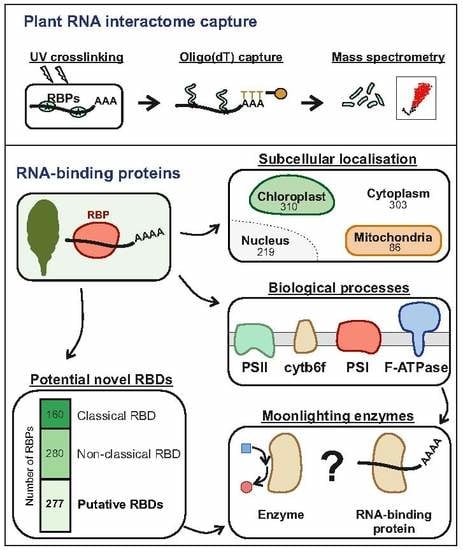
Discovering the RNA-Binding Proteome of Plant Leaves with an Improved RNA Interactome Capture Method
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Plant RNA Interactome Capture (ptRIC)
2.2.1. UV Crosslinking
2.2.2. Cell Lysis
2.2.3. Oligo(dT) Capture
2.3. Downstream Applications of Isolated RNA–Protein Complexes
2.3.1. RNA Quantification and Normalization
2.3.2. RNase Digestion
2.3.3. Protein Concentration and Western Blot or Silver Staining
2.4. Data Analysis
Statistical Analysis of the RBPome
2.5. Data Availability
3. Results and Discussion
3.1. Adapting RIC to Plant Leaves
3.2. ptRIC Captures Bona Fide RBPs
3.3. Building a High-Confidence Leaf RBPome for Arabidopsis
3.4. Function and Localization of Leaf RBPs
3.5. RNA-Binding Domains in Leaf RBPs
3.6. RBPs Uniquely Identified by ptRIC
3.7. Unconventional RNA-Binding Proteins in Plant Leaves
3.8. Proteins of the Photosynthetic Apparatus Moonlight as RNA Binders
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008, 582, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Sysoev, V.O.; Fischer, B.; Frese, C.K.; Gupta, I.; Krijgsveld, J.; Hentze, M.W.; Castello, A.; Ephrussi, A. Global changes of the RNA-bound proteome during the maternal-to-zygotic transition in Drosophila. Nat. Commun. 2016, 7, 12128. [Google Scholar] [CrossRef] [PubMed]
- Perez-Perri, J.I.; Rogell, B.; Schwarzl, T.; Stein, F.; Zhou, Y.; Rettel, M.; Brosig, A.; Hentze, M.W. Discovery of RNA-binding proteins and characterization of their dynamic responses by enhanced RNA interactome capture. Nat. Commun. 2018, 9, 4408. [Google Scholar] [CrossRef]
- Garcia-Moreno, M.; Noerenberg, M.; Ni, S.; Järvelin, A.I.; González-Almela, E.; Lenz, C.E.; Bach-Pages, M.; Cox, V.; Avolio, R.; Davis, T.; et al. System-wide profiling of RNA-binding proteins uncovers key regulators of virus infection. Mol. Cell 2019, 74, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Trendel, J.; Schwarzl, T.; Horos, R.; Prakash, A.; Bateman, A.; Hentze, M.W.; Krijgsveld, J. The human RNA-binding proteome and Its dynamics during translational arrest. Cell 2019, 176, 391–403. [Google Scholar] [CrossRef]
- Scherrer, T.; Mittal, N.; Janga, S.C.; Gerber, A.P. A screen for RNA-binding proteins in yeast indicates dual functions for many enzymes. PLoS ONE 2010, 5, e15499. [Google Scholar] [CrossRef]
- Tsvetanova, N.G.; Klass, D.M.; Salzman, J.; Brown, P.O. Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. PLoS ONE 2010, 5, e12671. [Google Scholar] [CrossRef]
- Butter, M.; Scheibe, M.; Mörl, M.; Mann, M. Unbiased RNA–Protein interaction screen by quantitative proteomics. Proc. Natl. Acad. Sci. USA 2009, 106, 10626–10631. [Google Scholar] [CrossRef]
- Castello, A.; Horos, R.; Strein, C.; Fischer, B.; Eichelbaum, K.; Steinmetz, L.M.; Krijgsveld, J.; Hentze, M.W. System-wide identification of RNA-binding proteins by interactome capture. Nat. Protoc. 2013, 8, 491–500. [Google Scholar] [CrossRef]
- Anantharaman, V.; Koonin, E.V.; Aravind, L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002, 30, 1427–1464. [Google Scholar] [CrossRef]
