The Anti-Inflammatory Protein TNIP1 Is Intrinsically Disordered with Structural Flexibility Contributed by Its AHD1-UBAN Domain
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Silico Analysis
2.2. Recombinant Protein Expression and Purification
2.3. Analytical Gel Filtration
2.4. Light Scattering
2.5. Analytical Ultracentrifugation (AUC)
2.6. Limited Proteolysis
2.7. Circular Dichroism
2.8. Protein Expression for Nuclear Magnetic Resonance (NMR) Studies
2.9. NMR Spectroscopy
3. Results
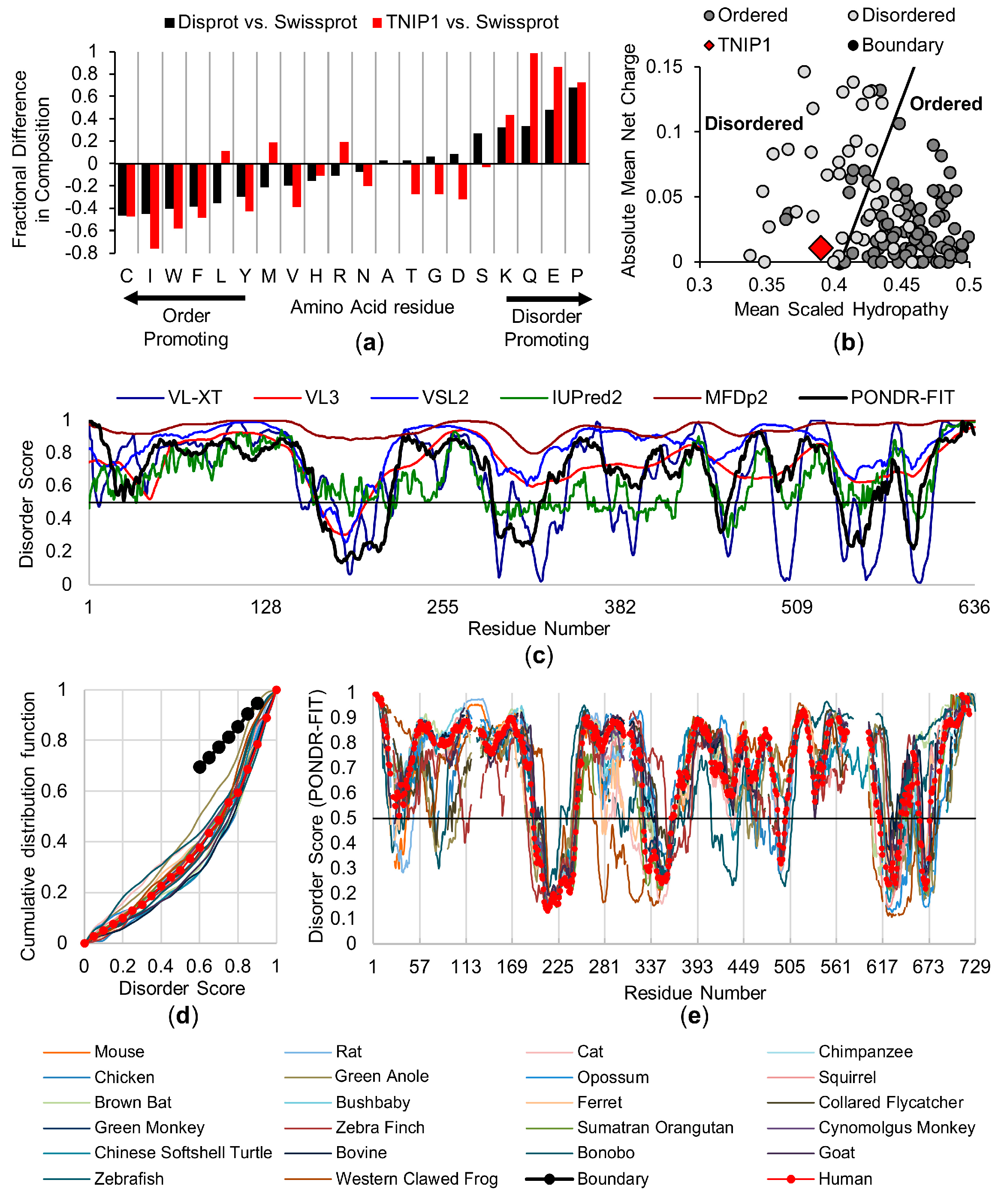
3.1. In Silico Analysis of Full-Length Human TNIP1 and Species Homologues
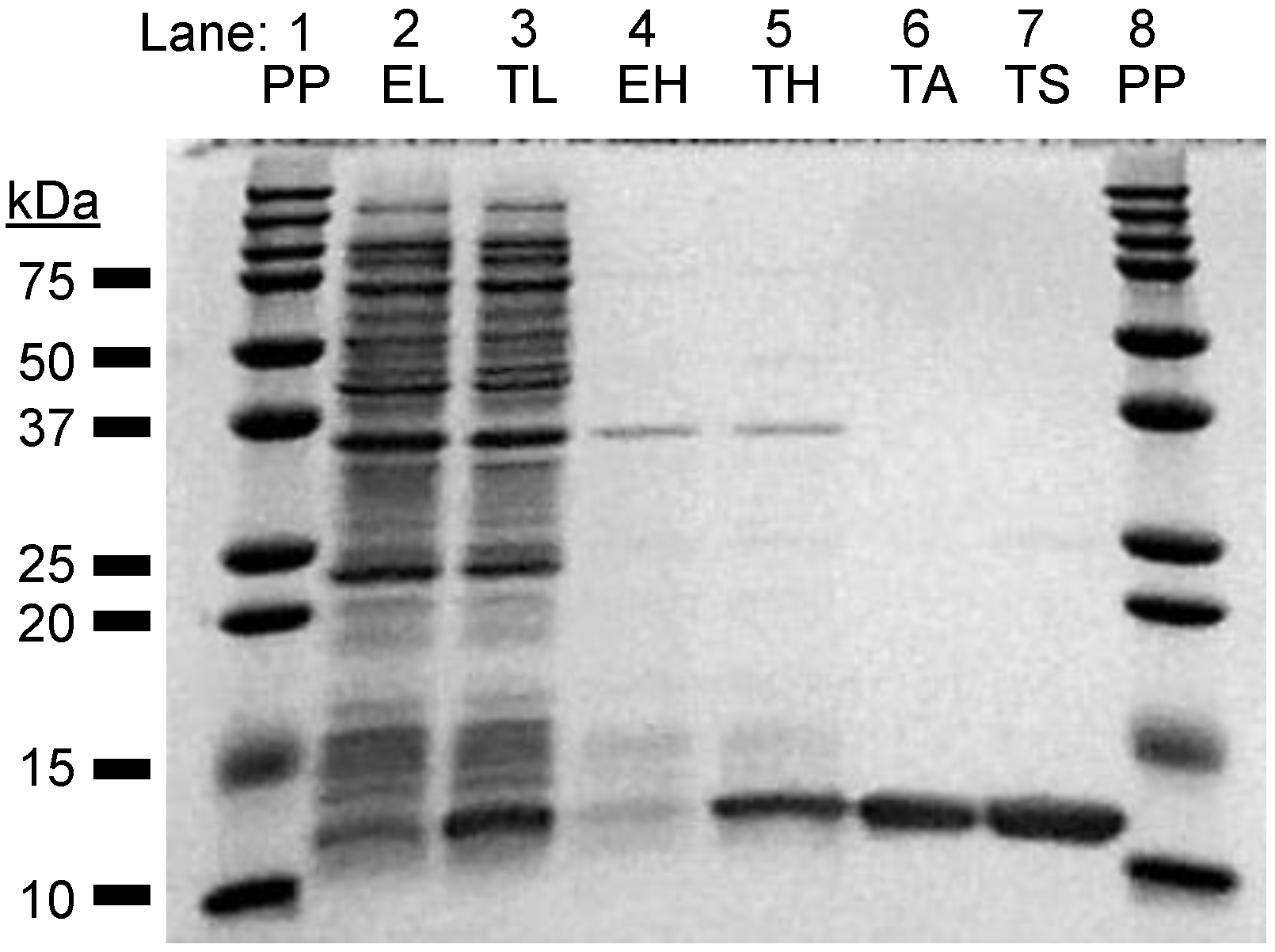
3.2. Full-Length TNIP1 Expression and Limited Proteolysis
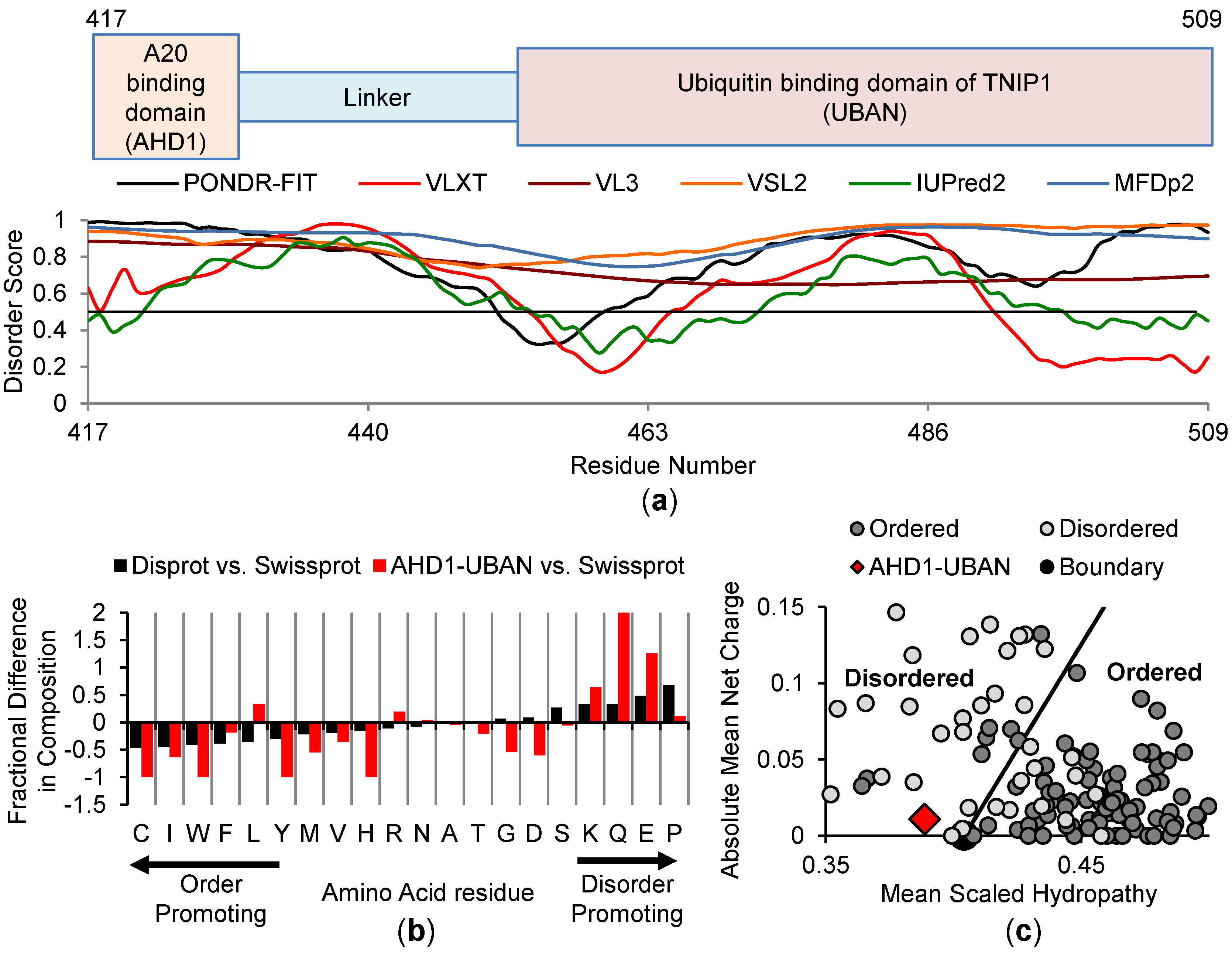
3.3. In Silico Analysis of the Anti-Inflammatory AHD1-UBAN Region of TNIP1
3.4. Expression and Limited Proteolysis of the TNIP1 AHD1-UBAN Domain
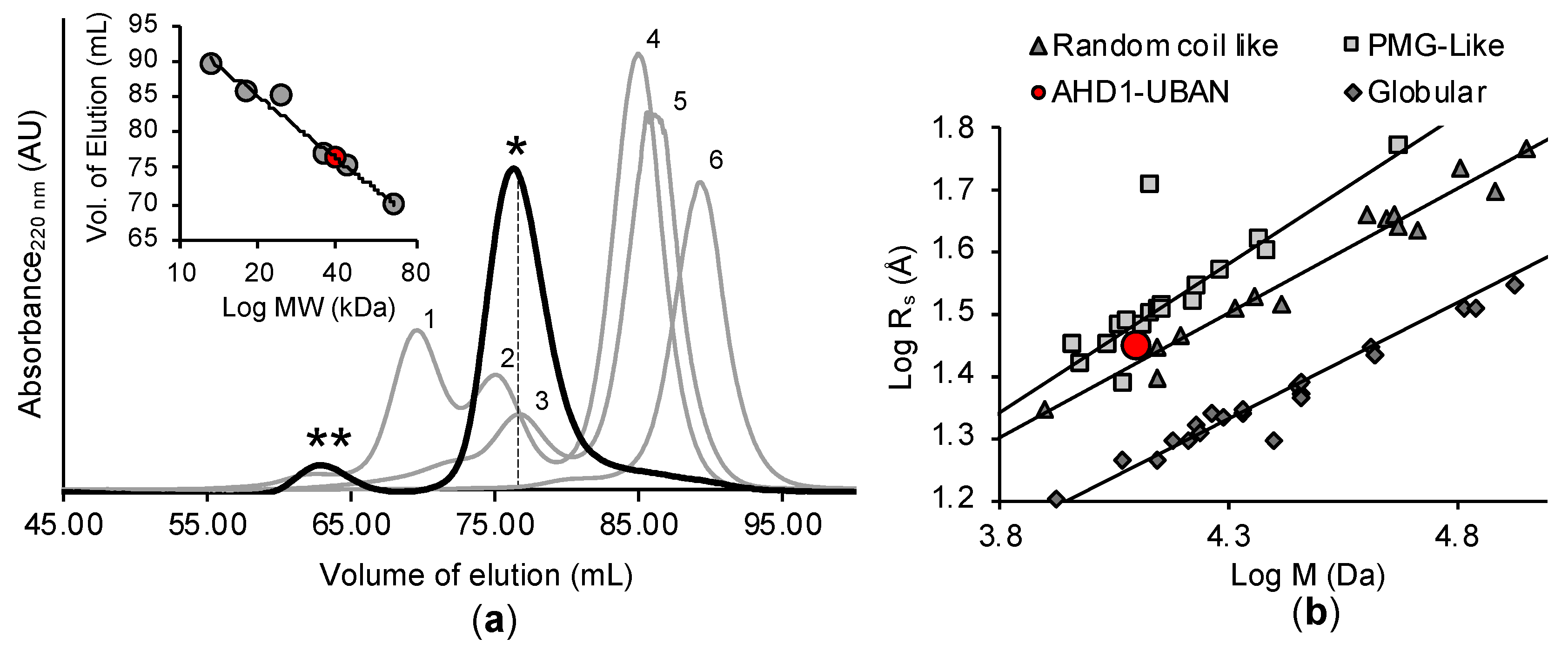
3.5. Analysis of Hydrodynamic and Oligomerization Properties of TNIP1 AHD1-UBAN Region
3.6. Estimation of Secondary Structure within the TNIP1 AHD1-UBAN Region by Circular Dichroism
3.7. 1H–15N HSQC NMR to Visualize Structure and Dynamics of TNIP1 AHD1-UBAN in Solution
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Heyninck, K.; Kreike, M.M.; Beyaert, R. Structure-function analysis of the A20-binding inhibitor of NF-kappa B activation, ABIN-1. FEBS Lett. 2003, 536, 135–140. [Google Scholar]
- Heyninck, K.; De Valck, D.; Vanden Berghe, W.; Van Criekinge, W.; Contreras, R.; Fiers, W.; Haegeman, G.; Beyaert, R. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J. Cell Biol. 1999, 145, 1471–1482. [Google Scholar]
- Fukushi, M.; Dixon, J.; Kimura, T.; Tsurutani, N.; Dixon, M.J.; Yamamoto, N. Identification and cloning of a novel cellular protein Naf1, nef-associated factor 1, that increases cell surface CD4 expression. FEBS Lett. 1999, 442, 83–88. [Google Scholar]
- Gupta, K.; Ott, D.; Hope, T.J.; Siliciano, R.F.; Boeke, J.D. A human nuclear shuttling protein that interacts with human immunodeficiency virus type 1 matrix is packaged into virions. J. Virol. 2000, 74, 11811–11824. [Google Scholar]
- Zhang, S.; Fukushi, M.; Hashimoto, S.; Gao, C.; Huang, L.; Fukuyo, Y.; Nakajima, T.; Amagasa, T.; Enomoto, S.; Koike, K.; et al. A new ERK2 binding protein, Naf1, attenuates the EGF/ERK2 nuclear signaling. Biochem. Biophys. Res. Commun. 2002, 297, 17–23. [Google Scholar] [CrossRef]
- Flores, A.M.; Gurevich, I.; Zhang, C.; Ramirez, V.P.; Devens, T.R.; Aneskievich, B.J. TNIP1 is a corepressor of agonist-bound PPARs. Arch. Biochem. Biophys. 2011, 516, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, I.; Aneskievich, B.J. Liganded RARalpha and RARgamma interact with but are repressed by TNIP1. Biochem. Biophys. Res. Commun. 2009, 389, 409–414. [Google Scholar] [CrossRef] [Green Version]
