The Antitumor Agent Ansamitocin P-3 Binds to Cell Division Protein FtsZ in Actinosynnema pretiosum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids and Media
2.2. Growth Determination of Actinosynnema pretiosum Using Microplate Reader
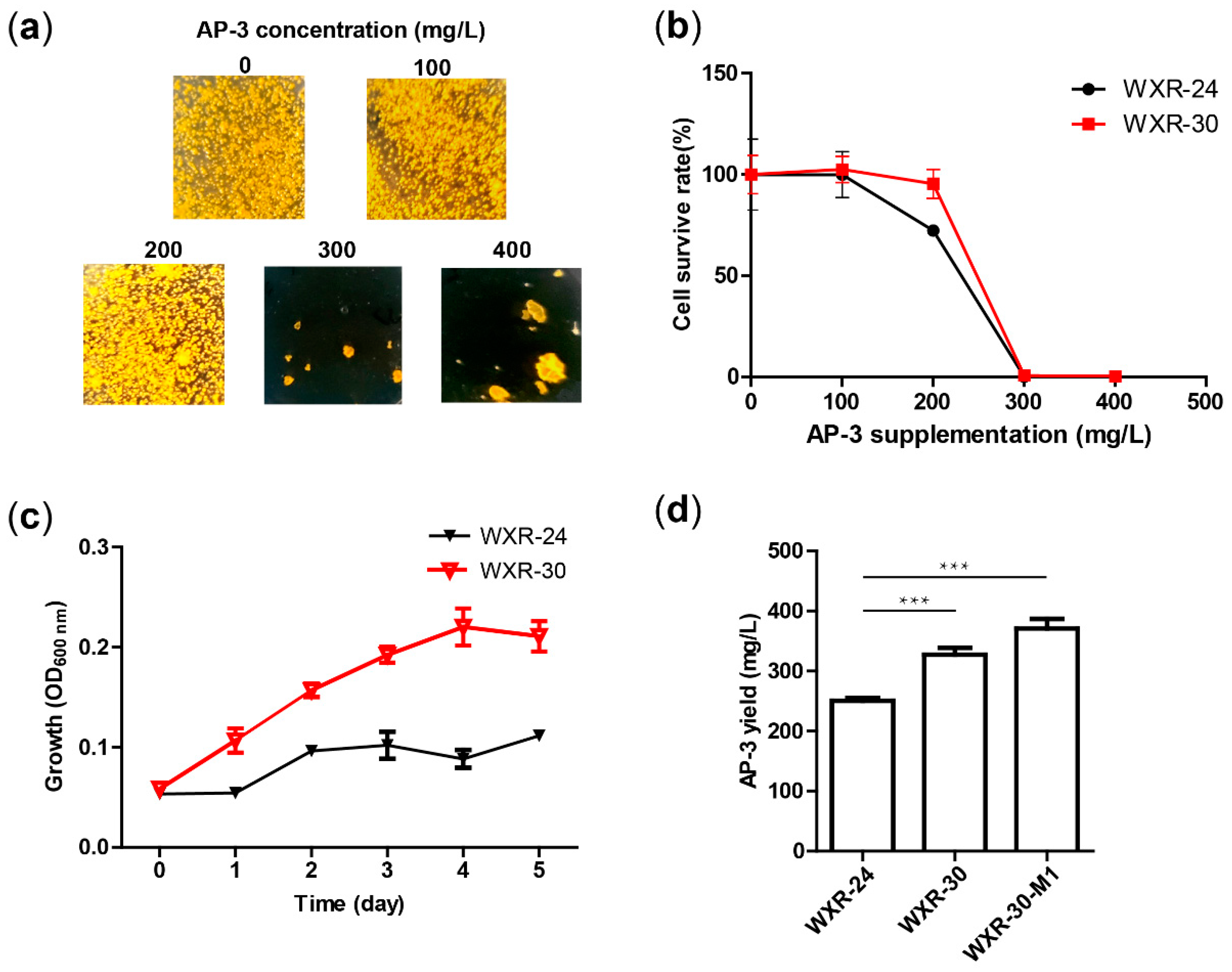
2.3. AP-3 Resistance Evaluation
2.4. Colony Counting and Average Colony Size
2.5. Homologous Modeling and Structure Comparison
2.6. Protein Expression and Purification
2.7. Surface Plasmon Resonance (SPR) Biosensor Analysis
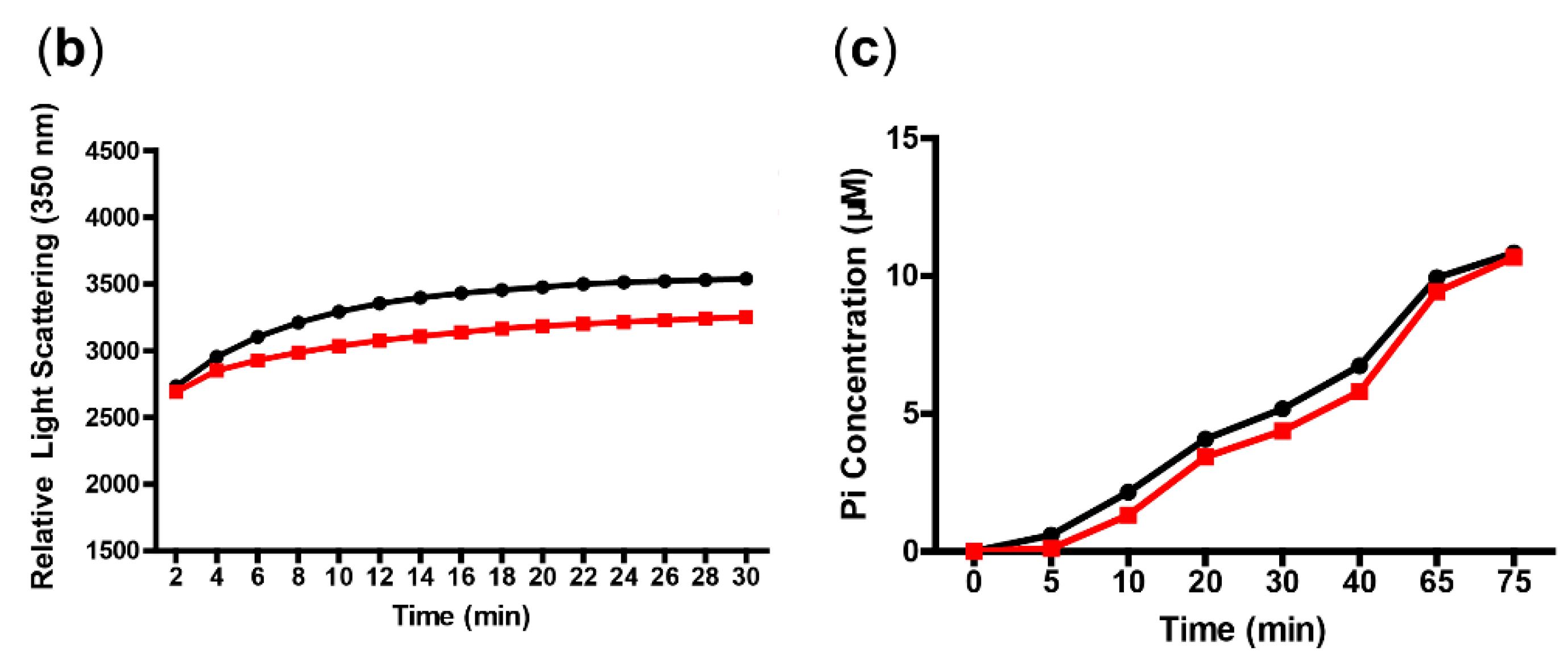
2.8. Light Scattering and GTPase Activity Assays
2.9. Gene Overexpression
2.10. AP-3 Yield Determination
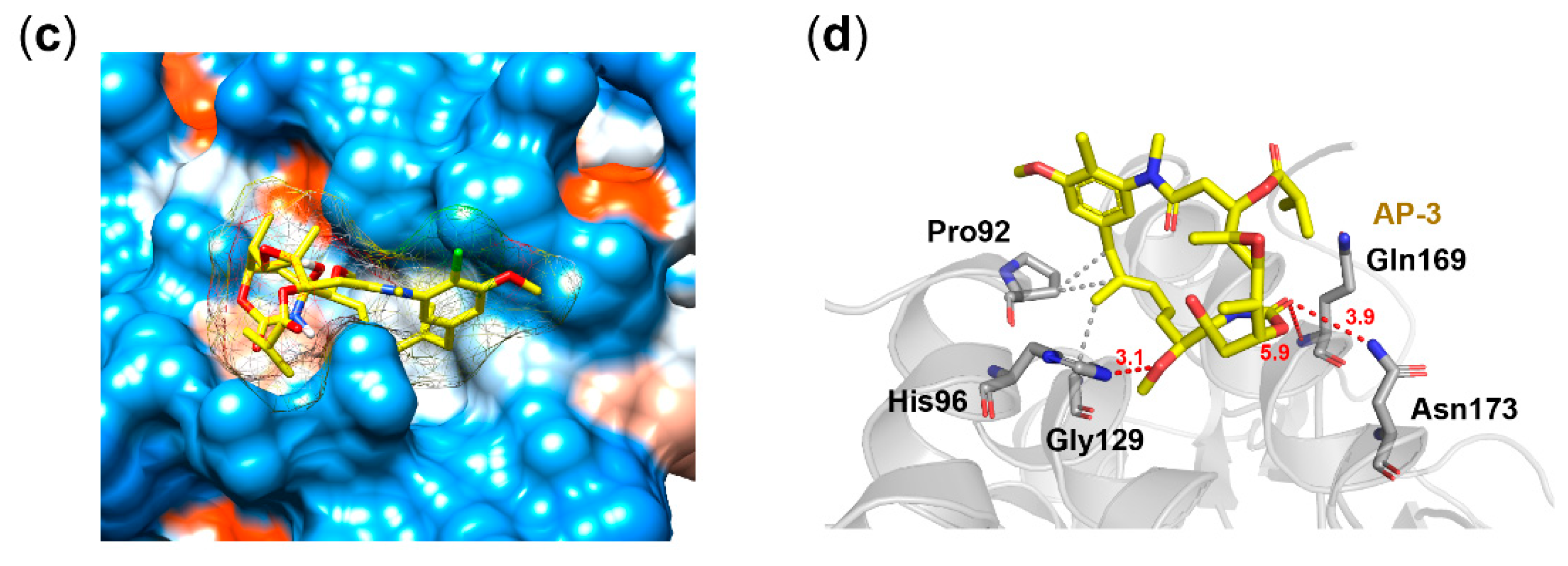
2.11. Docking Analysis
2.12. Microscopy Analysis
2.13. qRT-PCR Analysis
3. Results
3.1. Growth Inhibition of Actinosynnema pretiosum WXR-24 Under Exogenous AP-3 Supplementation
3.2. Cell Division Protein FtsZ is Predicted as a Target of AP-3
3.3. In Vitro Interaction of FtsZ and AP-3
3.4. Docking Analysis of AP-3 on FtsZ
3.5. Introducing an Extra Copy of ftsZ Improved AP-3 Resistance and Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Prelog, V.; Oppolzer, W. Ansamycins, a novel class of microbial metabolites. Helv. Chim. Acta. 1973, 56, 1179–1187. [Google Scholar] [PubMed]
- Kang, Q.; Shen, Y.; Bai, L. Biosynthesis of 3,5-AHBA-derived natural products. Nat. Prod. Rep. 2012, 29, 243–263. [Google Scholar] [CrossRef] [PubMed]
- Barok, M.; Joensuu, H.; Isola, J. Trastuzumab emtansine: Mechanisms of action and drug resistance. Breast Cancer Res. 2014, 16, 209. [Google Scholar] [PubMed] [Green Version]
- Li, B.T.; Shen, R.; Buonocore, D.; Olah, Z.T.; Ni, A.; Ginsberg, M.S.; Ulaner, G.A.; Offin, M.; Feldman, D.; Hembrough, T.; et al. Ado-trastuzumab emtansine for patients with HER2-mutant lung cancers: Results from a Phase II basket trial. J. Clin. Oncol. 2018, 36, 2532–2537. [Google Scholar] [CrossRef]
