
- Wingler, A. The function of trehalose biosynthesis in plants. Phytochemistry 2002, 60, 437–440. [Google Scholar] [CrossRef]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriaga, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Ramon, M.; Rolland, F. Plant development: Introducing trehalose metabolism. Trends Plant Sci. 2007, 12, 185–188. [Google Scholar] [CrossRef]
- Jang, I.-C.; Oh, S.-J.; Seo, J.-S.; Choi, W.-B.; Song, S.I.; Kim, C.H.; Kim, Y.S.; Seo, H.-S.; Choi, Y.D.; Nahm, B.H.; et al. Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance without stunting growth. Plant Physiol. 2003, 131, 516–524. [Google Scholar] [CrossRef]
- Eastmond, P.J.; Van Dijken, A.J.H.; Spielman, M.; Kerr, A.; Tissier, A.F.; Dickinson, H.G.; Jones, J.D.G.; Smeekens, S.C.; Graham, I.A. Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J. 2002, 29, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Avonce, N.; Leyman, B.; Mascorro-Gallardo, J.O.; Van Dijck, P.; Thevelein, J.M.; Iturriaga, G. The Arabidopsis Trehalose-6-P Synthase AtTPS1 Gene Is a Regulator of Glucose, Abscisic Acid, and Stress Signaling. Plant Physiol. 2004, 136, 3649–3659. [Google Scholar] [CrossRef]
- Yang, H.L.; Liu, Y.J.; Wang, C.L.; Zeng, Q.Y. Molecular evolution of trehalose-6-phosphate synthase (TPS) gene family in Populus, Arabidopsis and rice. PLoS ONE 2012, 7, e42438. [Google Scholar] [CrossRef]
- Vandesteene, L.; Ramon, M.; Le Roy, K.; Van Dijck, P.; Rolland, F. A Single Active Trehalose-6-P Synthase (TPS) and a Family of Putative Regulatory TPS-Like Proteins in Arabidopsis. Mol. Plant 2010, 3, 406–419. [Google Scholar] [CrossRef]
- Zang, B.; Li, H.; Li, W.; Deng, X.W.; Wang, X. Analysis of trehalose-6-phosphate synthase (TPS) gene family suggests the formation of TPS complexes in rice. Plant Mol. Biol. 2011, 76, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Delorge, I.; Janiak, M.; Carpentier, S.; Van Dijck, P. Fine tuning of trehalose biosynthesis and hydrolysis as novel tools for the generation of abiotic stress tolerant plants. Front. Plant Sci. 2014, 5, 147. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef] [PubMed]
- Vandesteene, L.; López-Galvis, L.; Vanneste, K.; Feil, R.; Maere, S.; Lammens, W.; Rolland, F.; Lunn, J.E.; Avonce, N.; Beeckman, T.; et al. Expansive Evolution of the TREHALOSE-6-PHOSPHATE PHOSPHATASE Gene Family in Arabidopsis. Plant Physiol. 2012, 160, 884. [Google Scholar] [CrossRef] [PubMed]
- Delorge, I.; Figueroa, C.M.; Feil, R.; Lunn, J.E.; Van Dijck, P. Trehalose-6-phosphate synthase 1 is not the only active TPS in Arabidopsis thaliana. Biochem. J. 2015, 466, 283–290. [Google Scholar] [CrossRef]
- Almeida, A.M.; Villalobos, E.; Araújo, S.S.; Leyman, B.; Van Dijck, P.; Alfaro-Cardoso, L.; Fevereiro, P.S.; Torné, J.M.; Santos, D.M. Transformation of tobacco with an Arabidopsis thaliana gene involved in trehalose biosynthesis increases tolerance to several abiotic stresses. Euphytica 2005, 146, 165–176. [Google Scholar] [CrossRef]