- Järvelin, A.I.; Noerenberg, M.; Davis, I.; Castello, A. The new (dis) order in RNA regulation. Cell Commun. Signal. 2016, 14, 9. [Google Scholar] [CrossRef] [PubMed]
- Castello, A.; Fischer, B.; Frese, C.K.; Horos, R.; Alleaume, A.M.; Foehr, S.; Curk, T.; Krijgsveld, J.; Hentze, M.W. Comprehensive identification of RNA-binding domains in human cells. Mol. Cell 2016, 63, 696–710. [Google Scholar] [CrossRef] [PubMed]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Baltz, A.G.; Munschauer, M.; Schwanhausser, B.; Vasile, A.; Murakawa, Y.; Schueler, M.; Youngs, N.; Penfold-Brown, D.; Drew, K.; Milek, M.; et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 2012, 46, 674–690. [Google Scholar] [CrossRef] [PubMed]
- Pashev, I.G.; Dimitrov, S.I.; Angelov, D. Crosslinking proteins to nucleic acids by ultraviolet laser irradiation. Trends Biochem. Sci. 1991, 16, 323–326. [Google Scholar] [CrossRef]
- Lueong, S.; Merce, C.; Fischer, B.; Hoheisel, J.D.; Erben, E.D. Gene expression regulatory networks in Trypanosoma brucei: Insights into the role of the mRNA-binding proteome. Mol. Microbiol. 2016, 100, 457–471. [Google Scholar] [CrossRef]
- Nandan, D.; Thomas, S.A.; Nguyen, A.; Moon, K.-M.; Foster, L.J.; Reiner, N.E. Comprehensive Identification of mRNA-Binding Proteins of Leishmania donovani by Interactome Capture. PLoS ONE 2017, 12, e0170068. [Google Scholar] [CrossRef]
- De Pablos, L.M.; Ferreira, T.R.; Dowle, A.; Forrester, S.; Parry, E.; Newling, K.; Walrad, P.B. The mRNA-bound proteome of Leishmania mexicana: Novel genetic insight into an ancient parasite. Mol. Cell. Proteom. 2019, 18, 1271–1284. [Google Scholar] [CrossRef]
- Bunnik, E.M.; Batugedara, G.; Saraf, A.; Prudhomme, J.; Florens, L.; Le Roch, K.G. The mRNA-bound proteome of the human malaria parasite Plasmodium falciparum. Genome Biol. 2016, 17, 147. [Google Scholar] [CrossRef]
- Beckmann, B.M. RNA interactome capture in yeast. Methods 2017, 118–119, 82–92. [Google Scholar] [CrossRef]
- Beckmann, B.M.; Horos, R.; Fischer, B.; Castello, A.; Eichelbaum, K.; Alleaume, A.-M.; Schwarzl, T.; Curk, T.; Foehr, S.; Huber, W.; et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015, 6, 10127. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.F.; Jain, S.; She, M.; Parker, R. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 2013, 20, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Matia-González, A.M.; Laing, E.E.; Gerber, A.P. Conserved mRNA-binding proteomes in eukaryotic organisms. Nat. Struct. Mol. Biol. 2015, 22, 1027–1033. [Google Scholar] [CrossRef] [PubMed]
- Wessels, H.-H.; Imami, K.; Baltz, A.G.; Kolinski, M.; Beldovskaya, A.; Selbach, M.; Small, S.; Ohler, U.; Landthaler, M. The mRNA-bound proteome of the early fly embryo. Genome Res. 2016, 26, 1000–1009. [Google Scholar] [CrossRef]
- Despic, V.; Dejung, M.; Gu, M.; Krishnan, J.; Zhang, J.; Herzel, L.; Straube, K.; Gerstein, M.B.; Butter, F.; Neugebauer, K.M. Dynamic RNA-protein interactions underlie the zebrafish maternal-to-zygotic transition. Genome Res. 2017, 27, 1184–1194. [Google Scholar] [CrossRef]
