Mapping of Recognition Sites of Monoclonal Antibodies Responsible for the Inhibition of Pneumolysin Functional Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Recombinant Cytolysins
2.2. Synthesis of Truncated PLY Fragments
2.3. Development of Monoclonal Antibodies (MAbs)
2.4. Enzyme-Linked Immunosorbent Assay (ELISA)
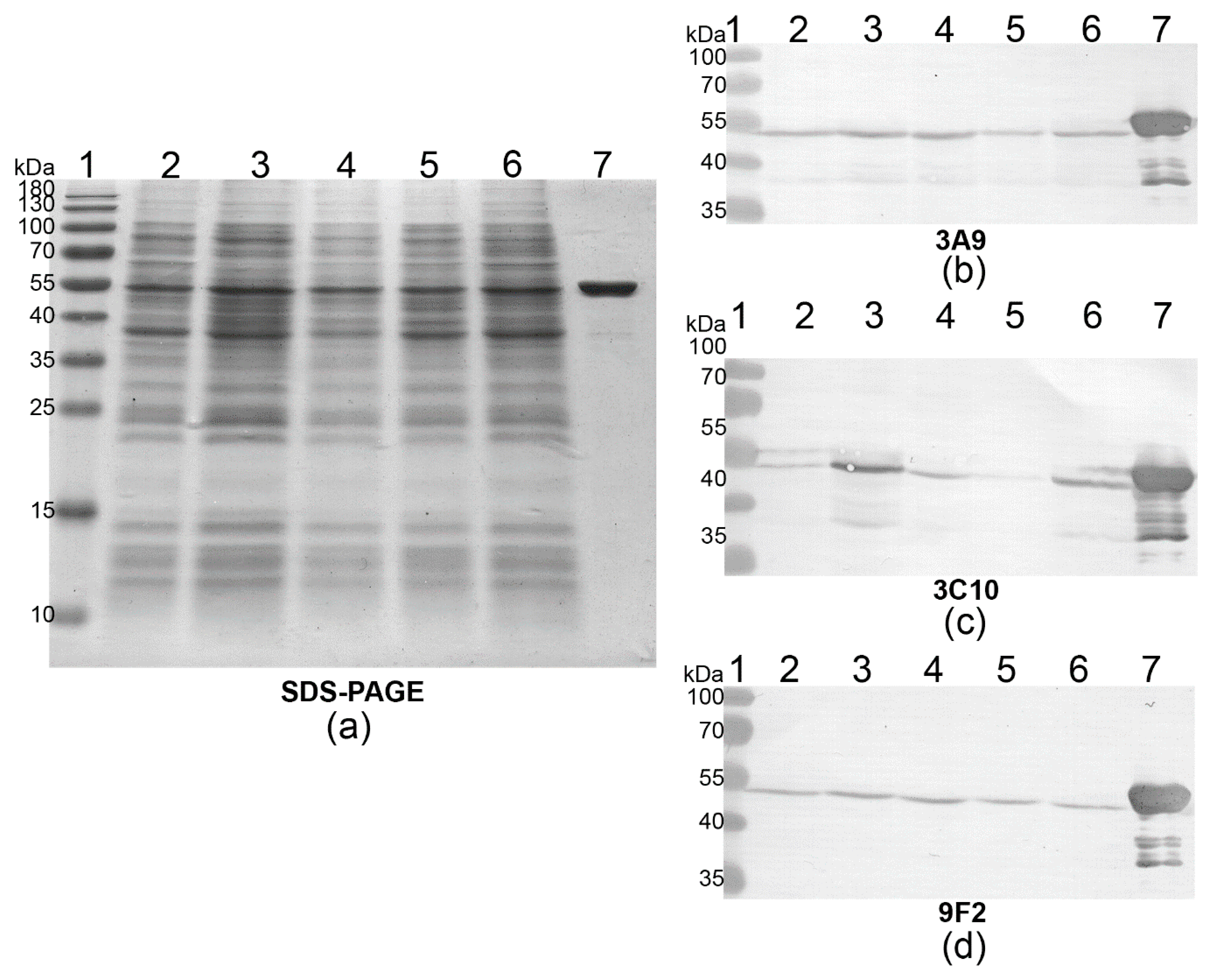
2.5. Western Blot (WB)
2.6. Cholesterol Binding Test by Dot Blot
2.7. Determination of MAb Neutralising Activity
2.8. Computional Analysis of PLY Epitopes Involved in Cytolytic Activity
3. Results
3.1. Development of the MAbs Against PLY
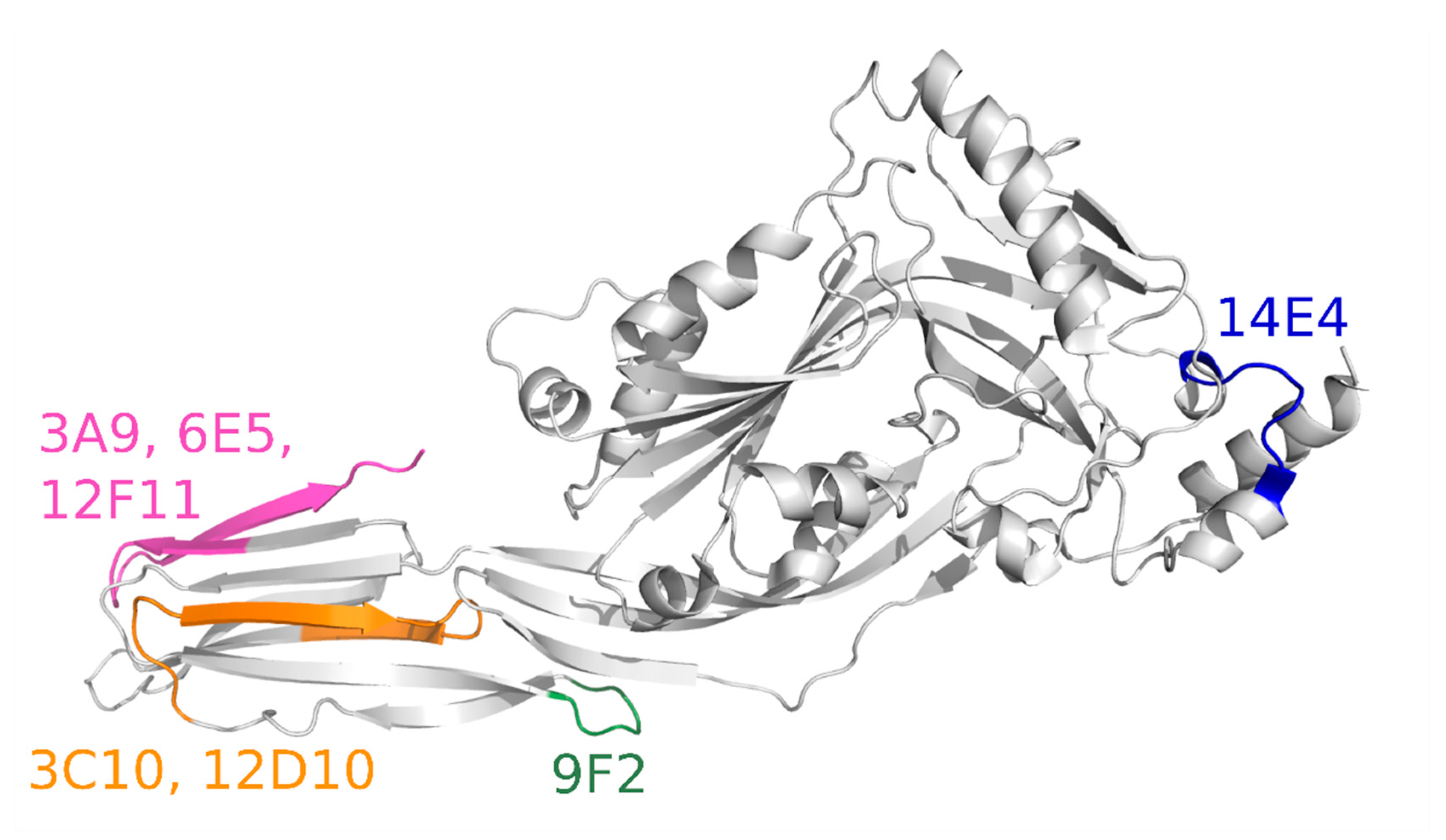
3.2. Mapping of MAb Epitopes
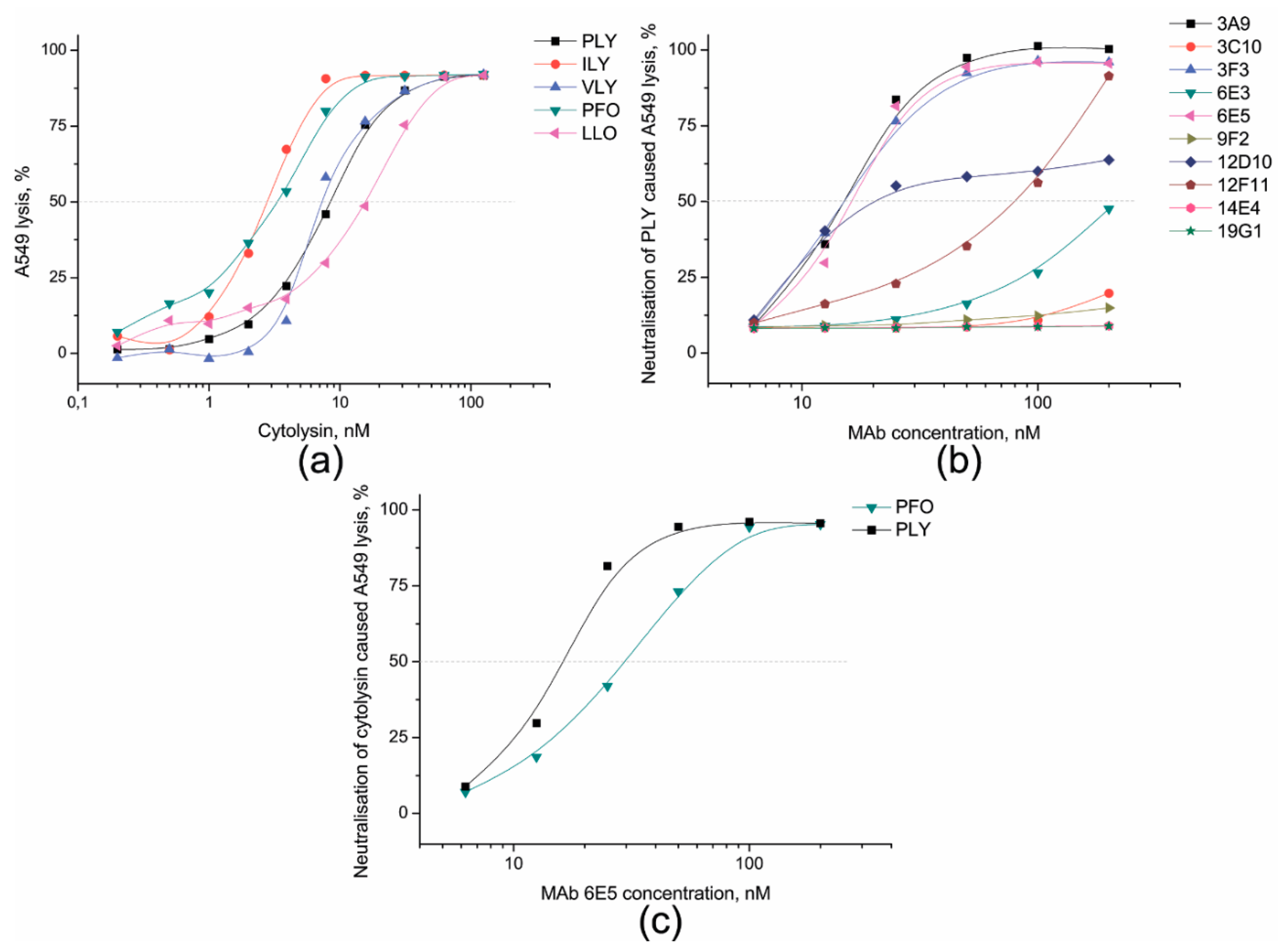
3.3. Identification of PLY-Neutralising MAbs and Determination of Their Neutralisation Potency
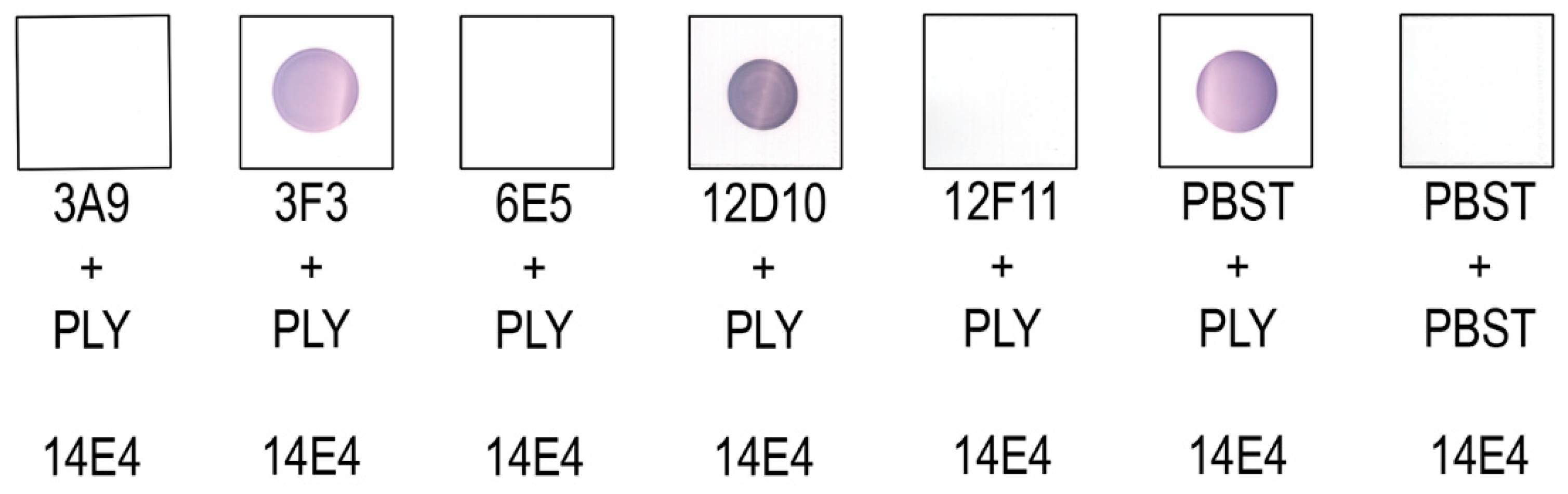
3.4. Investigation of MAb Interference with the PLY-Cholesterol Binding
3.5. Investigation of MAb Interference with the PLY-MRC-1 Binding
3.6. Computational Analysis of PLY Regions Recognised by the Neutralising MAbs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Anderson, R.; Feldman, C. Pneumolysin as a potential therapeutic target in severe pneumococcal disease. J. Infect. 2017, 74, 527–544. [Google Scholar] [CrossRef] [PubMed]
- Lucas, R.; Czikora, I.; Sridhar, S.; Zemskov, E.; Gorshkov, B.; Siddaramappa, U.; Oseghale, A.; Lawson, J.; Verin, A.; Rick, F.; et al. Mini-Review: Novel Therapeutic Strategies to Blunt Actions of Pneumolysin in the Lungs. Toxins 2013, 5, 1244–1260. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, J.L.; McDaniel, L.S. Selective pressure: Rise of the nonencapsulated pneumococcus. PLoS Pathog. 2019, 15, e1007911. [Google Scholar] [CrossRef]
