A Revised Structure for the Glycolipid Terminus of Escherichia coli K5 Heparosan Capsular Polysaccharide
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
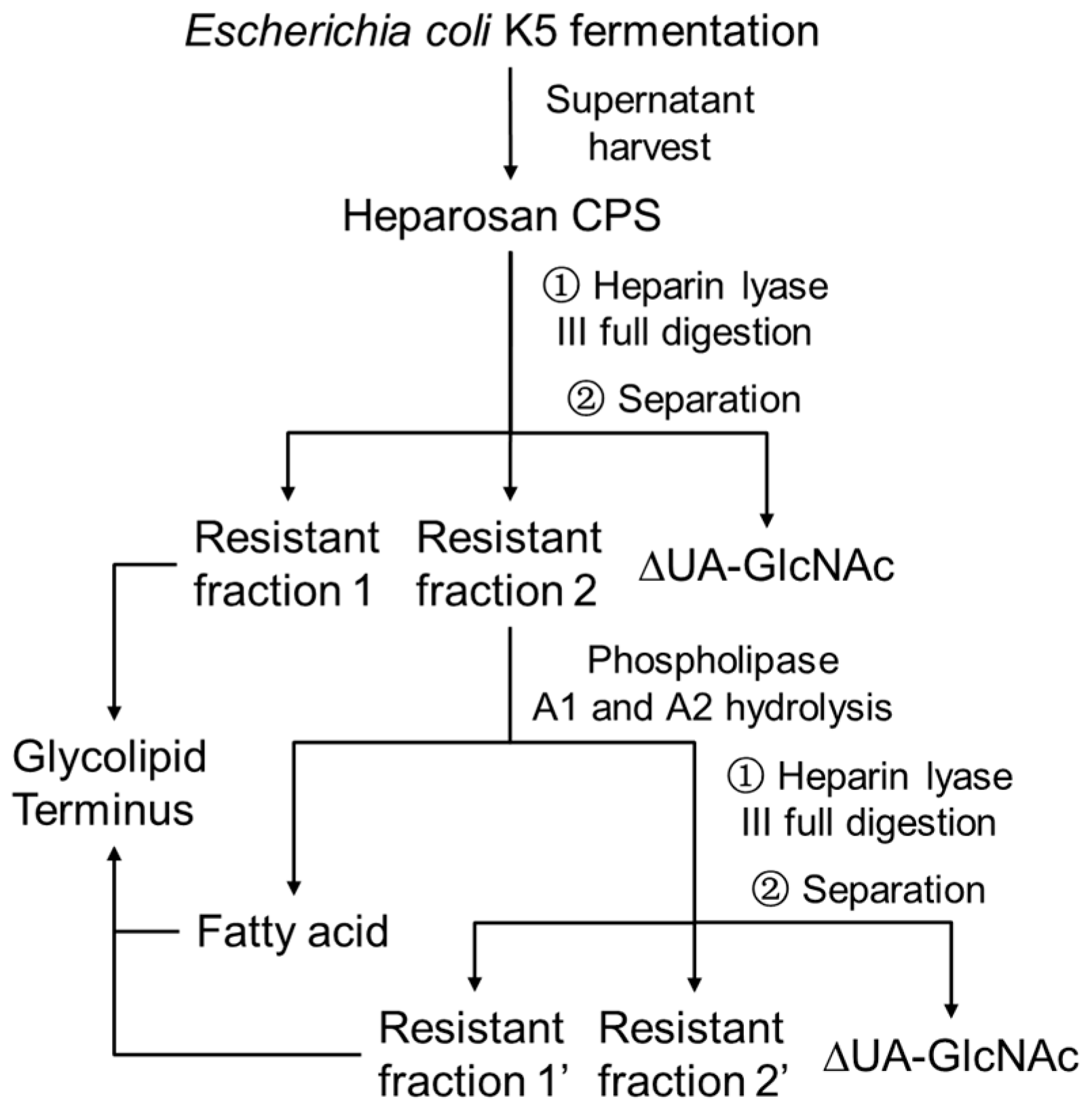
2.2. Heparin Lyase III Digestion
2.3. HPGPC Profiles
2.4. Phospholipids Hydrolysis
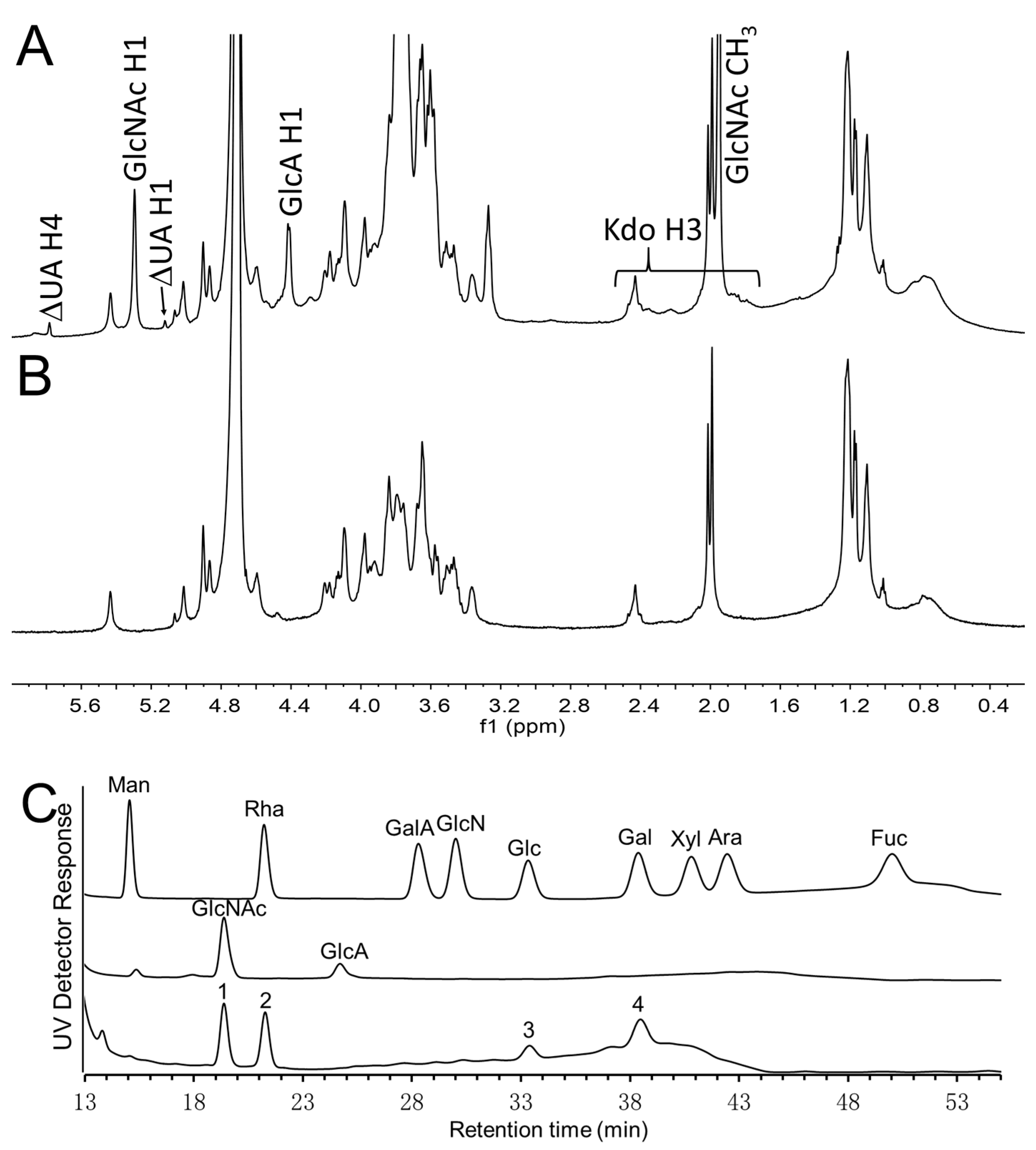
2.5. NMR Spectroscopy
2.6. Monosaccharide Analysis
2.7. Mass Spectroscopy
3. Results and Discussions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bayer, M.E.; Thurow, H. Polysaccharide capsule of Escherichia coli: Microscope study of its size, structure, and sites of synthesis. J. Bacteriol. 1977, 130, 911–936. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, M.A.; Silverstein, S.C. Influence of the Escherichia coli capsule on complement fixation and on phagocytosis and killing by human phagocytes. J. Clin. Investig. 1980, 65, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Goller, C.C.; Seed, P.C. High-Throughput Identification of Chemical Inhibitors of E. coli Group 2 Capsule Biogenesis as Anti-Virulence Agents. PLoS ONE 2010, 5, e11642. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cress, B.F.; Englaender, J.A.; He, W.; Kasper, D.; Linhardt, R.J.; Koffas, M.A.G. Masquerading microbial pathogens: Capsular polysaccharides mimic host-tissue molecules. Fems. Microbiol. Rev. 2014, 38, 660–697. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3, a004952. [Google Scholar] [CrossRef]
