Auxin-Abscisic Acid Interactions in Plant Growth and Development
Abstract
:1. Introduction
2. Auxin Homeostasis and Response
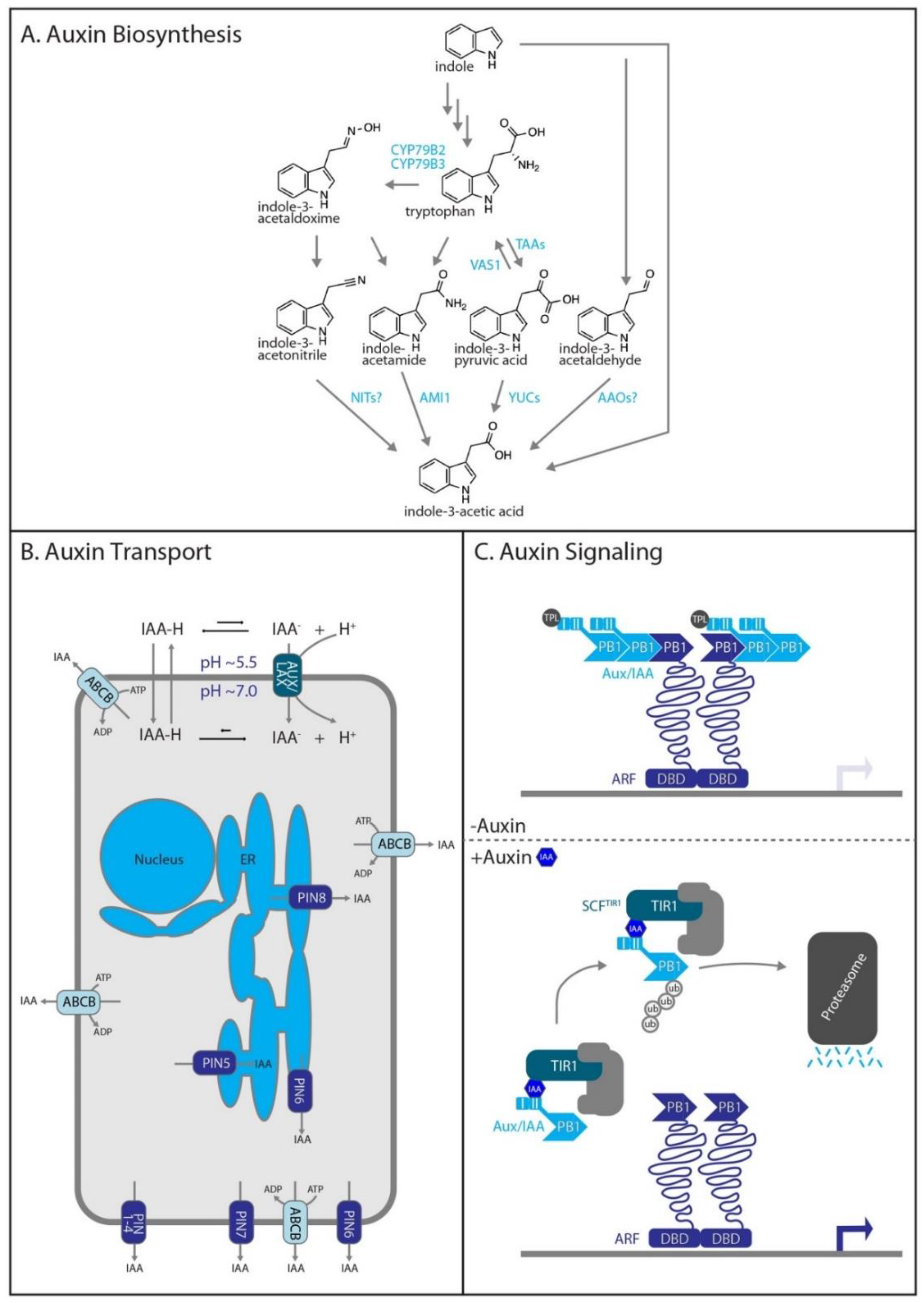
2.1. Auxin Biosynthesis
2.2. Auxin Transport
2.3. Auxin Signaling
3. ABA Homeostasis and Response
3.1. ABA Biosynthesis
3.2. ABA Transport
3.3. ABA Signaling
4. Auxin-ABA Interactions
4.1. Auxin -ABA Interactions in Seed Germination
4.2. Auxin–ABA Interactions in Cell Expansion and Hypocotyl Elongation
4.3. Auxin–ABA Interactions in Root Elongation
4.4. Auxin–ABA Interactions in Lateral Root Formation
4.5. Auxin–ABA Interactions in Cotyledon Growth
5. Discussion
6. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Weijers, D.; Wagner, D. Transcriptional Responses to the Auxin Hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Moore, S.; Chen, C.; Lindsey, K. Crosstalk Complexities between Auxin, Cytokinin, and Ethylene in Arabidopsis Root Development: From Experiments to Systems Modeling, and Back Again. Mol. Plant 2017, 10, 1480–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asghar, M.A.; Li, Y.; Jiang, H.; Sun, X.; Ahmad, B.; Imran, S.; Yu, L.; Liu, C.; Yang, W.; Du, J. Crosstalk between Abscisic Acid and Auxin under Osmotic Stress. Agron. J. 2019, 111. [Google Scholar] [CrossRef]
- Omoarelojie, L.O.; Kulkarni, M.G.; Finnie, J.F.; Van Staden, J. Strigolactones and their crosstalk with other phytohormones. Ann. Bot. 2019, 124, 749–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, D.; Ori, N. Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 2007, 144, 1240–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, H.; Strader, L.C. Interplay of Auxin and Cytokinin in Lateral Root Development. Int. J. Mol. Sci. 2019, 20, 486. [Google Scholar] [CrossRef] [Green Version]
- Perrot-Rechenmann, C. Cellular responses to auxin: Division versus expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic Acid Signaling and Abiotic Stress Tolerance in Plants: A Review on Current Knowledge and Future Prospects. Front Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Obata, T.; Feil, R.; Lunn, J.E.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Fernie, A.R. The Role of Abscisic Acid Signaling in Maintaining the Metabolic Balance Required for Arabidopsis Growth under Nonstress Conditions. Plant Cell 2019, 31, 84–105. [Google Scholar] [CrossRef] [Green Version]
- Gray, W.M.; Ostin, A.; Sandberg, G.; Romano, C.P.; Estelle, M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 7197–7202. [Google Scholar] [CrossRef] [Green Version]
- Rowe, J.H.; Topping, J.F.; Liu, J.; Lindsey, K. Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol. 2016, 211, 225–239. [Google Scholar] [CrossRef] [Green Version]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y. Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.; Yim, S.; Choi, H.; Kim, A.; Lee, K.P.; Lopez-Molina, L.; Martinoia, E.; Lee, Y. Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 2015, 6, 8113. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.; Ferrer, J.L.; Ljung, K.; Pojer, F.; Hong, F.; Long, J.A.; Li, L.; Moreno, J.E.; Bowman, M.E.; Ivans, L.J.; et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 2008, 133, 164–176. [Google Scholar] [CrossRef] [Green Version]