- Keller, L.E.; Robinson, D.A.; McDaniel, L.S. Nonencapsulated streptococcus pneumoniae: Emergence and pathogenesis. mBio 2016, 7, e01792. [Google Scholar] [CrossRef]
- Lagousi, T.; Basdeki, P.; Routsias, J.; Spoulou, V. Novel Protein-Based Pneumococcal Vaccines: Assessing the Use of Distinct Protein Fragments Instead of Full-Length Proteins as Vaccine Antigens. Vaccines 2019, 7, 9. [Google Scholar] [CrossRef]
- Anderson, R.; Nel, J.G.; Feldman, C. Multifaceted role of pneumolysin in the pathogenesis of myocardial injury in community-acquired pneumonia. Int. J. Mol. Sci. 2018, 19, 1147. [Google Scholar] [CrossRef]
- Rubins, J.B.; Charboneau, D.; Fasching, C.; Berry, A.M.; Paton, J.C.; Alexander, J.E.; Andrew, P.W.; Mitchell, T.J.; Janoff, E.N. Distinct roles for pneumolysin’s cytotoxic and complement activities in the pathogenesis of pneumococcal pneumonia. Am. J. Respir. Crit. Care Med. 1996, 153, 1339–1346. [Google Scholar] [CrossRef] [PubMed]
- Rubins, J.B.; Charboneau, D.; Paton, J.C.; Mitchell, T.J.; Andrew, P.W.; Janoff, E.N. Dual function of pneumolysin in the early pathogenesis of murine pneumococcal pneumonia. J. Clin. Invest. 1995, 95, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Del Mar García-Suárez, M.; Cima-Cabal, M.D.; Flórez, N.; García, P.; Cernuda-Cernuda, R.; Astudillo, A.; Vázquez, F.; De Los Toyos, J.R.; Méndez, F.J. Protection against pneumococcal pneumonia in mice by monoclonal antibodies to pneumolysin. Infect. Immun. 2004, 72, 4534–4540. [Google Scholar] [CrossRef]
- Cassidy, S.; O’Riordan, M. More Than a Pore: The Cellular Response to Cholesterol-Dependent Cytolysins. Toxins 2013, 5, 618–636. [Google Scholar] [CrossRef]
- Tweten, R.K. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infect. Immun. 2005, 73, 6199–6209. [Google Scholar] [CrossRef] [PubMed]