- Coombe, D.; Kett, W. Heparan sulfate-protein interactions: Therapeutic potential through structure-function insights. Cell Mol. Life Sci. 2005, 62, 410–424. [Google Scholar] [CrossRef]
- Willis, L.M.; Stupak, J.; Richards, M.R.; Lowary, T.L.; Li, J.; Whitfield, C. Conserved glycolipid termini in capsular polysaccharides synthesized by ATP-binding cassette transporter-dependent pathways in Gram-negative pathogens. Proc. Natl. Acad. Sci. USA 2013, 110, 7868. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ly, M.; Zhang, F.; Zhong, W.; Suen, A.; Hickey, A.M.; Dordick, J.S.; Linhardt, R.J. E. coli K5 fermentation and the preparation of heparosan, a bioengineered heparin precursor. Biotechnol. Bioeng. 2010, 107, 964–973. [Google Scholar] [CrossRef]
- Vaidyanathan, D.; Williams, A.; Dordick, J.S.; Koffas, M.A.G.; Linhardt, R.J. Engineered heparins as new anticoagulant drugs. Bioeng. Transl. Med. 2017, 2, 17–30. [Google Scholar] [CrossRef]
- Cress, B.F.; Bhaskar, U.; Vaidyanathan, D.; Williams, A.; Cai, C.; Liu, X.; Fu, L.; M-Chari, V.; Zhang, F.; Mousa, S.A.; et al. Heavy Heparin: A Stable Isotope-Enriched, Chemoenzymatically-Synthesized, Poly-Component Drug. Angew. Chem. Int. Ed. 2019, 58, 5962–5966. [Google Scholar] [CrossRef]