- Venghateri, J.B.; Gupta, T.K.; Verma, P.J.; Kunwar, A.; Panda, D. Ansamitocin P3 depolymerizes microtubules and induces apoptosis by binding to tubulin at the vinblastine site. PLoS ONE 2013, 8, e75182. [Google Scholar] [CrossRef]
- Prota, A.E.; Bargsten, K.; Diaz, J.F.; Marsh, M.; Cuevas, C.; Liniger, M.; Neuhaus, C.; Andreu, J.M.; Altmann, K.H.; Steinmetz, M.O. A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs. Proc. Natl. Acad. Sci. USA 2014, 111, 13817–13821. [Google Scholar] [CrossRef] [Green Version]
- Erickson, H.P. FtsZ, a prokaryotic homolog of tubulin? Cell 1995, 80, 367–370. [Google Scholar] [CrossRef] [Green Version]
- Amos, L.A.; Ent, F.V.D.; Löwe, J. Structural/functional homology between the bacterial and eukaryotic cytoskeletons. Curr. Opin. Cell Biol. 2004, 16, 24–31. [Google Scholar] [CrossRef]
- Haranahalli, K.; Tong, S.; Ojima, I. Recent advances in the discovery and development of antibacterial agents targeting the cell-division protein FtsZ. Bioorg. Med. Chem. 2016, 24, 6354–6369. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, A.; Lutkenhaus, J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol. 1994, 176, 2754–2758. [Google Scholar] [CrossRef] [Green Version]
- Du, S.; Lutkenhaus, J. At the heart of bacterial cytokinesis: The Z ring. Trends Microbiol. 2019, 27, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lu, C.; Chang, X.; Shen, Y. Constitutive overexpression of asm18 increases the production and diversity of maytansinoids in Actinosynnema pretiosum. Appl. Microbiol. Biotechnol. 2016, 100, 2641–2649. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.Q.; Zhang, Y.; Qian, Z.G.; Xiao, H.; Zhong, J.J. Combination of traditional mutation and metabolic engineering to enhance ansamitocin P-3 production in Actinosynnema pretiosum. Biotechnol. Bioeng. 2017, 114, 2794–2806. [Google Scholar] [CrossRef]