- Mauro, C.; Pacifico, F.; Lavorgna, A.; Mellone, S.; Iannetti, A.; Acquaviva, R.; Leonardi, A. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J. Biol. Chem. 2006, 281, 18482–18488. [Google Scholar]
- Ma, Y.; Yuan, S.; Tian, X.; Lin, S.; Wei, S.; Hu, T.; Chen, S.; Li, X.; Chen, S.; Wu, D.; et al. ABIN1 inhibits HDAC1 ubiquitination and protects it from both proteasome- and lysozyme-dependent degradation. J. Cell Biochem. 2018, 119, 3030–3043. [Google Scholar] [CrossRef]
- Mirza, N.; Sowa, A.S.; Lautz, K.; Kufer, T.A. NLRP10 affects the stability of abin-1 to control inflammatory responses. J. Immunol. 2019, 202, 218–227. [Google Scholar] [CrossRef] [Green Version]
- Ashida, H.; Kim, M.; Schmidt-Supprian, M.; Ma, A.; Ogawa, M.; Sasakawa, C. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB-mediated inflammatory response. Nat. Cell. Biol. 2010, 12, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Vandereyken, K.; Van Leene, J.; de Coninck, B.; Cammue, B.P.A. Hub protein controversy: Taking a closer look at plant stress response hubs. Front. Plant Sci. 2018, 9, 694. [Google Scholar] [CrossRef] [Green Version]
- Haynes, C.; Oldfield, C.J.; Ji, F.; Klitgord, N.; Cusick, M.E.; Radivojac, P.; Uversky, V.N.; Vidal, M.; Iakoucheva, L.M. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2006, 2, e100. [Google Scholar]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu. Rev. Biophys. 2008, 37, 215–246. [Google Scholar] [CrossRef]
- Hu, G.; Wu, Z.; Uversky, V.N.; Kurgan, L. Functional analysis of human hub proteins and their interactors involved in the intrinsic disorder-enriched interactions. Int. J. Mol. Sci. 2017, 18, 2761. [Google Scholar] [CrossRef] [Green Version]
- Shamilov, R.; Aneskievich, B.J. TNIP1 in autoimmune diseases: Regulation of toll-like receptor signaling. J. Immunol. Res. 2018, 2018, 3491269. [Google Scholar]
- G’Sell, R.T.; Gaffney, P.M.; Powell, D.W. A20-binding inhibitor of NF-kappaB activation 1 is a physiologic inhibitor of NF-kappaB: A molecular switch for inflammation and autoimmunity. Arthritis Rheumatol. 2015, 67, 2292–2302. [Google Scholar] [CrossRef] [Green Version]
- Allanore, Y.; Saad, M.; Dieude, P.; Avouac, J.; Distler, J.H.; Amouyel, P.; Matucci-Cerinic, M.; Riemekasten, G.; Airo, P.; Melchers, I.; et al. Genome-wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet. 2011, 7, e1002091. [Google Scholar] [CrossRef]