- Gallagher, J.T.; Turnbull, J.E.; Lyon, M. Patterns of sulphation in heparan sulphate: Polymorphism based on a common structural theme. Int. J. Biochem. 1992, 24, 553–560. [Google Scholar] [CrossRef]
- Rabenstein, D.L. Heparin and heparan sulfate: Structure and function. Nat. Prod. Rep. 2002, 19, 312–331. [Google Scholar] [CrossRef]
- Sansonetti, P. Host–pathogen interactions: The seduction of molecular cross talk. Gut 2002, 50, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Micoli, F.; Costantino, P.; Adamo, R. Potential targets for next generation antimicrobial glycoconjugate vaccines. FEMS Microbiol. Rev. 2018, 42, 388–423. [Google Scholar] [CrossRef]
- Wang, Z.; Dordick, J.S.; Linhardt, R.J. Escherichia coli K5 heparosan fermentation and improvement by genetic engineering. Bioeng. Bugs. 2011, 2, 63–67. [Google Scholar] [CrossRef]
- Lohse, D.L.; Linhardt, R.J. Purification and characterization of heparin lyases from Flavobacterium heparinum. J. Biol. Chem. 1992, 267, 24347–24355. [Google Scholar]
- Yan, L.; Xia, K.; Yu, Y.; Miliakos, A.; Chaturvedi, S.; Zhang, F.; Chen, S.; Chaturvedi, V.; Linhardt, R.J. Unique Cell Surface Mannan of Yeast Pathogen Candida auris with Selective Binding to IgG. Acs Infect. Dis. 2020, 6, 1018–1031. [Google Scholar] [CrossRef]