- Bhakdi, S.; Weller, U.; Walev, I.; Martin, E.; Jonas, D.; Palmer, M. A guide to the use of pore-forming toxins for controlled permeabilization of cell membranes. Med. Microbiol. Immunol. 1993, 182, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.E.; Faraj, B.H.A.; Gingras, A.R.; Lonnen, R.; Sheikh, M.A.; El-Mezgueldi, M.; Moody, P.C.E.; Andrew, P.W.; Wallis, R. The crystal structure of pneumolysin at 2.0Å resolution reveals the molecular packing of the pre-pore complex. Sci. Rep. 2015, 5, 13293. [Google Scholar] [CrossRef]
- Lawrence, S.L.; Feil, S.C.; Morton, C.J.; Farrand, A.J.; Mulhern, T.D.; Gorman, M.A.; Wade, K.R.; Tweten, R.K.; Parker, M.W. Crystal structure of Streptococcus pneumoniae pneumolysin provides key insights into early steps of pore formation. Sci. Rep. 2015, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Vögele, M.; Bhaskara, R.M.; Mulvihill, E.; van Pee, K.; Yildiz, Ö.; Kühlbrandt, W.; Müller, D.J.; Hummer, G. Membrane perforation by the pore-forming toxin pneumolysin. Proc. Natl. Acad. Sci. USA 2019, 116, 13352–13357. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, R.; Heuck, A.P.; Tweten, R.K.; Johnson, A.E. Structural insights into the membrane-anchoring mechanism of a cholesterol-dependent cytolysin. Nat. Struct. Biol. 2002, 9, 823–827. [Google Scholar] [CrossRef] [PubMed]
- Dowd, K.J.; Tweten, R.K. The cholesterol-dependent cytolysin signature motif: A critical element in the allosteric pathway that couples membrane binding to pore assembly. PLoS Pathog. 2012, 8, 44. [Google Scholar] [CrossRef]
- Farrand, A.J.; LaChapelle, S.; Hotze, E.M.; Johnson, A.E.; Tweten, R.K. Only two amino acids are essential for cytolytic toxin recognition of cholesterol at the membrane surface. Proc. Natl. Acad. Sci. USA 2010, 107, 4341–4346. [Google Scholar] [CrossRef]