- Li, H.-W.; Zang, B.-S.; Deng, X.-W.; Wang, X.-P. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 2011, 234, 1007–1018. [Google Scholar] [CrossRef] [PubMed]
- Romero, C.; Belles, J.M.; Vaya, J.L.; Serrano, R.; Culianez-Macia, F.A. Expression of the yeast trehalose-6-phosphate synthase gene in transgenic tobacco plants: Pleiotropic phenotypes include drought tolerance. Planta 1997, 201, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E. Gene families and evolution of trehalose metabolism in plants. Funct. Plant Biol. 2007, 34, 550–563. [Google Scholar] [CrossRef]
- Chary, S.N.; Hicks, G.R.; Choi, Y.G.; Carter, D.; Raikhel, N.V. Trehalose-6-Phosphate Synthase/Phosphatase Regulates Cell Shape and Plant Architecture in Arabidopsis. Plant Physiol. 2008, 146, 97. [Google Scholar] [CrossRef]
- Tian, L.; Xie, Z.; Lu, C.; Hao, X.; Wu, S.; Huang, Y.; Li, D.; Chen, L. The trehalose-6-phosphate synthase TPS5 negatively regulates ABA signaling in Arabidopsis thaliana. Plant Cell Rep. 2019, 38, 869–882. [Google Scholar] [CrossRef]
- Vishal, B.; Krishnamurthy, P.; Ramamoorthy, R.; Kumar, P.P. OsTPS8 controls yield-related traits and confers salt stress tolerance in rice by enhancing suberin deposition. New Phytol. 2019, 221, 1369–1386. [Google Scholar] [CrossRef]
- Liu, X.; Fu, L.; Qin, P.; Sun, Y.; Liu, J.; Wang, X. Overexpression of the wheat trehalose 6-phosphate synthase 11 gene enhances cold tolerance in Arabidopsis thaliana. Gene 2019, 710, 210–217. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, H.; Sun, L.; Fan, G.; Ye, M.; Jiang, L.; Liu, X.; Ma, K.; Shi, C.; Bao, F.; et al. The genetic architecture of floral traits in the woody plant Prunus mume. Nat. Commun. 2018, 9, 1702. [Google Scholar] [CrossRef]
- Li, C.; Tan, D.X.; Liang, D.; Chang, C.; Jia, D.; Ma, F. Melatonin mediates the regulation of ABA metabolism, free-radical scavenging, and stomatal behaviour in two Malus species under drought stress. J. Exp. Bot. 2015, 66, 669–680. [Google Scholar] [CrossRef]
- Li, W.; O’Kelly, B.; Fang, K.; Yang, M. Water Content Determinations for Peaty Soil Using the Oven-Drying Method. Environ. Geotech. 2018, 0, 1–34. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y.; Zhong, J.; Zhang, T.; Li, D.; Ba, T.; Xu, T.; Chang, L.; Zhang, Q.; Sun, M. Root Physiological Traits and Transcriptome Analyses Reveal that Root Zone Water Retention Confers Drought Tolerance to Opisthopappus taihangensis. Sci. Rep. 2020, 10, 2627. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, W.; Sun, L.; Zhao, F.; Huang, B.; Yang, W.; Tao, Y.; Wang, J.; Yuan, Z.; Fan, G.; et al. The genome of Prunus mume. Nat. Commun. 2012, 3, 1318. [Google Scholar] [CrossRef]