- Kasahara, H. Current aspects of auxin biosynthesis in plants. Biosci. Biotechnol. Biochem. 2016, 80, 34–42. [Google Scholar] [CrossRef]
- Zazimalova, E.; Murphy, A.S.; Yang, H.; Hoyerova, K.; Hosek, P. Auxin transporters--why so many? Cold Spring Harb. Perspect. Biol. 2010, 2, a001552. [Google Scholar] [CrossRef] [Green Version]
- Geilfus, C.M. The pH of the Apoplast: Dynamic Factor with Functional Impact Under Stress. Mol. Plant 2017, 10, 1371–1386. [Google Scholar] [CrossRef]
- Zazimalova, E.; Krecek, P.; Skupa, P.; Hoyerova, K.; Petrasek, J. Polar transport of the plant hormone auxin - the role of PIN-FORMED (PIN) proteins. Cell Mol. Life Sci. 2007, 64, 1621–1637. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Abd_Allah, E.F.; Khan, A.L.; Al-Harrasi, A.S. Molecular players of auxin transport systems: Advances in genomic and molecular events. J. Plant Interact. 2018, 13, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Salehin, M.; Bagchi, R.; Estelle, M. SCFTIR1/AFB-based auxin perception: Mechanism and role in plant growth and development. Plant Cell 2015, 27, 9–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strader, L.C.; Zhao, Y. Auxin perception and downstream events. Curr. Opin. Plant Biol. 2016, 33, 8–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powers, S.K.; Strader, L.C. Regulation of auxin transcriptional responses. Dev. Dyn. 2019. [Google Scholar] [CrossRef] [Green Version]
- Weijers, D.; Nemhauser, J.; Yang, Z. Auxin: Small molecule, big impact. J. Exp. Bot. 2018, 69, 133–136. [Google Scholar] [CrossRef]
- Simon, S.; Skupa, P.; Viaene, T.; Zwiewka, M.; Tejos, R.; Klima, P.; Carna, M.; Rolcik, J.; De Rycke, R.; Moreno, I.; et al. PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol. 2016, 211, 65–74. [Google Scholar] [CrossRef]
- Finkelstein, R. Abscisic Acid synthesis and response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef] [Green Version]
- Seiler, C.; Harshavardhan, V.T.; Rajesh, K.; Reddy, P.S.; Strickert, M.; Rolletschek, H.; Scholz, U.; Wobus, U.; Sreenivasulu, N. ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J. Exp. Bot. 2011, 62, 2615–2632. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Guzman, M.; Apostolova, N.; Belles, J.M.; Barrero, J.M.; Piqueras, P.; Ponce, M.R.; Micol, J.L.; Serrano, R.; Rodriguez, P.L. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 2002, 14, 1833–1846. [Google Scholar] [CrossRef] [Green Version]
- Seo, M.; Peeters, A.J.; Koiwai, H.; Oritani, T.; Marion-Poll, A.; Zeevaart, J.A.; Koornneef, M.; Kamiya, Y.; Koshiba, T. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA 2000, 97, 12908–12913. [Google Scholar] [CrossRef] [Green Version]
- Akira Endo, M.O.; Koshiba, T. Abscisic Acid: Metabolism, Transport and Signaling; Zhang, D.-P., Ed.; Springer: Dordrecht, The Netherlands, 2014. [Google Scholar]
- Boursiac, Y.; Leran, S.; Corratge-Faillie, C.; Gojon, A.; Krouk, G.; Lacombe, B. ABA transport and transporters. Trends Plant Sci. 2013, 18, 325–333. [Google Scholar] [CrossRef]
- Ikegami, K.; Okamoto, M.; Seo, M.; Koshiba, T. Activation of abscisic acid biosynthesis in the leaves of Arabidopsis thaliana in response to water deficit. J. Plant Res. 2009, 122, 235–243. [Google Scholar] [CrossRef]