- Ning, X.; Wang, X.; Wu, Y.; Kang, Q.; Bai, L. Identification and engineering of post-pks modification bottlenecks for ansamitocin P-3 titer improvement in Actinosynnema pretiosum subsp. pretiosum ATCC 31280. Biotechnol. J. 2017, 12. [Google Scholar] [CrossRef]
- Zhao, M.; Fan, Y.; Wei, L.; Hu, F.; Hua, Q. Effects of the methylmalonyl-COA metabolic pathway on ansamitocin production in Actinosynnema pretiosum. Appl. Biochem. Biotechnol. 2017, 181, 1167–1178. [Google Scholar] [CrossRef]
- Du, Z.Q.; Zhong, J.J. Rational approach to improve ansamitocin P-3 production by integrating pathway engineering and substrate feeding in Actinosynnema pretiosum. Biotechnol. Bioeng. 2018, 115, 2456–2466. [Google Scholar] [CrossRef]
- Li, J.; Sun, R.; Ning, X.; Wang, X.; Wang, Z. Genome-scale metabolic model of Actinosynnema pretiosum ATCC 31280 and its application for ansamitocin P-3 production improvement. Genes 2018, 9, 364. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Godzik, A. FATCAT: A web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004, 32, W582–W585. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Ji, H.; Ali, I.; Deng, Z.; Bai, L.; Zheng, J. Structural and biochemical insights to the recruitment of acyl carrier protein-linked extender units in ansamitocin biosynthesis. ChemBioChem 2020. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.J.; Butter, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Centre: Norwich, UK, 2000. [Google Scholar]
- Li, Y.; Zhao, P.; Kang, Q.; Ma, J.; Bai, L.; Deng, Z. Dual carbamoylations on the polyketide and glycosyl moiety by asm21 result in extended ansamitocin biosynthesis. Chem. Biol. 2011, 18, 1571–1580. [Google Scholar] [CrossRef] [Green Version]