- Subramanian, K.; Neill, D.R.; Malak, H.A.; Spelmink, L.; Khandaker, S.; Dalla Libera Marchiori, G.; Dearing, E.; Kirby, A.; Yang, M.; Achour, A.; et al. Pneumolysin binds to the mannose receptor C type 1 (MRC-1) leading to anti-inflammatory responses and enhanced pneumococcal survival. Nat. Microbiol. 2019, 4, 62–70. [Google Scholar] [CrossRef]
- Mitchell, T.J.; Andrew, P.W.; Saunders, F.K.; Smith, A.N.; Boulnois, G.J. Complement activation and antibody binding by pneumolysin via a region of the toxin homologous to a human acute-phase protein. Mol. Microbiol. 1991, 5, 1883–1888. [Google Scholar] [CrossRef]
- Paton, J.C.; Rowan Kelly, B.; Ferrante, A. Activation of human complement by the pneumococcal toxin pneumolysin. Infect. Immun. 1984, 43, 1085–1087. [Google Scholar] [CrossRef] [PubMed]
- Iliev, A.I.; Djannatian, J.R.; Opazo, F.; Gerber, J.; Nau, R.; Mitchell, T.J.; Wouters, F.S. Rapid microtubule bundling and stabilization by the Streptococcus pneumoniae neurotoxin pneumolysin in a cholesterol-dependent, non-lytic and Src-kinase dependent manner inhibits intracellular trafficking. Mol. Microbiol. 2009, 71, 461–477. [Google Scholar] [CrossRef] [PubMed]
- Iliev, A.I.; Djannatian, J.R.; Nau, R.; Mitchell, T.J.; Wouters, F.S. Cholesterol-dependent actin remodeling via RhoA and Rac1 activation by the Streptococcus pneumoniae toxin pneumolysin. Proc. Natl. Acad. Sci. USA 2007, 104, 2897–2902. [Google Scholar] [CrossRef]
- Hugo, F.; Reichwein, J.; Arvand, M.; Krämer, S.; Bhakdi, S. Use of a monoclonal antibody to determine the mode of transmembrane pore formation by streptolysin O. Infect. Immun. 1986, 54, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Nato, F.; Reich, K.; Lhopital, S.; Rouyre, S.; Geoffroy, C.; Mazie, J.C.; Cossart, P. Production and characterization of neutralizing and nonneutralizing monoclonal antibodies against listeriolysin O. Infect. Immun. 1991, 59, 4641–4646. [Google Scholar] [CrossRef] [PubMed]