- Kuromori, T.; Miyaji, T.; Yabuuchi, H.; Shimizu, H.; Sugimoto, E.; Kamiya, A.; Moriyama, Y.; Shinozaki, K. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. USA 2010, 107, 2361–2366. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhu, H.; Pan, Y.; Yu, Y.; Luan, S.; Li, L. A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol. Plant 2014, 7, 1522–1532. [Google Scholar] [CrossRef] [Green Version]
- Borghi, L.; Kang, J.; Ko, D.; Lee, Y.; Martinoia, E. The role of ABCG-type ABC transporters in phytohormone transport. Biochem. Soc. Trans. 2015, 43, 924–930. [Google Scholar] [CrossRef]
- Kuromori, T.; Sugimoto, E.; Shinozaki, K. Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J. 2011, 67, 885–894. [Google Scholar] [CrossRef]
- Merilo, E.; Jalakas, P.; Laanemets, K.; Mohammadi, O.; Horak, H.; Kollist, H.; Brosche, M. Abscisic Acid Transport and Homeostasis in the Context of Stomatal Regulation. Mol. Plant 2015, 8, 1321–1333. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Cao, J.; He, J.; Chen, Q.; Li, X.; Yang, Y. Molecular Mechanism for the Regulation of ABA Homeostasis During Plant Development and Stress Responses. Int. J. Mol. Sci. 2018, 19, 3643. [Google Scholar] [CrossRef] [Green Version]
- Ng, L.M.; Melcher, K.; Teh, B.T.; Xu, H.E. Abscisic acid perception and signaling: Structural mechanisms and applications. Acta Pharmacol. Sin. 2014, 35, 567–584. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Zhang, W.; Wang, X. Post-translational control of ABA signalling: The roles of protein phosphorylation and ubiquitination. Plant Biotechnol. J. 2017, 15, 4–14. [Google Scholar] [CrossRef]
- Brandt, B.; Brodsky, D.E.; Xue, S.; Negi, J.; Iba, K.; Kangasjarvi, J.; Ghassemian, M.; Stephan, A.B.; Hu, H.; Schroeder, J.I. Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. USA 2012, 109, 10593–10598. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Murata, M.; Minami, H.; Yamamoto, S.; Kagaya, Y.; Hobo, T.; Yamamoto, A.; Hattori, T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005, 44, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Sato, Y.; Fukao, Y.; Fujiwara, M.; Umezawa, T.; Shinozaki, K.; Hibi, T.; Taniguchi, M.; Miyake, H.; Goto, D.B.; et al. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 2009, 424, 439–448. [Google Scholar] [CrossRef] [Green Version]
- Weiner, J.J.; Peterson, F.C.; Volkman, B.F.; Cutler, S.R. Structural and functional insights into core ABA signaling. Curr. Opin. Plant Biol. 2010, 13, 495–502. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, Y.; Takahashi, K.; Inoue, S.; Kinoshita, T. Abscisic acid suppresses hypocotyl elongation by dephosphorylating plasma membrane H(+)-ATPase in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 845–853. [Google Scholar] [CrossRef] [Green Version]
- Strader, L.C.; Wheeler, D.L.; Christensen, S.E.; Berens, J.C.; Cohen, J.D.; Rampey, R.A.; Bartel, B. Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 2011, 23, 984–999. [Google Scholar] [CrossRef] [Green Version]