- Rao, S.N.; Head, M.S.; Kulkarni, A.; LaLonde, J.M. Validation studies of the site-directed docking program LibDock. J. Chem. Inf. Model. 2007, 47, 2159–2171. [Google Scholar] [CrossRef] [Green Version]
- Nogales, E.; Downing, K.H.; Amos, L.A.; Löwe, J. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol. 1998, 5, 451–458. [Google Scholar] [CrossRef]
- Zhang, Y.; Wen, W.; Pu, J.; Tang, M.; Zhang, L.; Peng, C.; Xu, Y.; Tang, G. Extracellularly oxidative activation and inactivation of matured prodrug for cryptic self-resistance in naphthyridinomycin biosynthesis. Proc. Natl. Acad. Sci. USA 2018, 115, 11232–11237. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Huang, W.; He, Q.L.; Zhao, Z.X.; Zhang, F.; Wang, R.; Kang, J.; Tang, G.L. Self-resistance to an antitumor antibiotic: A DNA glycosylase triggers the base-excision repair system in yatakemycin biosynthesis. Angew. Chem. Int. Ed. Engl. 2012, 51, 10532–10536. [Google Scholar] [CrossRef]
- Chang, C.Y.; Yan, X.; Crnovcic, I.; Annaval, T.; Chang, C.; Nocek, B.; Rudolf, J.D.; Yang, D.; Hindra; Babnigg, G.; et al. Resistance to enediyne antitumor antibiotics by sequestration. Cell Chem. Biol. 2018, 25, 1075–1085. [Google Scholar] [CrossRef]
- Liu, L.; Pan, J.; Wang, Z.; Yan, X.; Yang, D.; Zhu, X.; Shen, B.; Duan, Y.; Huang, Y. Ribosome engineering and fermentation optimization leads to overproduction of tiancimycin A, a new enediyne natural product from Streptomyces sp. CB03234. J. Ind. Microbiol. Biotechnol. 2018, 45, 141–151. [Google Scholar] [CrossRef]
- Yin, S.; Wang, X.; Shi, M.; Yuan, F.; Wang, H.; Jia, X.; Yuan, F.; Sun, J.; Liu, T.; Yang, K.; et al. Improvement of oxytetracycline production mediated via cooperation of resistance genes in Streptomyces rimosus. Sci. China Life Sci. 2017, 60, 992–999. [Google Scholar] [CrossRef]
- Malla, S.; Niraula, N.P.; Liou, K.; Sohng, J.K. Self-resistance mechanism in Streptomyces peucetius: Overexpression of drrA, drrB and drrC for doxorubicin enhancement. Microbiol. Res. 2010, 165, 259–267. [Google Scholar] [CrossRef]