- Thompson, J.E.; Pourhossein, M.; Waterhouse, A.; Hudson, T.; Goldrick, M.; Derrick, J.P.; Roberts, I.S. The K5 lyase KflA combines a viral tail spike structure with a bacterial polysaccharide lyase mechanism. J. Biol. Chem. 2010, 285, 23963–23969. [Google Scholar] [CrossRef] [PubMed]
- Willis, L.M.; Whitfield, C. Structure, biosynthesis, and function of bacterial capsular polysaccharides synthesized by ABC transporter-dependent pathways. Carbohydr. Res. 2013, 378, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Willis, L.M.; Whitfield, C. KpsC and KpsS are retaining KpsC and KpsS are retaining 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) transferases involved in synthesis of bacterial capsules. Proc. Natl. Acad. Sci. USA 2013, 110, 20753. [Google Scholar] [CrossRef]
- Ovchinnikova, O.G.; Doyle, L.; Huang, B.-S.; Kimber, M.S.; Lowary, T.L.; Whitfield, C. Biochemical Characterization of Bifunctional 3-Deoxy-β-d-manno-oct-2-ulosonic Acid (β-Kdo) Transferase KpsC from Escherichia coli Involved in Capsule Biosynthesis. J. Biol. Chem. 2016, 291, 21519–21530. [Google Scholar] [CrossRef]
- Fraysse, N.; Lindner, B.; Kaczynski, Z.; Sharypova, L.; Holst, O.; Niehaus, K.; Poinsot, V. Sinorhizobium meliloti strain 1021 produces a low-molecular-mass capsular polysaccharide that is a homopolymer of 3-deoxy-d-manno-oct-2-ulosonic acid harboring a phospholipid anchor. Glycobiology 2004, 15, 101–108. [Google Scholar] [CrossRef]