- Chen, Y.; Yan, H.; Song, Z.; Chen, F.; Wang, H.; Niu, J.; Shi, X.; Zhang, D.; Zhang, N.; Zhai, Z.; et al. Downregulation of TNIP1 expression leads to increased proliferation of human keratinocytes and severer psoriasis-like conditions in an imiquimod-induced mouse model of dermatitis. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Cruz, J.A.; Childs, E.E.; Amatya, N.; Garg, A.V.; Beyaert, R.; Kane, L.P.; Aneskievich, B.J.; Ma, A.; Gaffen, S.L. Interleukin-17 signaling triggers degradation of the constitutive NF-kappaB inhibitor ABIN-1. Immunohorizons 2017, 1, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Herhaus, L.; van den Bedem, H.; Tang, S.; Maslennikov, I.; Wakatsuki, S.; Dikic, I.; Rahighi, S. Molecular recognition of M1-linked ubiquitin chains by native and phosphorylated UBAN domains. J. Mol. Biol. 2019, 431, 3146–3156. [Google Scholar]
- Lin, S.M.; Lin, S.C.; Hong, J.Y.; Su, T.W.; Kuo, B.J.; Chang, W.H.; Tu, Y.F.; Lo, Y.-C. Structural insights into linear tri-ubiquitin recognition by A20-binding inhibitor of NF-kappaB, ABIN-2. Structure 2017, 25, 66–78. [Google Scholar]
- Babu, M.M. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem. Soc. Trans. 2016, 44, 1185–1200. [Google Scholar]
- van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.W.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Wagner, S.; Carpentier, I.; Rogov, V.; Kreike, M.; Ikeda, F.; Lohr, F.; Wu, C.-J.; Ashwell, J.D.; Dotsch, V.; Dikic, I.; et al. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene 2008, 27, 3739–3745. [Google Scholar] [CrossRef] [Green Version]
- Nanda, S.K.; Venigalla, R.K.; Ordureau, A.; Patterson-Kane, J.C.; Powell, D.W.; Toth, R.; Arthur, J.S.C.; Cohen, P. Polyubiquitin binding to ABIN1 is required to prevent autoimmunity. J. Exp. Med. 2011, 208, 1215–1228. [Google Scholar] [CrossRef] [Green Version]
- Kattah, M.G.; Shao, L.; Rosli, Y.Y.; Shimizu, H.; Whang, M.I.; Advincula, R.; Achacoso, P.; Shah, S.; Duong, B.H.; Onizawa, M.; et al. A20 and ABIN-1 synergistically preserve intestinal epithelial cell survival. J. Exp. Med. 2018, 215, 1839–1852. [Google Scholar] [CrossRef]
- Reed, B.J.; Locke, M.N.; Gardner, R.G. A conserved deubiquitinating enzyme uses intrinsically disordered regions to scaffold multiple protein interaction sites. J. Biol. Chem. 2015, 290, 20601–20612. [Google Scholar] [CrossRef] [Green Version]
- Sigalov, A.B. Unusual biophysics of immune signaling-related intrinsically disordered proteins. Self Nonself 2010, 1, 271–281. [Google Scholar] [CrossRef] [Green Version]
- Sigalov, A.B.; Zhuravleva, A.V.; Orekhov, V.Y. Binding of intrinsically disordered proteins is not necessarily accompanied by a structural transition to a folded form. Biochimie 2007, 89, 419–421. [Google Scholar]