- Zhao, Y.; Song, Z.; Ma, Z.; Bechthold, A.; Yu, X. Sequential improvement of rimocidin production in Streptomyces rimosus M527 by introduction of cumulative drug-resistance mutations. J. Ind. Microbiol. Biotechnol. 2019, 46, 697–708. [Google Scholar] [CrossRef]
- Lee, S.K.; Kim, H.R.; Jin, Y.Y.; Yang, S.H.; Suh, J.W. Improvement of daptomycin production via increased resistance to decanoic acid in Streptomyces roseosporus. J. Biosci. Bioeng. 2016, 122, 427–433. [Google Scholar] [CrossRef]
- Shentu, X.P.; Cao, Z.Y.; Xiao, Y.; Tang, G.; Ochi, K.; Yu, X.P. Substantial improvement of toyocamycin production in Streptomyces diastatochromogenes by cumulative drug-resistance mutations. PLoS ONE 2018, 13, e0203006. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.C.; Kishony, R. Opposing effects of target overexpression reveal drug mechanisms. Nat. Commun. 2014, 5, 4296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belanger, A.E.; Besra, G.S.; Ford, M.E.; Mikusova, K.; Belisle, J.T.; Brennan, P.J.; Inamine, J.M. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc Natl. Acad. Sci. USA 1996, 93, 11919–11924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grove, J.E.; Brown, R.J.; Watts, D.J. The intracellular target for the antiresorptive aminobisphosphonate drugs in Dictyostelium discoideum is the enzyme farnesyl diphosphate synthase. J. Bone Miner. Res. 2000, 15, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.E., Jr.; Lutkenhaus, J. Overproduction of FtsZ induces minicell formation in E. coli. Cell 1985, 42, 941–949. [Google Scholar] [CrossRef]
- Weart, R.B.; Levin, P.A. Growth rate-dependent regulation of medial FtsZ ring formation. J. Bacteriol. 2003, 185, 2826–2834. [Google Scholar] [CrossRef] [Green Version]