- Lorrai, R.; Boccaccini, A.; Ruta, V.; Possenti, M.; Costantino, P.; Vittorioso, P. Abscisic acid inhibits hypocotyl elongation acting on gibberellins, DELLA proteins and auxin. AoB Plants 2018, 10, ply061. [Google Scholar] [CrossRef]
- Chapman, E.J.; Greenham, K.; Castillejo, C.; Sartor, R.; Bialy, A.; Sun, T.P.; Estelle, M. Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS ONE 2012, 7, e36210. [Google Scholar] [CrossRef] [Green Version]
- Reed, R.C.; Brady, S.R.; Muday, G.K. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol. 1998, 118, 1369–1378. [Google Scholar] [CrossRef] [Green Version]
- Thole, J.M.; Beisner, E.R.; Liu, J.; Venkova, S.V.; Strader, L.C. Abscisic acid regulates root elongation through the activities of auxin and ethylene in Arabidopsis. G3 (Bethesda) 2014, 4, 1259–1274. [Google Scholar] [CrossRef] [Green Version]
- Rymen, B.; Kawamura, A.; Schafer, S.; Breuer, C.; Iwase, A.; Shibata, M.; Ikeda, M.; Mitsuda, N.; Koncz, C.; Ohme-Takagi, M.; et al. ABA Suppresses Root Hair Growth via the OBP4 Transcriptional Regulator. Plant Physiol. 2017, 173, 1750–1762. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Huang, L.; Yan, A.; Liu, Y.; Liu, B.; Yu, C.; Zhang, A.; Schiefelbein, J.; Gan, Y. Multiple phytohormones promote root hair elongation by regulating a similar set of genes in the root epidermis in Arabidopsis. J. Exp. Bot. 2016, 67, 6363–6372. [Google Scholar] [CrossRef] [PubMed]
- Garciarrubio, A.; Legaria, J.P.; Covarrubias, A.A. Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta 1997, 203, 182–187. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, H.; Zhao, Y.; Feng, Z.; Li, Q.; Yang, H.Q.; Luan, S.; Li, J.; He, Z.H. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15485–15490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinaldi, M.A.; Liu, J.; Enders, T.A.; Bartel, B.; Strader, L.C. A gain-of-function mutation in IAA16 confers reduced responses to auxin and abscisic acid and impedes plant growth and fertility. Plant Mol. Biol. 2012, 79, 359–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enders, T.A.; Frick, E.M.; Strader, L.C. An Arabidopsis kinase cascade influences auxin-responsive cell expansion. Plant J. 2017, 92, 68–81. [Google Scholar] [CrossRef]
- De Smet, I.; Signora, L.; Beeckman, T.; Inze, D.; Foyer, C.H.; Zhang, H. An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 2003, 33, 543–555. [Google Scholar] [CrossRef] [Green Version]
- Casimiro, I.; Marchant, A.; Bhalerao, R.P.; Beeckman, T.; Dhooge, S.; Swarup, R.; Graham, N.; Inze, D.; Sandberg, G.; Casero, P.J.; et al. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 2001, 13, 843–852. [Google Scholar] [CrossRef] [Green Version]
- Shkolnik-Inbar, D.; Bar-Zvi, D. Expression of ABSCISIC ACID INSENSITIVE 4 (ABI4) in developing Arabidopsis seedlings. Plant Signal Behav. 2011, 6, 694–696. [Google Scholar] [CrossRef] [Green Version]
- Brady, S.M.; Sarkar, S.F.; Bonetta, D.; McCourt, P. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 2003, 34, 67–75. [Google Scholar] [CrossRef]
- Kermode, A.R. Role of Abscisic Acid in Seed Dormancy. J. Plant Growth Regul. 2005, 24, 319–344. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Yang, P. Studies on the molecular mechanisms of seed germination. Proteomics 2015, 15, 1671–1679. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strader, L.C.; Monroe-Augustus, M.; Bartel, B. The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol. 2008, 8, 41. [Google Scholar] [CrossRef] [Green Version]