- Darji, A.; Niebuhr, K.; Hense, M.; Ju, J.; Wehland, J.; Chakraborty, T.; Weiss, S. Neutralizing Monoclonal Antibodies against Listeriolysin: Mapping of Epitopes Involved in Pore Formation. Infect. Immun. 1996, 64, 2356–2358. [Google Scholar] [CrossRef] [PubMed]
- Zvirbliene, A.; Pleckaityte, M.; Lasickiene, R.; Kucinskaite-Kodze Indre, I.; Zvirblis, G. Production and characterization of monoclonal antibodies against vaginolysin: Mapping of a region critical for its cytolytic activity. Toxicon 2010, 56, 19–28. [Google Scholar] [CrossRef]
- De Los Toyos, J.R.; Méndez, F.J.; Aparicio, J.F.; Vázquez, F.; Suárez, M.D.M.G.; Fleites, A.; Hardisson, C.; Morgan, P.J.; Andrew, P.W.; Mitchell, T.J. Functional analysis of pneumolysin by use of monoclonal antibodies. Infect. Immun. 1996, 64, 480–484. [Google Scholar] [CrossRef]
- Jacobs, T.; Cima-Cabal, M.D.; Darji, A.; Méndez, F.J.; Vázquez, F.; Jacobs, A.A.C.; Shimada, Y.; Ohno-Iwashita, Y.; Weiss, S.; De Los Toyos, J.R. The conserved undecapeptide shared by thiol-activated cytolysins is involved in membrane binding. FEBS Lett. 1999, 459, 463–466. [Google Scholar] [CrossRef]
- Erdenlig, S.; Ainsworth, A.J.; Austin, F.W. Production of monoclonal antibodies to Listeria monocytogenes and their application to determine the virulence of isolates from channel catfish. Appl. Environ. Microbiol. 1999, 65, 2827–2832. [Google Scholar] [CrossRef]
- Pleckaityte, M.; Mistiniene, E.; Lasickiene, R.; Zvirblis, G.; Zvirbliene, A. Generation of recombinant single-chain antibodies neutralizing the cytolytic activity of vaginolysin, the main virulence factor of Gardnerella vaginalis. BMC Biotechnol. 2011, 11, 100. [Google Scholar] [CrossRef] [PubMed]