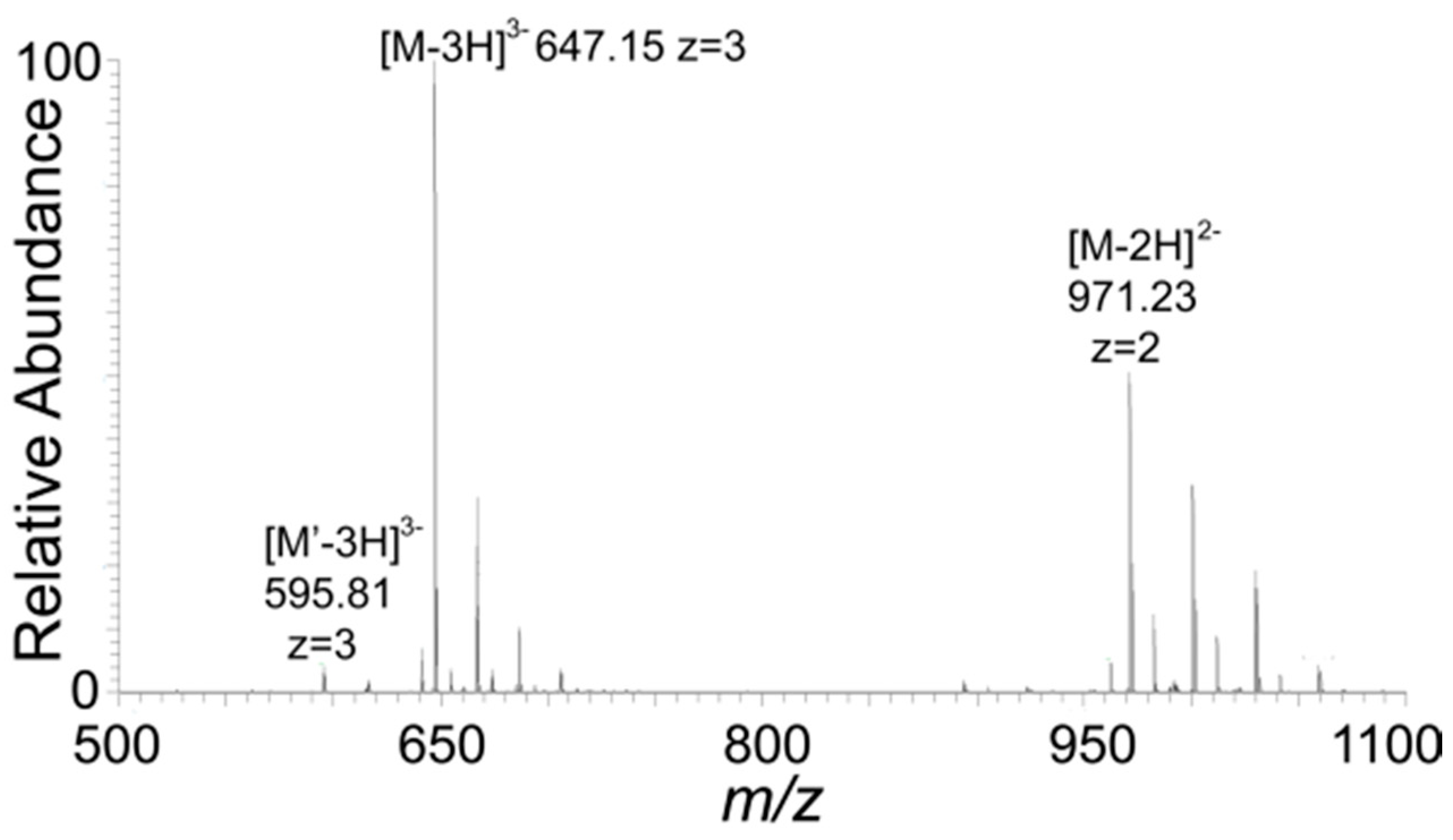
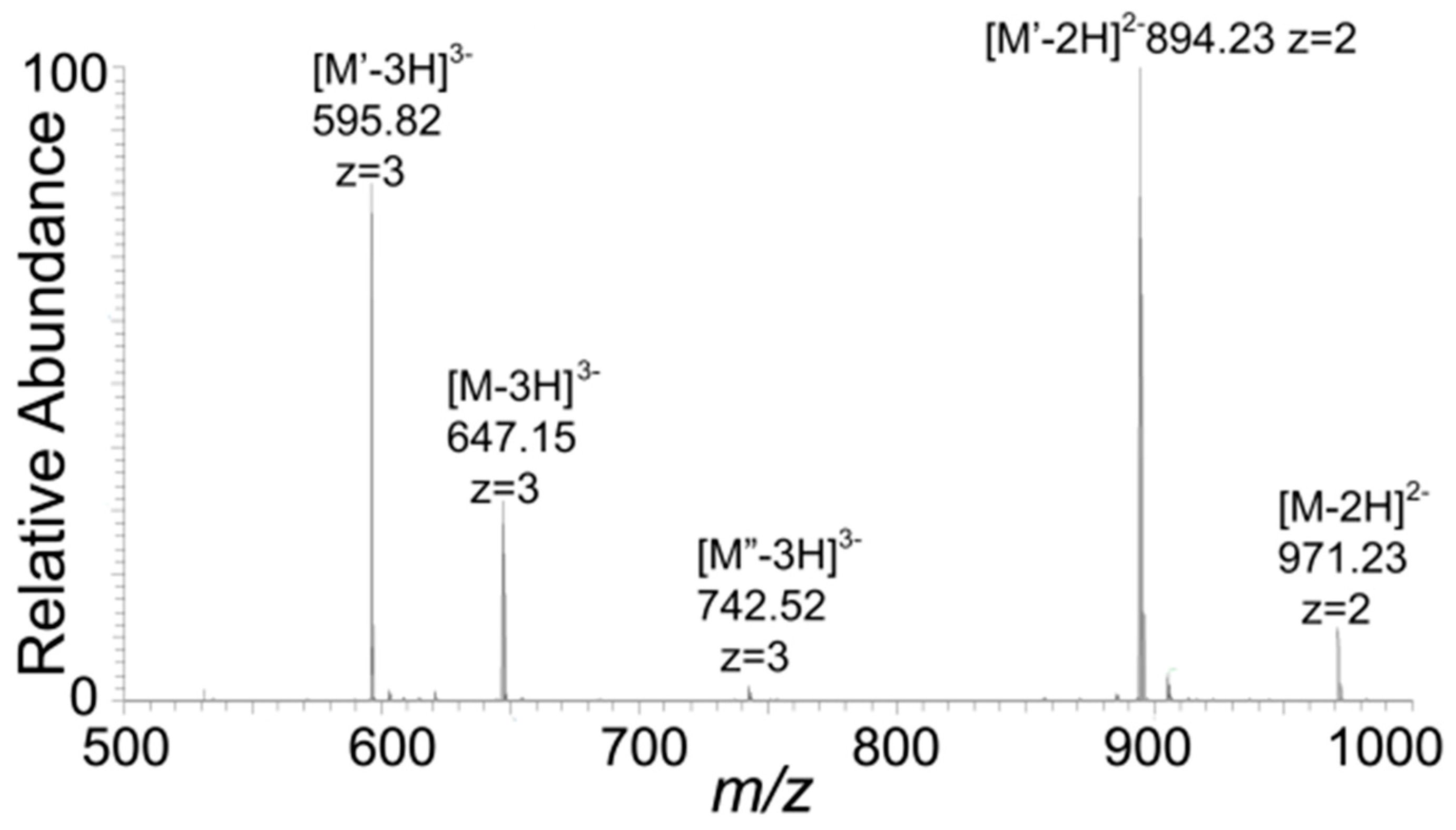
| Observed m/z a | Species a | Reassigned Species (Monoisotopic m/z) |
|---|---|---|
| 853.23− | l-PG(C16:0)-Kdo6-(GlcNAc-GlcA)2 | l-PG(C16:0)-Kdo7-GlcA-GlcNAc-ΔUA (852.933−) |
| 862.13− | l-PG(C18:1)-Kdo6-(GlcNAc-GlcA)2 | l-PG(C18:0)-Kdo7-GlcA-GlcNAc-ΔUA (862.283−) |
| 979.63− | l-PG(C16:0)-Kdo6-(GlcNAc-GlcA)3 | l-PG(C16:0)-Kdo7-(GlcA-GlcNAc)2-ΔUA (979.303−) |
| 988.93− | l-PG(C18:1)-Kdo6-(GlcNAc-GlcA)3 | l-PG(C18:0)-Kdo7-(GlcA-GlcNAc)2-ΔUA (988.653−) |
| 829.34−/1106.03− | l-PG(C16:0)-Kdo6-(GlcNAc-GlcA)4 | l-PG(C16:0)-Kdo7-(GlcA-GlcNAc)3-ΔUA (829.004−/1105.673−) |
| 1115.23− | l-PG(C18:1)-Kdo6-(GlcNAc-GlcA)4 | l-PG(C18:0)-Kdo7-(GlcA-GlcNAc)3-ΔUA (1115.023−) |
| 1000.23− | l-PG(C16:0)-Kdo8-(GlcNAc-GlcA)2 | l-PG(C16:0)-Kdo9-GlcA-GlcNAc-ΔUA (999.643−) |
| 1126.73− | l-PG(C16:0)-Kdo8-(GlcNAc-GlcA)3 | l-PG(C16:0)-Kdo9-(GlcA-GlcNAc)2-ΔUA (1126.013−) |
| 1135.73− | l-PG(C18:1)-Kdo8-(GlcNAc-GlcA)3 | l-PG(C18:0)-Kdo9-(GlcA-GlcNAc)2-ΔUA (1135.353−) |
| 939.64−/1252.93− | l-PG(C16:0)-Kdo8-(GlcNAc-GlcA)4 | l-PG(C16:0)-Kdo9-(GlcA-GlcNAc)3-ΔUA (939.034−/1252.383−) |
| Kdo Residues | H3 ax. (C3) | H3 eq. (C3) | H4 (C4) | H5 (C5) |
|---|---|---|---|---|
| →4)-β-Kdop-(→2 | 1.89 (32.9) | 2.51 (32.9) | 3.87 (71.9) | 4.11 (64.9) |