- Kwon, S.C.; Yi, H.; Eichelbaum, K.; Fohr, S.; Fischer, B.; You, K.T.; Castello, A.; Krijgsveld, J.; Hentze, M.W.; Kim, V.N. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1122–1130. [Google Scholar] [CrossRef]
- Liao, Y.; Castello, A.; Fischer, B.; Leicht, S.; Föehr, S.; Frese, C.K.; Ragan, C.; Kurscheid, S.; Pagler, E.; Yang, H.; et al. The cardiomyocyte RNA-binding proteome: Links to intermediary metabolism and heart disease. Cell Rep. 2016, 16, 1456–1469. [Google Scholar] [CrossRef]
- Liepelt, A.; Naarmann-de Vries, I.S.; Simons, N.; Eichelbaum, K.; Föhr, S.; Archer, S.K.; Castello, A.; Usadel, B.; Krijgsveld, J.; Preiss, T.; et al. Identification of RNA-binding proteins in macrophages by interactome capture. Mol. Cell. Proteom. 2016, 15, 2699–2714. [Google Scholar] [CrossRef]
- Boucas, J.; Fritz, C.; Schmitt, A.; Riabinska, A.; Thelen, L.; Peifer, M.; Leeser, U.; Nuernberg, P.; Altmueller, J.; Gaestel, M.; et al. Label-free protein-RNA interactome analysis identifies Khsrp signaling downstream of the p38/Mk2 kinase complex as a critical modulator of cell cycle progression. PLoS ONE 2015, 10, e0125745. [Google Scholar] [CrossRef]
- Kramer, K.; Sachsenberg, T.; Beckmann, B.M.; Qamar, S.; Boon, K.-L.; Hentze, M.W.; Kohlbacher, O.; Urlaub, H. Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat. Methods 2014, 11, 1064–1070. [Google Scholar] [CrossRef]
- Conrad, T.; Albrecht, A.-S.; De Melo Costa, V.R.; Sauer, S.; Meierhofer, D.; Ørom, U.A. Serial interactome capture of the human cell nucleus. Nat. Commun. 2016, 7, 11212. [Google Scholar] [CrossRef]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Bach-Pages, M.; Castello, A.; Preston, G.M. Plant RNA interactome capture: Revealing the plant RBPome. Trends Plant Sci. 2017, 22, 449–451. [Google Scholar] [CrossRef] [PubMed]
- Reichel, M.; Liao, Y.; Rettel, M.; Ragan, C.; Evers, M.; Alleaume, A.-M.; Horos, R.; Hentze, M.W.; Preiss, T.; Millar, A.A. In planta determination of the mRNA-binding proteome of Arabidopsis etiolated seedlings. Plant Cell 2016, 28, 2435–2452. [Google Scholar] [CrossRef] [PubMed]
- Marondedze, C.; Thomas, L.; Serrano, N.L.; Lilley, K.S.; Gehring, C. The RNA-binding protein repertoire of Arabidopsis thaliana. Sci. Rep. 2016, 6, 29766. [Google Scholar] [CrossRef]
- Marondedze, C.; Thomas, L.; Gehring, C.; Lilley, K.S. Changes in the Arabidopsis RNA-binding proteome reveal novel stress response mechanisms. BMC Plant Biol. 2019, 19, 1–11. [Google Scholar] [CrossRef]
- Zhang, Z.; Boonen, K.; Ferrari, P.; Schoofs, L.; Janssens, E.; Van Noort, V.; Rolland, F.; Geuten, K. UV crosslinked mRNA-binding proteins captured from leaf mesophyll protoplasts. Plant Methods 2016, 12, 42. [Google Scholar] [CrossRef]
- Zhang, Z.; Boonen, K.; Li, M.; Geuten, K. mRNA interactome capture from plant protoplasts. J. Vis. Exp. 2017, 125, e56011. [Google Scholar] [CrossRef]
- Köster, T.; Reichel, M.; Staiger, D. CLIP and RNA interactome studies to unravel genome-wide RNA-protein interactions in vivo in Arabidopsis thaliana. Methods 2019. in Press. [Google Scholar]