- Zilnyte, M.; Venclovas, Č.; Zvirbliene, A.; Pleckaityte, M. The cytolytic activity of vaginolysin strictly depends on cholesterol and is potentiated by human CD59. Toxins 2015, 7, 110–128. [Google Scholar] [CrossRef] [PubMed]
- Simanavicius, M.; Tamosiunas, P.L.; Petraityte-Burneikiene, R.; Johne, R.; Ulrich, R.G.; Zvirbliene, A.; Kucinskaite-Kodze, I. Generation in yeast and antigenic characterization of hepatitis E virus capsid protein virus-like particles. Appl. Microbiol. Biotechnol. 2018, 102, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V.A. Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Šali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Olechnovič, K.; Kulberkyte, E.; Venclovas, Č. CAD-score: A new contact area difference-based function for evaluation of protein structural models. Proteins Struct. Funct. Bioinforma. 2013, 81, 149–162. [Google Scholar] [CrossRef]
- Olechnovič, K.; Venclovas, C. Voronota: A fast and reliable tool for computing the vertices of the Voronoi diagram of atomic balls. J. Comput. Chem. 2014, 35, 672–681. [Google Scholar] [CrossRef]
- Van Pee, K.; Neuhaus, A.; D’Imprima, E.; Mills, D.J.; Kühlbrandt, W.; Yildiz, Ö. CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin. Elife 2017, 6, e23644. [Google Scholar] [CrossRef]
- Larpin, Y.; Besançon, H.; Iacovache, M.I.; Babiychuk, V.S.; Babiychuk, E.B.; Zuber, B.; Draeger, A.; Köffel, R. Bacterial pore-forming toxin pneumolysin: Cell membrane structure and microvesicle shedding capacity determines differential survival of immune cell types. FASEB J. 2020, 34, 1665–1678. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Elkayam, T.; Wolfson, H.; Nussinov, R. Protein-protein interactions: Structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc. Natl. Acad. Sci. USA 2003, 100, 5772–5777. [Google Scholar] [CrossRef]