- Wang, W.; Wang, D. Extreme fuzziness: Direct interactions between two IDPs. Biomolecules 2019, 9, 81. [Google Scholar] [CrossRef] [Green Version]
- Borgia, A.; Borgia, M.B.; Bugge, K.; Kissling, V.M.; Heidarsson, P.O.; Fernandes, C.B.; Sottini, A.; Soranno, A.; Buholzer, K.J.; Nettels, D.; et al. Extreme disorder in an ultrahigh-affinity protein complex. Nature 2018, 555, 61–66. [Google Scholar] [CrossRef] [Green Version]
- Verstrepen, L.; Carpentier, I.; Verhelst, K.; Beyaert, R. ABINs: A20 binding inhibitors of NF-kappa B and apoptosis signaling. Biochem. Pharmacol. 2009, 78, 105–114. [Google Scholar] [CrossRef] [Green Version]
- Oldfield, C.J.; Xue, B.; Van, Y.Y.; Ulrich, E.L.; Markley, J.L.; Dunker, A.K.; Uversky, V.N. Utilization of protein intrinsic disorder knowledge in structural proteomics. Biochim. Biophys. Acta 2013, 1834, 487–498. [Google Scholar] [CrossRef] [Green Version]
- Fontana, A.; de Laureto, P.P.; Spolaore, B.; Frare, E. Identifying disordered regions in proteins by limited proteolysis. Methods Mol. Biol. 2012, 896, 297–318. [Google Scholar] [CrossRef]
- Suskiewicz, M.J.; Sussman, J.L.; Silman, I.; Shaul, Y. Context-dependent resistance to proteolysis of intrinsically disordered proteins. Protein Sci. 2011, 20, 1285–1297. [Google Scholar] [CrossRef] [Green Version]
- Piovesan, D.; Tabaro, F.; Mičetić, I.; Necci, M.; Quaglia, F.; Oldfield, C.J.; Aspromonte, M.C.; Davey, N.E.; Davidović, R.; Dosztányi, Z.; et al. DisProt 7.0: A major update of the database of disordered proteins. Nucleic Acids Res. 2017, 45, D219–D227. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Feng, Y.; Wang, X.; Li, J.; Liu, W.; Rong, L.; Zhou, S. An overview of predictors for intrinsically disordered proteins over 2010–2014. Int. J. Mol. Sci. 2015, 16, 23446–23462. [Google Scholar] [CrossRef]
- Martinelli, A.H.S.; Lopes, F.C.; John, E.B.O.; Carlini, C.R.; Ligabue-Braun, R. Modulation of disordered proteins with a focus on neurodegenerative diseases and other pathologies. Int. J. Mol. Sci. 2019, 20, 1322. [Google Scholar] [CrossRef] [Green Version]
- UniProt Consortium, T. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef] [Green Version]
- Meng, F.; Uversky, V.N.; Kurgan, L. Comprehensive review of methods for prediction of intrinsic disorder and its molecular functions. Cell. Mol. Life Sci. 2017, 74, 3069–3090. [Google Scholar] [CrossRef]
- Nielsen, J.T.; Mulder, F.A.A. Quality and bias of protein disorder predictors. Sci. Rep. 2019, 9, 5137. [Google Scholar] [CrossRef] [Green Version]
- Katuwawala, A.; Oldfield, C.J.; Kurgan, L. Accuracy of protein-level disorder predictions. Brief. Bioinform. 2019, 21, 1509–1522. [Google Scholar] [CrossRef]