- Jiang, F.; Zhang, J.; Wang, S.; Yang, L.; Luo, Y.; Gao, S.; Zhang, M.; Wu, S.; Hu, S.; Sun, H.; et al. The apricot (Prunus armeniaca L.) genome elucidates Rosaceae evolution and beta-carotenoid synthesis. Hortic. Res. 2019, 6, 128. [Google Scholar] [CrossRef]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L.; et al. The Peach v2.0 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genom. 2017, 18, 225. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.; Choi, K.; Kim, G.B.; Yu, H.J.; Cho, A.; Jang, H.; Kim, C.; Kim, H.J.; Chang, K.S.; Kim, J.H.; et al. Draft genome sequence of wild Prunus yedoensis reveals massive inter-specific hybridization between sympatric flowering cherries. Genome Biol. 2018, 19, 127. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; VanBuren, R.; Colle, M.; Poorten, T.J.; Wai, C.M.; Niederhuth, C.E.; Alger, E.I.; Ou, S.; Acharya, C.B.; Wang, J.; et al. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. GigaScience 2018, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef]
- Raymond, O.; Gouzy, J.; Just, J.; Badouin, H.; Verdenaud, M.; Lemainque, A.; Vergne, P.; Moja, S.; Choisne, N.; Pont, C.; et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 2018, 50, 772–777. [Google Scholar] [CrossRef] [PubMed]
- VanBuren, R.; Wai, C.M.; Colle, M.; Wang, J.; Sullivan, S.; Bushakra, J.M.; Liachko, I.; Vining, K.J.; Dossett, M.; Finn, C.E.; et al. A near complete, chromosome-scale assembly of the black raspberry (Rubus occidentalis) genome. GigaScience 2018, 7, giy094. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Lin, M.F.; Jia, R.H.; Li, J.C.; Zhang, M.J.; Chen, H.B.; Zhang, D.; Zhang, J.J.; Chen, X.Y. Evolution and expression patterns of the trehalose-6-phosphate synthase gene family in drumstick tree (Moringa oleifera Lam.). Planta 2018, 248, 999–1015. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Lee, T.H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2013, 41, D1152–D1158. [Google Scholar] [CrossRef]
- Wang, L.; Guo, K.; Li, Y.; Tu, Y.; Hu, H.; Wang, B.; Cui, X.; Peng, L. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 2010, 10, 282. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zou, Y.; Su, Z.; Huang, W.; Zhou, Z.; Arendsee, Z.; Zeng, Y. An update of DIVERGE software for functional divergence analysis of protein family. Mol. Biol. Evol. 2013, 30, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Gu, X. Maximum-Likelihood Approach for Gene Family Evolution Under Functional Divergence. Mol. Biol. Evol. 2001, 18, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Gu, X. Functional divergence in protein (family) sequence evolution. Genetica 2003, 118, 133–141. [Google Scholar] [CrossRef]
- Gu, X. A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol. Biol. Evol. 2006, 23, 1937–1945. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; DePamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef]
- Goddijn, O.J.; van Dun, K. Trehalose metabolism in plants. Trends Plant Sci. 1999, 4, 315–319. [Google Scholar] [CrossRef]