- Bogan, A.A.; Thorn, K.S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998, 280, 1–9. [Google Scholar] [CrossRef]
- Moreira, I.S.; Fernandes, P.A.; Ramos, M.J. Hot spots—A review of the protein-protein interface determinant amino-acid residues. Proteins Struct. Funct. Genet. 2007, 68, 803–812. [Google Scholar] [CrossRef]
- Kalin, M.; Kanclerski, K.; Granstrom, M.; Mollby, R. Diagnosis of pneumococcal pneumonia by enzyme-linked immunosorbent assay of antibodies to pneumococcal hemolysin (pneumolysin). J. Clin. Microbiol. 1987, 25, 226–229. [Google Scholar] [CrossRef] [PubMed]
- Jefferies, J.M.C.; Johnston, C.H.G.; Kirkham, L.S.; Cowan, G.J.M.; Ross, K.S.; Smith, A.; Clarke, S.C.; Brueggemann, A.B.; George, R.C.; Pichon, B.; et al. Presence of Nonhemolytic Pneumolysin in Serotypes of Streptococcus pneumoniae Associated with Disease Outbreaks. J. Infect. Dis. 2007, 196, 936–944. [Google Scholar] [CrossRef]
- Cima-Cabal, M.D.; Méndez, F.J.; Vázquez, F.; García-Suárez, M.; de los Toyos, J.R. A specific and ultrasensitive chemiluminescent sandwich elisa test for the detection and quantitation of pneumolysin. J. Immunoass. Immunochem. 2001, 22, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Álvarez, B.; García-Suárez, M.D.M.; Méndez, F.J.; De Los Toyos, J.R. Characterisation of mouse monoclonal antibodies for pneumolysin: Fine epitope mapping and V gene usage. Immunol. Lett. 2003, 88, 227–239. [Google Scholar] [CrossRef]
- Imaizumi, K.; Serizawa, A.; Hashimoto, N.; Kaidoh, T.; Takeuchi, S. Analysis of the functional domains of Arcanobacterium pyogenes pyolysin using monoclonal antibodies. Vet. Microbiol. 2001, 81, 235–242. [Google Scholar] [CrossRef]