- Ludwiczak, J.; Winski, A.; Szczepaniak, K.; Alva, V.; Dunin-Horkawicz, S. DeepCoil-a fast and accurate prediction of coiled-coil domains in protein sequences. Bioinformatics 2019, 35, 2790–2795. [Google Scholar] [CrossRef]
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Livernois, A.M.; Hnatchuk, D.J.; Findlater, E.E.; Graether, S.P. Obtaining highly purified intrinsically disordered protein by boiling lysis and single step ion exchange. Anal. Biochem. 2009, 392, 70–76. [Google Scholar] [CrossRef]
- Ferrari, L.; Rudiger, S.G.D. Recombinant production and purification of the human protein tau. Protein Eng. Des. Sel. 2018, 31, 447–455. [Google Scholar] [CrossRef]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar]
- KrishnaKumar, V.G.; Gupta, S. Simplified method to obtain enhanced expression of tau protein from E. coli and one-step purification by direct boiling. Prep. Biochem. Biotechnol. 2017, 47, 530–538. [Google Scholar] [CrossRef]
- Kalthoff, C. A novel strategy for the purification of recombinantly expressed unstructured protein domains. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2003, 786, 247–254. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Uversky, V.N. What does it mean to be natively unfolded? Eur. J. Biochem. 2002, 269, 2649. [Google Scholar]
- Tcherkasskaya, O.; Uversky, V.N. Denatured collapsed states in protein folding: Example of apomyoglobin. Proteins 2001, 44, 244–254. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- Maciejewski, M.W.; Schuyler, A.D.; Gryk, M.R.; Moraru, I.I.; Romero, P.R.; Ulrich, E.L.; Eghbalnia, H.R.; Livny, M.; Delaglio, F.; Hoch, J.C. NMRbox: A resource for biomolecular NMR computation. Biophys. J. 2017, 112, 1529–1534. [Google Scholar]
- Dunker, A.K.; Oh, J.; Oldfield, C.; Romero, P.; Ratliff, C.; Hipps, K.W.; Reeves, R.; Ausió, J.; Kissinger, C.; Bailey, R.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar]
- Uversky, V.N. Intrinsically disordered proteins from A to Z. Int. J. Biochem. Cell Biol. 2011, 43, 1090–1103. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Huang, J.Z.; Gao, X. LigandRFs: Random forest ensemble to identify ligand-binding residues from sequence information alone. BMC Bioinform. 2014, 15 (Suppl. S15). [Google Scholar] [CrossRef] [Green Version]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Oldfield, C.J.; Yan, W.; Shen, B.; Dunker, A.K. Intrinsically disordered domains: Sequence disorder function relationships. Protein Sci. 2019, 28, 1652–1663. [Google Scholar] [CrossRef]