- Belin, C.; Megies, C.; Hauserova, E.; Lopez-Molina, L. Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 2009, 21, 2253–2268. [Google Scholar] [CrossRef] [Green Version]
- Monroe-Augustus, M.; Zolman, B.K.; Bartel, B. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 2003, 15, 2979–2991. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Hua, D.; He, J.; Duan, Y.; Chen, Z.; Hong, X.; Gong, Z. Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet 2011, 7, e1002172. [Google Scholar] [CrossRef] [Green Version]
- Gendreau, E.; Traas, J.; Desnos, T.; Grandjean, O.; Caboche, M.; Hofte, H. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 1997, 114, 295–305. [Google Scholar] [CrossRef] [Green Version]
- Haruta, M.; Gray, W.M.; Sussman, M.R. Regulation of the plasma membrane proton pump (H(+)-ATPase) by phosphorylation. Curr. Opin. Plant Biol. 2015, 28, 68–75. [Google Scholar] [CrossRef] [Green Version]
- Rayle, D.L.; Cleland, R.E. The Acid Growth Theory of auxin-induced cell elongation is alive and well. Plant Physiol. 1992, 99, 1271–1274. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Hayashi, K.; Kinoshita, T. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 2012, 159, 632–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, H.; Gray, W.M. SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef] [Green Version]
- Spartz, A.K.; Ren, H.; Park, M.Y.; Grandt, K.N.; Lee, S.H.; Murphy, A.S.; Sussman, M.R.; Overvoorde, P.J.; Gray, W.M. SAUR Inhibition of PP2C-D Phosphatases Activates Plasma Membrane H+-ATPases to Promote Cell Expansion in Arabidopsis. Plant Cell 2014, 26, 2129–2142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, H.; Park, M.Y.; Spartz, A.K.; Wong, J.H.; Gray, W.M. A subset of plasma membrane-localized PP2C.D phosphatases negatively regulate SAUR-mediated cell expansion in Arabidopsis. PLoS Genet 2018, 14, e1007455. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Spartz, A.K.; Park, M.Y.; Du, M.; Gray, W.M. Mutation of a Conserved Motif of PP2C.D Phosphatases Confers SAUR Immunity and Constitutive Activity. Plant Physiol. 2019, 181, 353–366. [Google Scholar] [CrossRef] [Green Version]
- Spartz, A.K.; Lor, V.S.; Ren, H.; Olszewski, N.E.; Miller, N.D.; Wu, G.; Spalding, E.P.; Gray, W.M. Constitutive Expression of Arabidopsis SMALL AUXIN UP RNA19 (SAUR19) in Tomato Confers Auxin-Independent Hypocotyl Elongation. Plant Physiol. 2017, 173, 1453–1462. [Google Scholar] [CrossRef] [Green Version]
- Qin, H.; Huang, R. Auxin Controlled by Ethylene Steers Root Development. Int. J. Mol. Sci. 2018, 19, 3656. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.R.; Wang, Y.B.; He, S.B.; Hao, F.S. Mechanisms for Abscisic Acid Inhibition of Primary Root Growth. Plant Signal Behav. 2018, 13, e1500069. [Google Scholar] [CrossRef] [Green Version]