- Bass, J.; Wilkinson, D.; Rankin, D.; Phillips, B.; Szewczyk, N.; Smith, K.; Atherton, P. An overview of technical considerations for Western blotting applications to physiological research. Scand. J. Med. Sci. Sport. 2017, 27, 4–25. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 1–25. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Csordas, A.; Del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef] [PubMed]
- Urdaneta, E.C.; Vieira-Vieira, C.H.; Hick, T.; Wessels, H.H.; Figini, D.; Moschall, R.; Medenbach, J.; Ohler, U.; Granneman, S.; Selbach, M.; et al. Purification of cross-linked RNA-protein complexes by phenol-toluol extraction. Nat. Commun. 2019, 10, 990. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Qi, Y. RNAi in plants: An argonaute-centered view. Plant Cell 2016, 28, 272–285. [Google Scholar] [CrossRef]
- Rocha, P.S.C.F.; Sheikh, M.; Melchiorre, R.; Fagard, M.; Boutet, S.; Loach, R.; Moffatt, B.; Wagner, C.; Vaucheret, H.; Furner, I. The Arabidopsis Homology-Dependent Gene Silencing1 gene codes for an S-adenosyl-L-homocysteine hydrolase required for DNA methylation-dependent gene silencing. Plant Cell 2005, 17, 404–417. [Google Scholar] [CrossRef]
- Schuster, G.; Stern, D. RNA Polyadenylation and Decay in Mitochondria and Chloroplasts. In Progress in Molecular Biology and Translational Science; Academic Press: Cambridge, MA, USA, 2009; Volume 85, pp. 393–422. [Google Scholar]
- Kudla, J.; Hayes, R.; Gruissem, W. Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J. 1996, 15, 7137–7146. [Google Scholar] [CrossRef]
- Lunde, B.M.; Moore, C.; Varani, G. RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007, 8, 479–490. [Google Scholar] [CrossRef]
- Pisarev, A.V.; Kolupaeva, V.G.; Yusupov, M.M.; Hellen, C.U.T.; Pestova, T.V. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008, 27, 1609–1621. [Google Scholar] [CrossRef]
- Kilchert, C.; Kecman, T.; Priest, E.; Hester, S.; Kus, K.; Castello, A.; Mohammed, S.; Vasiljeva, L. System-wide analyses of the fission yeast poly(A)+ RNA interactome reveal insights into organisation and function of RNA-protein complexes. bioRxiv 2019. [Google Scholar] [CrossRef]
- Warner, J.R.; McIntosh, K.B. How common are extraribosomal functions of ribosomal proteins? Mol. Cell 2009, 34, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, H.; Hong, Y.; Huang, L.; Li, X.; Zhang, Y.; Ouyang, Z.; Song, F. Genome-wide identification, biochemical characterization, and expression analyses of the YTH domain-containing RNA-binding protein family in Arabidopsis and rice. Plant Mol. Biol. Rep. 2014, 32, 1169–1186. [Google Scholar] [CrossRef]
- Liao, S.; Sun, H.; Xu, C. YTH domain: A family of N6-methyladenosine (m6A) readers. Genom. Proteom. Bioinform. 2018, 16, 99–107. [Google Scholar] [CrossRef]
- Silverman, I.M.; Li, F.; Gregory, B.D. Genomic era analyses of RNA secondary structure and RNA-binding proteins reveal their significance to post-transcriptional regulation in plants. Plant Sci. 2013, 0, 55–62. [Google Scholar] [CrossRef]
- O’Toole, N.; Hattori, M.; Andres, C.; Iida, K.; Lurin, C.; Schmitz-Linneweber, C.; Sugita, M.; Small, I. On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 2008, 25, 1120–1128. [Google Scholar] [CrossRef]