- Chen, J.W.; Romero, P.; Uversky, V.N.; Dunker, A.K. Conservation of intrinsic disorder in protein domains and families: II. functions of conserved disorder. J. Proteome Res. 2006, 5, 888–898. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N.; El-Baky, N.A.; El-Fakharany, E.M.; Sabry, A.; Mattar, E.H.; Uversky, A.V.; Redwan, E.M. Functionality of intrinsic disorder in tumor necrosis factor-alpha and its receptors. FEBS J. 2017, 284, 3589–3618. [Google Scholar] [CrossRef] [Green Version]
- . Moncoq, K.; Broutin, I.; LaRue, V.; Perdereau, M.; Cailliau, K.; Browaeys-Poly, E.; Burnol, A.-F.; Ducruix, A. The PIR domain of Grb14 is an intrinsically unstructured protein: Implication in insulin signaling. FEBS Lett. 2003, 554, 240–246. [Google Scholar]
- Csokova, N.; Skrabana, R.; Liebig, H.D.; Mederlyova, A.; Kontsek, P.; Novak, M. Rapid purification of truncated tau proteins: Model approach to purification of functionally active fragments of disordered proteins, implication for neurodegenerative diseases. Protein Expr. Purif. 2004, 35, 366–372. [Google Scholar] [CrossRef]
- Schramm, A.; Bignon, C.; Brocca, S.; Grandori, R.; Santambrogio, C.; Longhi, S. An arsenal of methods for the experimental characterization of intrinsically disordered proteins—How to choose and combine them? Arch. Biochem. Biophys. 2019, 676, 108055. [Google Scholar]
- Kim, T.D.; Ryu, H.J.; Cho, H.I.; Yang, C.H.; Kim, J. Thermal behavior of proteins: Heat-resistant proteins and their heat-induced secondary structural changes. Biochemistry 2000, 39, 14839–14846. [Google Scholar]
- Verstrepen, L.; Carpentier, I.; Beyaert, R. The biology of A20-binding inhibitors of NF-kappaB activation (ABINs). Adv. Exp. Med. Biol. 2014, 809, 13–31. [Google Scholar]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar]
- Salvay, A.G.; Communie, G.; Ebel, C. Sedimentation velocity analytical ultracentrifugation for intrinsically disordered proteins. Methods Mol. Biol. 2012, 896, 91–105. [Google Scholar] [CrossRef]
- Buck, M. Trifluoroethanol and colleagues: Cosolvents come of age. recent studies with peptides and proteins. Q. Rev. Biophys. 1998, 31, 297–355. [Google Scholar] [CrossRef]
- Chemes, L.B.; Alonso, L.G.; Noval, M.G.; de Prat-Gay, G. Circular dichroism techniques for the analysis of intrinsically disordered proteins and domains. Methods Mol. Biol. 2012, 895, 387–404. [Google Scholar] [CrossRef]
- Konrat, R. NMR contributions to structural dynamics studies of intrinsically disordered proteins. J. Magn. Reson. 2014, 241, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N.; Davé, V.; Iakoucheva, L.M.; Malaney, P.; Metallo, S.J.; Pathak, R.R.; Joerger, A.C. Pathological unfoldomics of uncontrolled chaos: Intrinsically disordered proteins and human diseases. Chem. Rev. 2014, 114, 6844–6879. [Google Scholar] [CrossRef] [Green Version]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar]