- Slovak, R.; Ogura, T.; Satbhai, S.B.; Ristova, D.; Busch, W. Genetic control of root growth: From genes to networks. Ann. Bot. 2016, 117, 9–24. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Chen, L.; Forde, B.G.; Davies, W.J. The Biphasic Root Growth Response to Abscisic Acid in Arabidopsis Involves Interaction with Ethylene and Auxin Signalling Pathways. Front. Plant Sci. 2017, 8, 1493. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Scheres, B. Lateral root formation and the multiple roles of auxin. J. Exp. Bot. 2018, 69, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Chen, M.X.; Liu, R.; Zhang, L.; Hou, X.; Liu, S.; Ding, X.; Jiang, Y.; Xu, J.; Zhang, J.; et al. Abscisic Acid Regulates Auxin Distribution to Mediate Maize Lateral Root Development Under Salt Stress. Front. Plant. Sci. 2019, 10, 716. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Zhao, Y.; Gao, J.; Xiang, C.; Zhu, J.K. The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci. Rep. 2016, 6, 27177. [Google Scholar] [CrossRef] [PubMed]
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 2007, 19, 2440–2453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shkolnik-Inbar, D.; Bar-Zvi, D. ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 2010, 22, 3560–3573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoynova-Bakalova, E.; Karanov, E.; Petrov, P.; Hall, M.A. Cell division and cell expansion in cotyledons of Arabidopsis seedlings. New Phytologist 2004, 162, 471–479. [Google Scholar] [CrossRef]
- Mansfield, S.G.; Briarty, L.G. The Dynamics of Seedling and Cotyledon Cell Development in Arabidopsis thaliana During Reserve Mobilization. Int. J. Plant Sci. 1996, 157, 280–295. [Google Scholar] [CrossRef]
- Strader, L.C.; Culler, A.H.; Cohen, J.D.; Bartel, B. Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol. 2010, 153, 1577–1586. [Google Scholar] [CrossRef] [Green Version]


| Treatment Effect | |||
|---|---|---|---|
| Tissue | ABA | Auxin | ABA-Auxin Interaction |
| Dark-grown hypocotyls | Inhibits growth [46] | Inhibits growth [47] | unknown |
| Light-grown hypocotyls | Inhibits growth (grown under red light) [48] | Promotes growth [49] | unknown |
| Dark-grown roots | unknown | Inhibits growth [50] | unknown |
| Light-grown roots | Inhibits growth [51] | Inhibits growth [51] | Auxin downstream [51] |
| Root hairs | Inhibits expansion [52] | Promotes expansion [53] | unknown |
| Seed germination | Inhibits [54] | Inhibits [55] | Auxin downstream [51,56] |
| Cotyledon expansion | Inhibits [48] | Inhibits [57] | unknown |
| Lateral root formation | Inhibits [58] | Promotes [59] | Auxin downstream [60,61] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emenecker, R.J.; Strader, L.C. Auxin-Abscisic Acid Interactions in Plant Growth and Development. Biomolecules 2020, 10, 281. https://doi.org/10.3390/biom10020281
Emenecker RJ, Strader LC. Auxin-Abscisic Acid Interactions in Plant Growth and Development. Biomolecules. 2020; 10(2):281. https://doi.org/10.3390/biom10020281
Chicago/Turabian StyleEmenecker, Ryan J., and Lucia C. Strader. 2020. "Auxin-Abscisic Acid Interactions in Plant Growth and Development" Biomolecules 10, no. 2: 281. https://doi.org/10.3390/biom10020281
APA StyleEmenecker, R. J., & Strader, L. C. (2020). Auxin-Abscisic Acid Interactions in Plant Growth and Development. Biomolecules, 10(2), 281. https://doi.org/10.3390/biom10020281