- Lurin, C.; Andres, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyere, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 2004, 16, 2089–2103. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Ichinose, M.; Sugita, M. The DYW domains of pentatricopeptide repeat RNA editing factors contribute to discriminate target and non-target editing sites. Plant Cell Physiol. 2018, 59, 1652–1659. [Google Scholar] [CrossRef]
- Salone, V.; Rüdinger, M.; Polsakiewicz, M.; Hoffmann, B.; Groth-Malonek, M.; Szurek, B.; Small, I.; Knoop, V.; Lurin, C. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007, 581, 4132–4138. [Google Scholar] [CrossRef]
- Boussardon, C.; Avon, A.; Kindgren, P.; Bond, C.S.; Challenor, M.; Lurin, C.; Small, I. The cytidine deaminase signature HxE(x)nCxxC of DYW1 binds zinc and is necessary for RNA editing of ndhD-1. New Phytol. 2014, 203, 1090–1095. [Google Scholar] [CrossRef] [PubMed]
- Jaimes-Miranda, F.; Chávez Montes, R.A. The plant MBF1 protein family: A bridge between stress and transcription. J. Exp. Bot. 2020, 71, 1782–1791. [Google Scholar] [CrossRef] [PubMed]
- Klass, D.M.; Scheibe, M.; Butter, F.; Hogan, G.J.; Mann, M.; Brown, P.O. Quantitative proteomic analysis reveals concurrent RNA–protein interactions and identifies new RNA-binding proteins in Saccharomyces cerevisiae. Genome Res. 2013, 23, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, K.; Wang, X.F.; Miyazaki, H.; Soejima, H.; Hirose, S. Mbf1 ensures polycomb silencing by protecting E(Z) mRNA from degradation by Pacman. Development 2018, 145, dev162461. [Google Scholar] [CrossRef]
- Gaspar, M.; Bousser, A.; Sissoëff, I.; Roche, O.; Hoarau, J.; Mahé, A. Cloning and characterization of ZmPIP1-5b, an aquaporin transporting water and urea. Plant Sci. 2003, 165, 21–31. [Google Scholar] [CrossRef]
- Holm, L.M.; Jahn, T.P.; Møller, A.L.B.; Schjoerring, J.K.; Ferri, D.; Klaerke, D.A.; Zeuthen, T. NH3 and NH4+ permeability in aquaporin-expressing Xenopus oocytes. Pflugers Arch. Eur. J. Physiol. 2005, 450, 415–428. [Google Scholar] [CrossRef]
- Quigley, F.; Rosenberg, J.M.; Shachar-hill, Y.; Bohnert, H.J. From genome to function: The Arabidopsis aquaporins. Genome Biol. 2001, 3, 1–17. [Google Scholar] [CrossRef]
- Clerch, L.B.; Wright, A.; Massaro, D. Dinucleotide-binding site of bovine liver catalase mimics a catalase mRNA-binding protein domain. Am. J. Physiol. 1996, 270, 790–794. [Google Scholar] [CrossRef]
- Castello, A.; Hentze, M.W.; Preiss, T. Metabolic enzymes enjoying new partnerships as RNA-binding proteins. Trends Endocrinol. Metab. 2015, 26, 746–757. [Google Scholar] [CrossRef]
- Chang, C.H.; Curtis, J.D.; Maggi, L.B.; Faubert, B.; Villarino, A.V.; O’Sullivan, D.; Huang, S.C.C.; Van Der Windt, G.J.W.; Blagih, J.; Qiu, J.; et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 2013, 153, 1239. [Google Scholar] [CrossRef]