- Letek, M.; Ordonez, E.; Fiuza, M.; Honrubia-Marcos, P.; Vaquera, J.; Gil, J.A.; Castro, D.; Mateos, L.M. Characterization of the promoter region of ftsZ from Corynebacterium glutamicum and controlled overexpression of FtsZ. Int Microbiol. 2007, 10, 271–282. [Google Scholar]
- Van Wezel, G.P.; van der Meulen, J.; Taal, E.; Koerten, H.; Kraal, B. Effects of increased and deregulated expression of cell division genes on the morphology and on antibiotic production of streptomycetes. Antonie Van Leeuwenhoek 2000, 78, 269–276. [Google Scholar] [CrossRef]
- Santos-Beneit, F.; Roberts, D.M.; Cantlay, S.; McCormick, J.R.; Errington, J. A mechanism for FtsZ-independent proliferation in Streptomyces. Nat. Commun. 2017, 8, 1378. [Google Scholar] [CrossRef] [Green Version]
- Sin, L.T.; Rahman, W.A.W.A.; Rahmat, A.R.; Samad, A.A. Computational modeling and experimental infrared spectroscopy of hydrogen bonding interactions in polyvinyl alcohol-starch blends. Polymer 2010, 51, 1206–1211. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Wang, R.; Kang, Q.; Bai, L. The Antitumor Agent Ansamitocin P-3 Binds to Cell Division Protein FtsZ in Actinosynnema pretiosum. Biomolecules 2020, 10, 699. https://doi.org/10.3390/biom10050699
Wang X, Wang R, Kang Q, Bai L. The Antitumor Agent Ansamitocin P-3 Binds to Cell Division Protein FtsZ in Actinosynnema pretiosum. Biomolecules. 2020; 10(5):699. https://doi.org/10.3390/biom10050699
Chicago/Turabian StyleWang, Xinran, Rufan Wang, Qianjin Kang, and Linquan Bai. 2020. "The Antitumor Agent Ansamitocin P-3 Binds to Cell Division Protein FtsZ in Actinosynnema pretiosum" Biomolecules 10, no. 5: 699. https://doi.org/10.3390/biom10050699
APA StyleWang, X., Wang, R., Kang, Q., & Bai, L. (2020). The Antitumor Agent Ansamitocin P-3 Binds to Cell Division Protein FtsZ in Actinosynnema pretiosum. Biomolecules, 10(5), 699. https://doi.org/10.3390/biom10050699