- Theillet, F.X.; Kalmar, L.; Tompa, P.; Han, K.H.; Selenko, P.; Dunker, A.K.; Daughdrill, G.W.; Uversky, N.V. The alphabet of intrinsic disorder: I. act like a pro: On the abundance and roles of proline residues in intrinsically disordered proteins. Intrinsic. Disord. Proteins 2013, 1, e24360. [Google Scholar]
- Bah, A.; Forman-Kay, J.D. Modulation of intrinsically disordered protein function by post-translational modifications. J. Biol. Chem. 2016, 291, 6696–6705. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.W.; Romero, P.; Uversky, V.N.; Dunker, A.K. Conservation of intrinsic disorder in protein domains and families: I. A database of conserved predicted disordered regions. J. Proteome Res. 2006, 5, 879–887. [Google Scholar] [CrossRef] [Green Version]
- Yoshizawa, T.; Nozawa, R.S.; Jia, T.Z.; Saio, T.; Mori, E. Biological phase separation: Cell biology meets biophysics. Biophys. Rev. 2020, 12, 519–539. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Yang, X.; Cheng, W.; Ma, Y.; Shang, Y.; Cao, L.; Chen, S.; Chen, Y.; Wang, M.; Guo, D. Immune regulator ABIN1 suppresses HIV-1 transcription by negatively regulating the ubiquitination of tat. Retrovirology 2017, 14, 12. [Google Scholar] [CrossRef] [Green Version]
- Renaud, L.; Picher-Martel, V.; Codron, P.; Julien, J.P. Key role of UBQLN2 in pathogenesis of amyotrophic lateral sclerosis and frontotemporal dementia. Acta Neuropathol. Commun. 2019, 7, 103. [Google Scholar] [CrossRef]
- Kanaan, N.M.; Hamel, C.; Grabinski, T.; Combs, B. Liquid-liquid phase separation induces pathogenic tau conformations in vitro. Nat. Commun. 2020, 11, 2809. [Google Scholar] [CrossRef]
- Uversky, V.N. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015, 282, 1182–1189. [Google Scholar] [CrossRef]
- Uversky, V.N. Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta 2013, 1834, 932–951. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and combining predictors of mostly disordered proteins. Biochemistry 2005, 44, 1989–2000. [Google Scholar] [CrossRef]
- Choi, U.B.; Sanabria, H.; Smirnova, T.; Bowen, M.E.; Weninger, K.R. Spontaneous switching among conformational ensembles in intrinsically disordered proteins. Biomolecules 2019, 9, 114. [Google Scholar] [CrossRef] [Green Version]
- Dyson, H.J.; Wright, P.E. Unfolded proteins and protein folding studied by NMR. Chem. Rev. 2004, 104, 3607–3622. [Google Scholar] [CrossRef]