- Karim, S.; Aronsson, H.; Ericson, H.; Pirhonen, M.; Leyman, B.; Welin, B.; Mantyla, E.; Palva, E.T.; Van Dijck, P.; Holmstrom, K.O. Improved drought tolerance without undesired side effects in transgenic plants producing trehalose. Plant Mol. Biol. 2007, 64, 371–386. [Google Scholar] [CrossRef]
- Han, B.; Fu, L.; Zhang, D.; He, X.; Chen, Q.; Peng, M.; Zhang, J. Interspecies and Intraspecies Analysis of Trehalose Contents and the Biosynthesis Pathway Gene Family Reveals Crucial Roles of Trehalose in Osmotic-Stress Tolerance in Cassava. Int. J. Mol. Sci. 2016, 17, 1077. [Google Scholar] [CrossRef]
- Farooq, M.; Ullah, A.; Lee, D.-J.; Alghamdi, S.S.; Siddique, K.H.M. Desi chickpea genotypes tolerate drought stress better than kabuli types by modulating germination metabolism, trehalose accumulation, and carbon assimilation. Plant Physiol. Biochem. 2018, 126, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, Y.; Mattson, N.; Yang, L.; Jin, Q. Genome-wide analysis of the Solanum tuberosum (potato) trehalose-6-phosphate synthase (TPS) gene family: Evolution and differential expression during development and stress. BMC Genom. 2017, 18, 926. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Hu, X.; Li, X.; Wang, B.; Wang, Y.; Jiang, H.; Mattson, N.; Xu, Y. Genome-Wide Identification and Evolution Analysis of Trehalose-6-Phosphate Synthase Gene Family in Nelumbo nucifera. Front. Plant Sci. 2016, 7, 1445. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Qi, S.; Ma, J.; Xing, L.; Fan, S.; Zhang, S.; Li, Y.; Shen, Y.; Zhang, D.; Han, M. Identification of TPS family members in apple (Malus x domestica Borkh.) and the effect of sucrose sprays on TPS expression and floral induction. Plant Physiol. Biochem. PPB 2017, 120, 10–23. [Google Scholar] [CrossRef]
- Mu, M.; Lu, X.-K.; Wang, J.-J.; Wang, D.-L.; Yin, Z.-J.; Wang, S.; Fan, W.-L.; Ye, W.-W. Genome-wide Identification and analysis of the stress-resistance function of the TPS (Trehalose-6-Phosphate Synthase) gene family in cotton. BMC Genet. 2016, 17, 54. [Google Scholar] [CrossRef]
- Xie, D.W.; Wang, X.N.; Fu, L.S.; Sun, J.; Zheng, W.; Li, Z.F. Identification of the trehalose-6-phosphate synthase gene family in winter wheat and expression analysis under conditions of freezing stress. J. Genet. 2015, 94, 55–65. [Google Scholar] [CrossRef]
- Chapman, M.A.; Leebens-Mack, J.H.; Burke, J.M. Positive Selection and Expression Divergence Following Gene Duplication in the Sunflower CYCLOIDEA Gene Family. Mol. Biol. Evol. 2008, 25, 1260–1273. [Google Scholar] [CrossRef]
- Yin, G.; Xu, H.; Xiao, S.; Qin, Y.; Li, Y.; Yan, Y.; Hu, Y. The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 2013, 13, 148. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Wei, S.; Pan, H.; Zhuge, Q.; Yin, T. Uneven selection pressure accelerating divergence of Populus and Salix. Hortic. Res. 2019, 6, 37. [Google Scholar] [CrossRef] [PubMed]