- Ross, S.; Giglione, C.; Pierre, M.; Espagne, C.; Meinnel, T. Functional and developmental impact of cytosolic protein N-terminal methionine excision in Arabidopsis. Plant Physiol. 2005, 137, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Lu, B.; Lee, I.; Ondrovičová, G.; Kutejová, E.; Suzuki, C.K. DNA and RNA Binding by the Mitochondrial Lon Protease Is Regulated by Nucleotide and Protein Substrate. J. Biol. Chem. 2004, 279, 13902–13910. [Google Scholar] [CrossRef] [PubMed]
- Volz, K. The functional duality of iron regulatory protein 1. Curr. Opin. Struct. Biol. 2008, 18, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, H.; Yuan, Q.; Feng, Y. Structure and function of the photosystem supercomplexes. Front. Plant Sci. 2018, 9, 1–7. [Google Scholar] [CrossRef]
- Yosef, I.; Irihimovitch, V.; Knopf, J.A.; Cohen, I.; Orr-Dahan, I.; Nahum, E.; Keasar, C.; Shapira, M. RNA binding activity of the bibulose-1,5-bisphosphate carboxylase/oxygenase large subunit from Chlamydomonas reinhardtii. J. Biol. Chem. 2004, 279, 10148–10156. [Google Scholar] [CrossRef]
- Cohen, I.; Sapir, Y.; Shapira, M. A conserved mechanism controls translation of Rubisco large subunit in different photosynthetic organisms. Plant Physiol. 2006, 141, 1089–1097. [Google Scholar] [CrossRef]
- Cohen, I.; Knopf, J.A.; Irihimovitch, V.; Shapira, M. A proposed mechanism for the inhibitory effects of oxidative stress on Rubisco assembly and its subunit expression. Plant Physiol. 2005, 137, 738–746. [Google Scholar] [CrossRef]
- Choquet, Y.; Zito, F.; Wostrikoff, K.; Wollman, F.A. Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell 2003, 15, 1443–1454. [Google Scholar] [CrossRef]
- Horos, R.; Büscher, M.; Kleinendorst, R.; Alleaume, A.M.; Tarafder, A.K.; Schwarzl, T.; Dziuba, D.; Tischer, C.; Zielonka, E.M.; Adak, A.; et al. The small non-coding vault RNA1-1 acts as a riboregulator of autophagy. Cell 2019, 176, 1054–1067.e12. [Google Scholar] [CrossRef]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bach-Pages, M.; Homma, F.; Kourelis, J.; Kaschani, F.; Mohammed, S.; Kaiser, M.; van der Hoorn, R.A.L.; Castello, A.; Preston, G.M. Discovering the RNA-Binding Proteome of Plant Leaves with an Improved RNA Interactome Capture Method. Biomolecules 2020, 10, 661. https://doi.org/10.3390/biom10040661
Bach-Pages M, Homma F, Kourelis J, Kaschani F, Mohammed S, Kaiser M, van der Hoorn RAL, Castello A, Preston GM. Discovering the RNA-Binding Proteome of Plant Leaves with an Improved RNA Interactome Capture Method. Biomolecules. 2020; 10(4):661. https://doi.org/10.3390/biom10040661
Chicago/Turabian StyleBach-Pages, Marcel, Felix Homma, Jiorgos Kourelis, Farnusch Kaschani, Shabaz Mohammed, Markus Kaiser, Renier A. L. van der Hoorn, Alfredo Castello, and Gail M. Preston. 2020. "Discovering the RNA-Binding Proteome of Plant Leaves with an Improved RNA Interactome Capture Method" Biomolecules 10, no. 4: 661. https://doi.org/10.3390/biom10040661
APA StyleBach-Pages, M., Homma, F., Kourelis, J., Kaschani, F., Mohammed, S., Kaiser, M., van der Hoorn, R. A. L., Castello, A., & Preston, G. M. (2020). Discovering the RNA-Binding Proteome of Plant Leaves with an Improved RNA Interactome Capture Method. Biomolecules, 10(4), 661. https://doi.org/10.3390/biom10040661