- Cohen, P.; Strickson, S. The role of hybrid ubiquitin chains in the MyD88 and other innate immune signalling pathways. Cell Death Differ. 2017, 24, 1153–1159. [Google Scholar] [CrossRef] [Green Version]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Romero, P.; Uversky, V.N.; Dunker, A.K. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry 2005, 44, 12454–12470. [Google Scholar] [CrossRef]
- Li, J.; McQuade, T.; Siemer, A.B.; Napetschnig, J.; Moriwaki, K.; Hsiao, Y.-S.; Damko, E.; Moquin, D.; Walz, T.; McDermott, A.; et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 2012, 150, 339–350. [Google Scholar] [CrossRef] [Green Version]
- Cumberworth, A.; Lamour, G.; Babu, M.M.; Gsponer, J. Promiscuity as a functional trait: Intrinsically disordered regions as central players of interactomes. Biochem. J. 2013, 454, 361–369. [Google Scholar] [CrossRef] [Green Version]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]


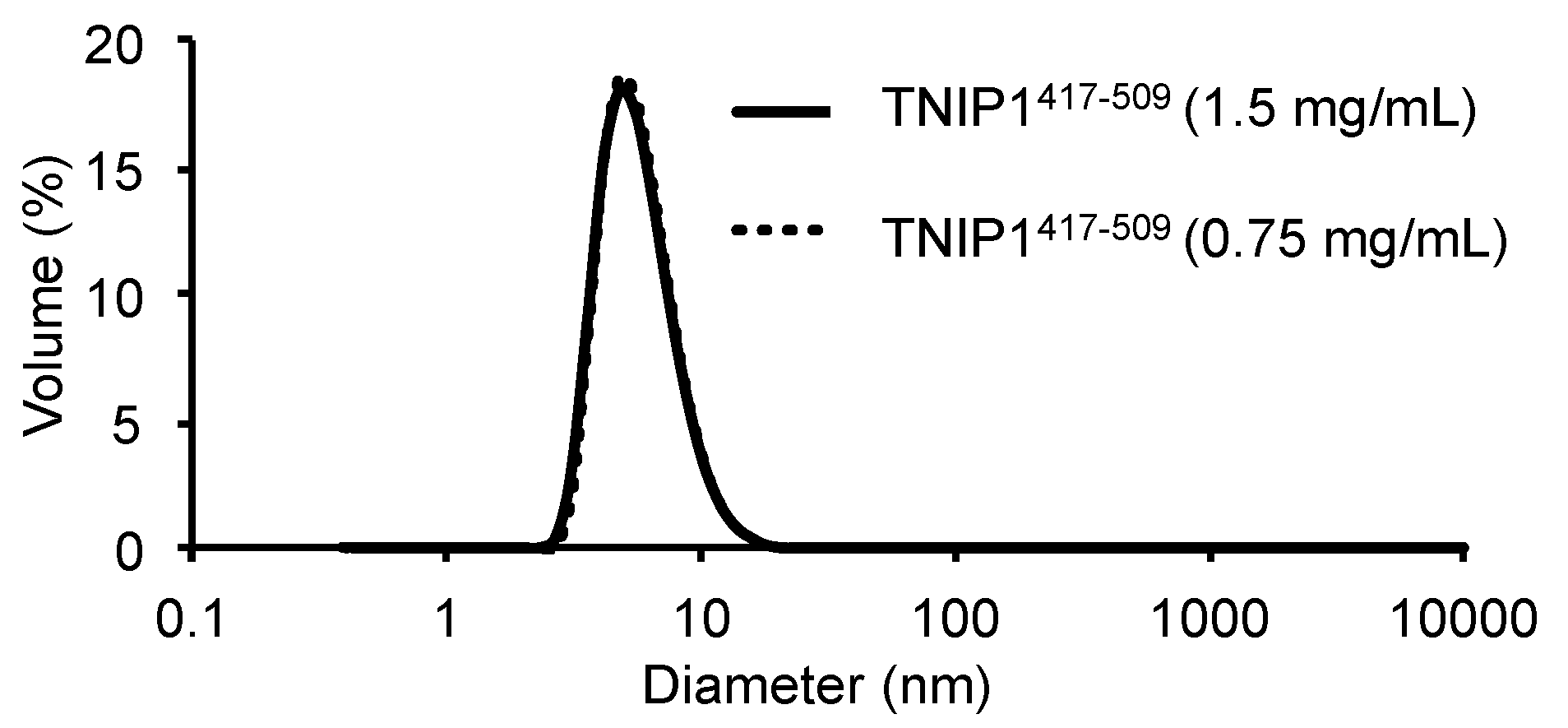
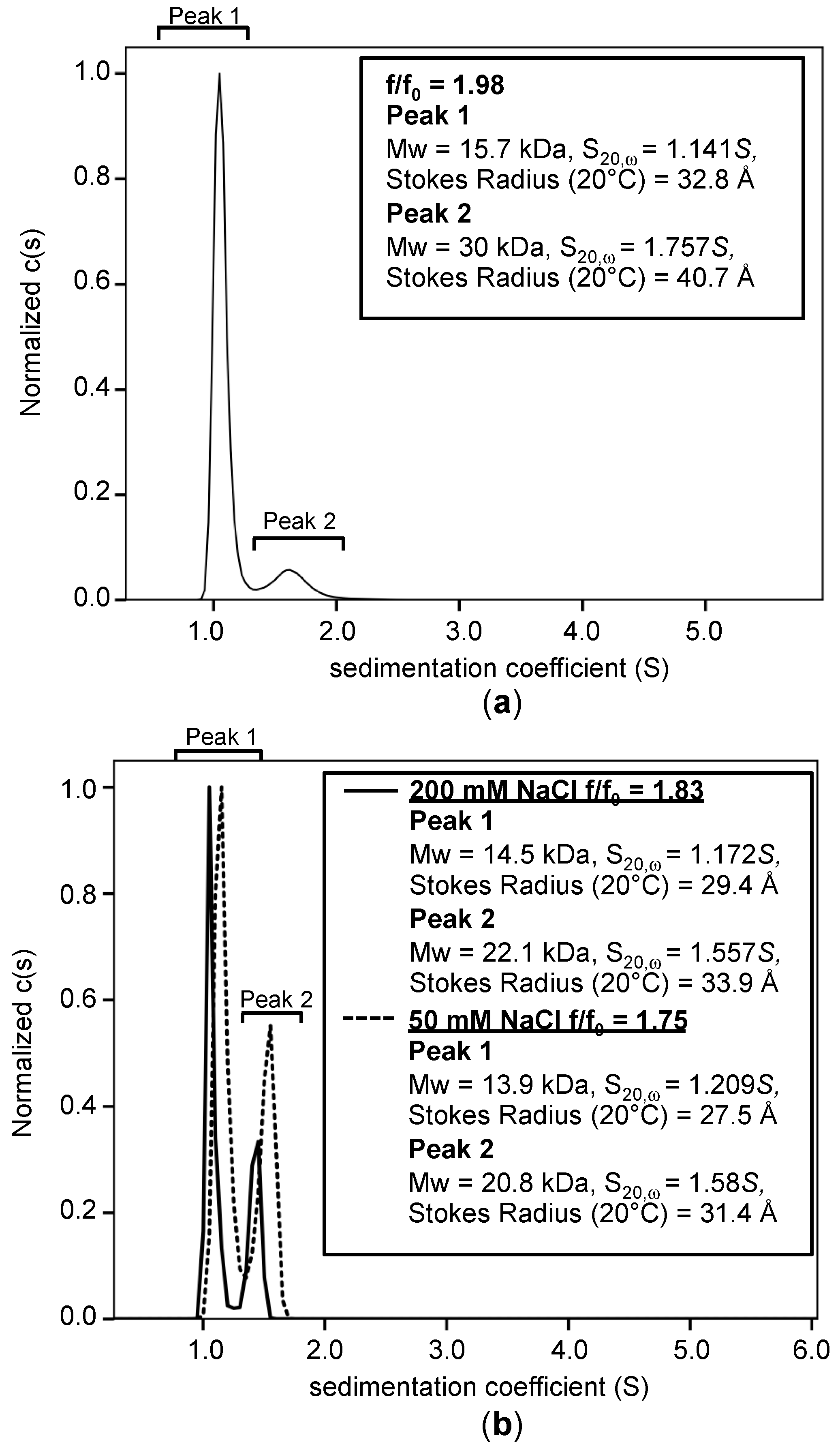
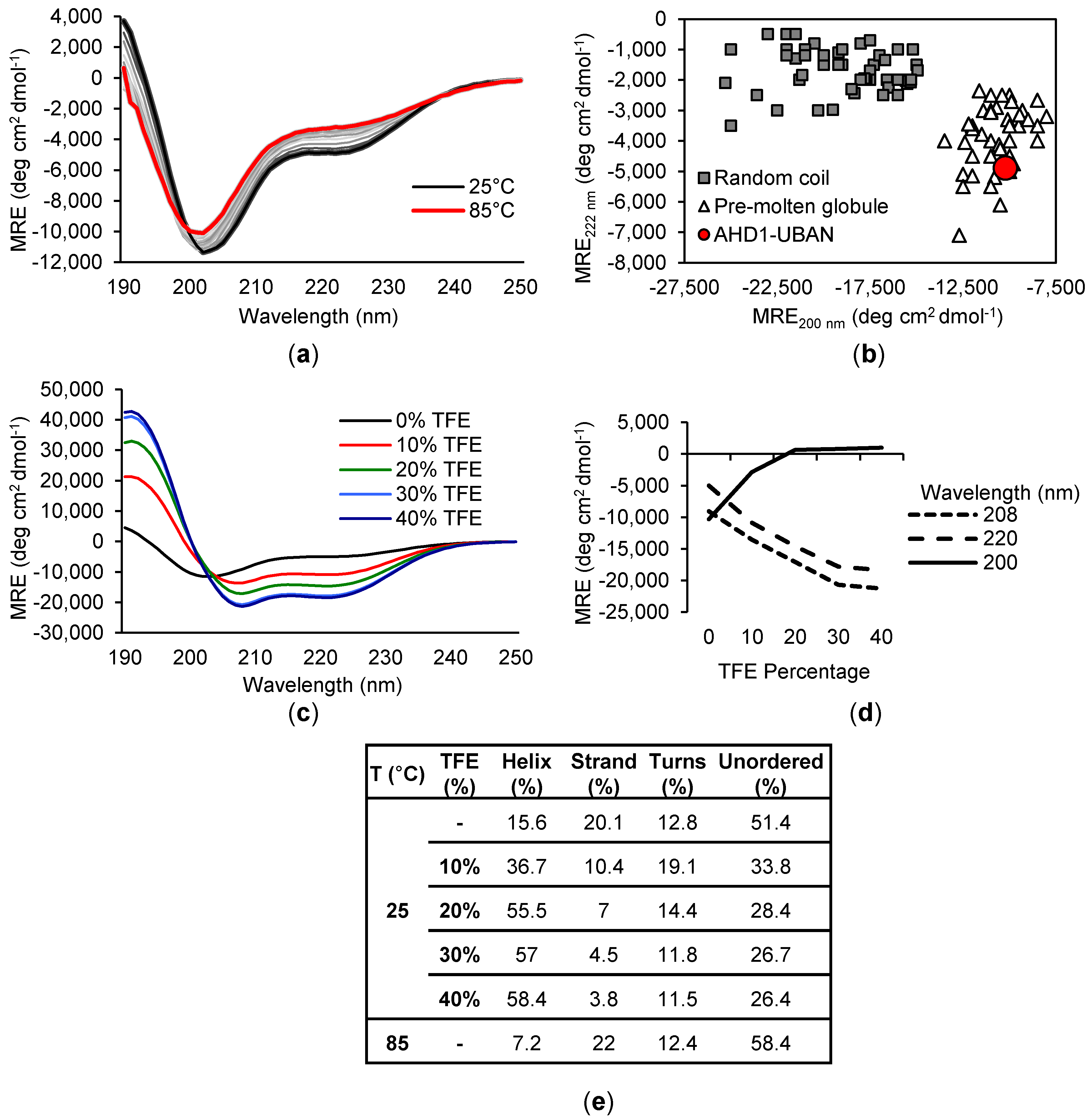
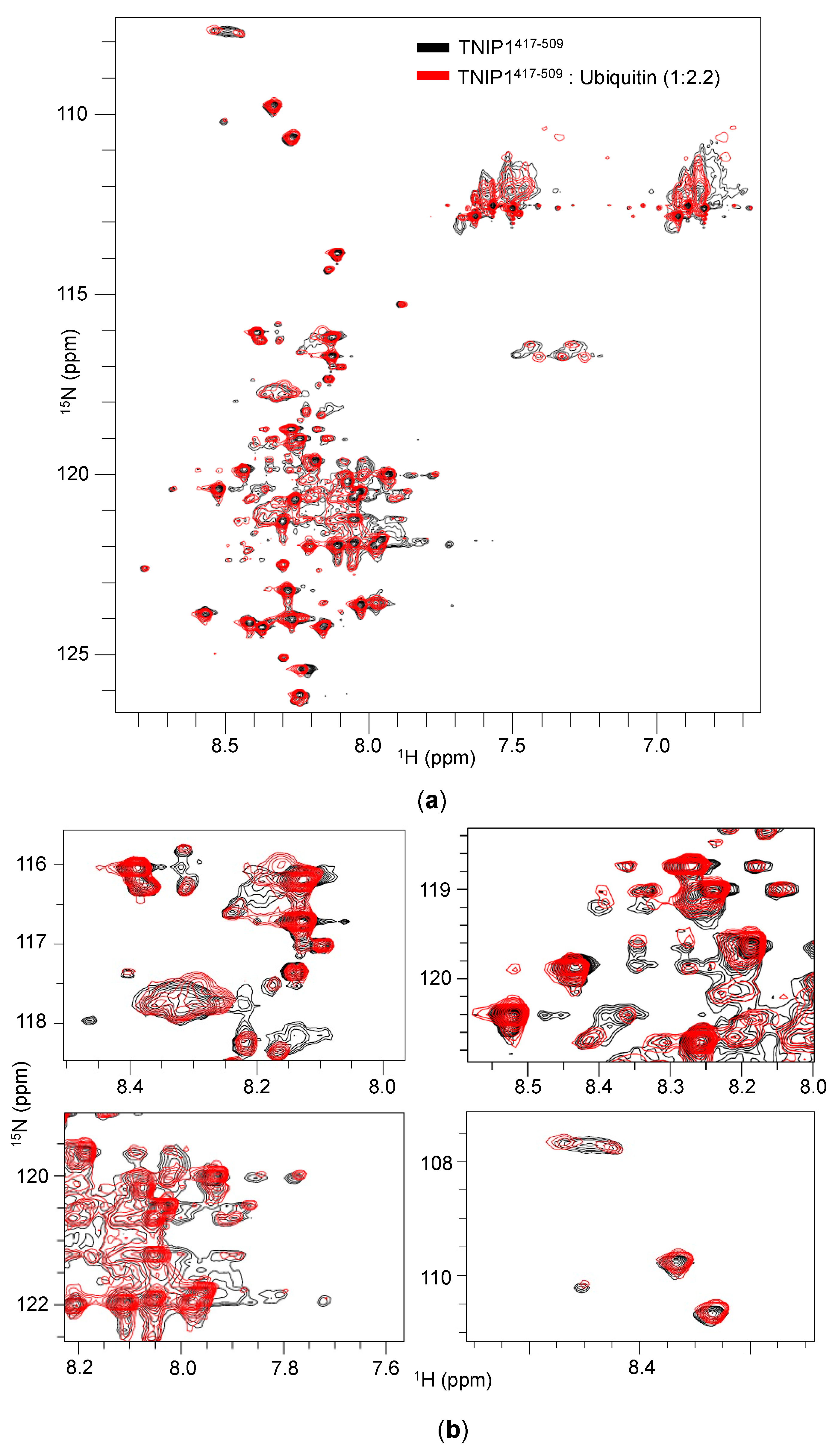
| pH | NaCl Concentration | Spectra (Total %) | Protein Concentration | f/f0 | MW (kDa) | s-Value (20, ω) | Stokes Radius (Å) | Figure |
|---|---|---|---|---|---|---|---|---|
| 8.0 | 200 mM | Peak 1 (90%) | 0.25 mg/mL (20 μM) | 1.98 | 15.7 | 1.141S | 32.8 | Figure 8a |
| Peak 2 (10%) | 30 | 1.757S | 40.7 | |||||
| 5.8 | 200 mM | Peak 1 (70%) | 0.25 mg/mL (20 μM) | 1.83 | 14.5 | 1.172S | 29.4 | Figure 8b (solid line) |
| Peak 2 (30%) | 22.1 | 1.557S | 33.9 | |||||
| 5.8 | 50 mM | Peak 1 (62%) | 0.25 mg/mL (20 μM) | 1.75 | 13.9 | 1.209S | 27.5 | Figure 8b (dotted line) |
| Peak 2 (38%) | 20.8 | 1.582S | 31.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shamilov, R.; Vinogradova, O.; Aneskievich, B.J. The Anti-Inflammatory Protein TNIP1 Is Intrinsically Disordered with Structural Flexibility Contributed by Its AHD1-UBAN Domain. Biomolecules 2020, 10, 1531. https://doi.org/10.3390/biom10111531
Shamilov R, Vinogradova O, Aneskievich BJ. The Anti-Inflammatory Protein TNIP1 Is Intrinsically Disordered with Structural Flexibility Contributed by Its AHD1-UBAN Domain. Biomolecules. 2020; 10(11):1531. https://doi.org/10.3390/biom10111531
Chicago/Turabian StyleShamilov, Rambon, Olga Vinogradova, and Brian J. Aneskievich. 2020. "The Anti-Inflammatory Protein TNIP1 Is Intrinsically Disordered with Structural Flexibility Contributed by Its AHD1-UBAN Domain" Biomolecules 10, no. 11: 1531. https://doi.org/10.3390/biom10111531
APA StyleShamilov, R., Vinogradova, O., & Aneskievich, B. J. (2020). The Anti-Inflammatory Protein TNIP1 Is Intrinsically Disordered with Structural Flexibility Contributed by Its AHD1-UBAN Domain. Biomolecules, 10(11), 1531. https://doi.org/10.3390/biom10111531