- Massingham, T.; Goldman, N. Detecting Amino Acid Sites Under Positive Selection and Purifying Selection. Genetics 2005, 169, 1753. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Rico-Medina, A.; Cano-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Barraza, A.; Contreras-Cubas, C.; Estrada-Navarrete, G.; Reyes, J.L.; Juárez-Verdayes, M.A.; Avonce, N.; Quinto, C.; Díaz-Camino, C.; Sanchez, F. The Class II Trehalose 6-phosphate Synthase Gene PvTPS9 Modulates Trehalose Metabolism in Phaseolus vulgaris Nodules. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]

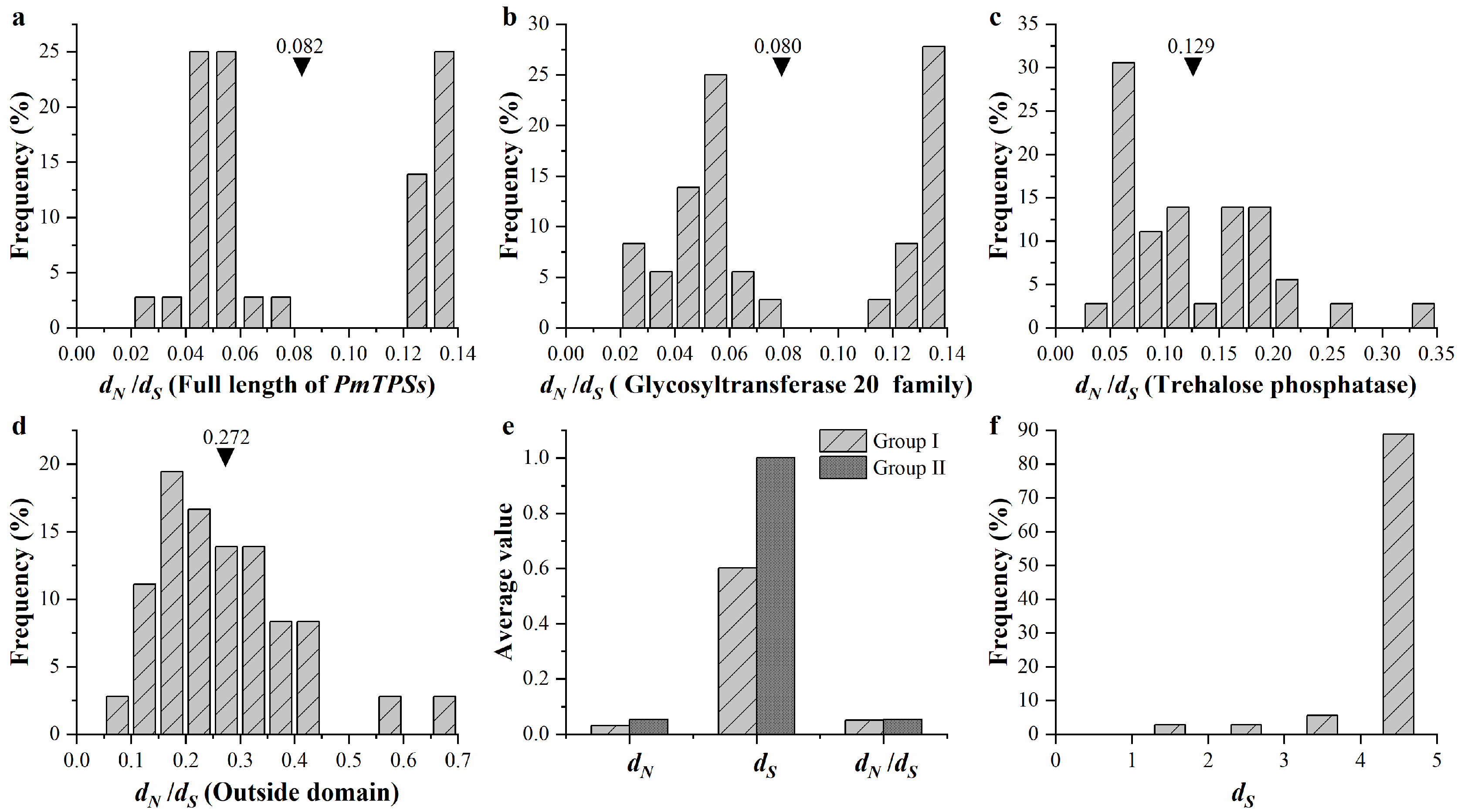

| Group | N a | dN/dS(ω) under M0 b | 2Δ/M3vs.M0 c | 2Δ/M8vs.M7 | M8 Estimates d | Selective Position e |
|---|---|---|---|---|---|---|
| I | 21 | 0.11642 | 333.308 ** | 4.551342 | p1 = 0.00414 ω = 2.40953 (p = 0.22333 q = 1.52122) | 5V,13Y,32K,37L,424S,437A,545K*,549K,673T,674D,675T,700P,701V,707N |
| II1 | 20 | 0.06074 | 184.367 ** | 0.01504 | p1 = 0.00001 w = 42.36549 (p = 0.28727 q = 3.69465) | 68S,302S,317M,321R |
| II2 | 11 | 0.09378 | 282.973 ** | 0.045086 | p1 = 0.00376 w = 1 (p = 0.27992 q = 2.27746) | 3L,54S,305I,314Q,411L,435S,497G |
| II3 | 20 | 0.12158 | 276.959 ** | 0.01508 | p1 = 0.00001 w = 16.38964 (p = 0.28482 q = 1.78922) | 10A,29S,30T,240Q,251S,332P,373L,378L,380I,382E,455A,709S,712S |
| II4 | 8 | 0.15098 | 92.617 ** | 6.04069 | p1 = 0.00808 w = 3.5522 (p = 0.29967 q = 1.6545) | 42S,43H,67S,84K,109S,311Q,357F,387Q,455D,456R,651A,785S,790V,834L,837V,843G |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Ma, K.; Zhang, T.; Li, L.; Wang, J.; Cheng, T.; Zhang, Q. Characteristics and Expression Analyses of Trehalose-6-Phosphate Synthase Family in Prunus mume Reveal Genes Involved in Trehalose Biosynthesis and Drought Response. Biomolecules 2020, 10, 1358. https://doi.org/10.3390/biom10101358
Yang Y, Ma K, Zhang T, Li L, Wang J, Cheng T, Zhang Q. Characteristics and Expression Analyses of Trehalose-6-Phosphate Synthase Family in Prunus mume Reveal Genes Involved in Trehalose Biosynthesis and Drought Response. Biomolecules. 2020; 10(10):1358. https://doi.org/10.3390/biom10101358
Chicago/Turabian StyleYang, Yongjuan, Kaifeng Ma, Tengxun Zhang, Lulu Li, Jia Wang, Tangren Cheng, and Qixiang Zhang. 2020. "Characteristics and Expression Analyses of Trehalose-6-Phosphate Synthase Family in Prunus mume Reveal Genes Involved in Trehalose Biosynthesis and Drought Response" Biomolecules 10, no. 10: 1358. https://doi.org/10.3390/biom10101358
APA StyleYang, Y., Ma, K., Zhang, T., Li, L., Wang, J., Cheng, T., & Zhang, Q. (2020). Characteristics and Expression Analyses of Trehalose-6-Phosphate Synthase Family in Prunus mume Reveal Genes Involved in Trehalose Biosynthesis and Drought Response. Biomolecules, 10(10), 1358. https://doi.org/10.3390/biom10101358