| 1.93 (32.3) | 2.23 (32.3) | 3.84 (71.9) | 4.09 (65.3) | |
| →7)-β-Kdop-(→2 | 1.80 (34.1) | 2.36 (34.1) | 3.64 (67.2) | 3.90 (64.9) |
| 1.82 (34.9) | 2.38 (34.9) | 3.66 (67.2) | 3.91 (64.9) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, L.; Fu, L.; Xia, K.; Chen, S.; Zhang, F.; Dordick, J.S.; Linhardt, R.J. A Revised Structure for the Glycolipid Terminus of Escherichia coli K5 Heparosan Capsular Polysaccharide. Biomolecules 2020, 10, 1516. https://doi.org/10.3390/biom10111516
Yan L, Fu L, Xia K, Chen S, Zhang F, Dordick JS, Linhardt RJ. A Revised Structure for the Glycolipid Terminus of Escherichia coli K5 Heparosan Capsular Polysaccharide. Biomolecules. 2020; 10(11):1516. https://doi.org/10.3390/biom10111516
Chicago/Turabian StyleYan, Lufeng, Li Fu, Ke Xia, Shiguo Chen, Fuming Zhang, Jonathan S. Dordick, and Robert J. Linhardt. 2020. "A Revised Structure for the Glycolipid Terminus of Escherichia coli K5 Heparosan Capsular Polysaccharide" Biomolecules 10, no. 11: 1516. https://doi.org/10.3390/biom10111516
APA StyleYan, L., Fu, L., Xia, K., Chen, S., Zhang, F., Dordick, J. S., & Linhardt, R. J. (2020). A Revised Structure for the Glycolipid Terminus of Escherichia coli K5 Heparosan Capsular Polysaccharide. Biomolecules, 10(11), 1516. https://doi.org/10.3390/biom10111516