- Imaizumi, K.; Matsunaga, K.; Higuchi, H.; Kaidoh, T.; Takeuchi, S. Effect of amino acid substitutions in the epitope regions of pyolysin from Arcanobacterium pyogenes. Vet. Microbiol. 2003, 91, 205–213. [Google Scholar] [CrossRef]
- Soltani, C.E.; Hotze, E.M.; Johnson, A.E.; Tweten, R.K. Structural elements of the cholesterol-dependent cytolysins that are responsible for their cholesterol-sensitive membrane interactions. Proc. Natl. Acad. Sci. USA 2007, 104, 20226–20231. [Google Scholar] [CrossRef] [PubMed]


| Truncated PLY Fragment | DNA Sequence, bp | Protein Sequence, aa |
|---|---|---|
| PLY1 | 1–468 | 1–156 |
| PLY2 | 331–810 | 111–270 |
| PLY3 | 661–1140 | 221–380 |
| PLY4 | 961–1413 | 321–471 |
| PLY5 | 1–1293 | 1–431 |
| PLY6 | 676–1206 | 226–402 |
| PLY7 | 961–1359 | 321–453 |
| PLY8 | 586–1269 | 196–423 |
| Full-length PLY (accession number: WP_001284359) | 1–1416 | 1–471 |
| Primer | Primer Sequence |
|---|---|
| PLY1_F | 5′-GAGCATATGTTAGTGAGCCGTGATTTTT-3′ |
| PLY1_5_R | 5′-GCCCATATGATGGCAAATAAAGCAGTAA-3′ |
| PLY2_F | 5′-GGCCATATGTTAGACTCCTTTTATCAAAGC-3′ |
| PLY2_R | 5′-GATCATATGAGCTTTCTCCAAGTGGAAGAC-3′ |
| PLY3_F | 5′-GGCCATATGTTAATCCCAAGTAATATAATA-3′ |
| PLY3_R | 5′-ATTCATATGGAGGATTTAAAACAGAGAGGA-3′ |
| PLY4_F | 5′-GCGCATATGTTAGTCATTTTCTACCTTATC-3′ |
| PLY4_R/PLY_7R | 5′-ATACATATGACAGCAGATCATCCAGGCTTG-3′ |
| PLY5_F | 5′-ATTCATATGTTAAAGCCCGGTACACTCTCT-3′ |
| PLY6_F | 5′-ATAGGATCCTTACTGCCCATTTCTGTCCCA-3′ |
| PLY6_R | 5′-ATCCATATGAGAGGAATTTCTGCAGAGCGT-3′ |
| PLY7_F | 5′-ATAGGATCCTTAATTCGTCCGCTTACGCAC-3′ |
| PLY8_F | 5′-ATAGGATCCTTAGACAGAGAGATTACGAAC-3′ |
| PLY8_R | 5′-ACGCATATGAAGCAGATTTATTATACAGTC-3′ |
| I recognition site within PLY | 196-KQIYYTVSVDAVKNPGDVFQDTVTV-220 |
| P1 | 194-NFKQIYYTVSVD-205 |
| P2 | 197-QIYYTVSVDAVK-208 |
| P3 | 200-YTVSVDAVKNPG-211 |
| P4 | 203-SVDAVKNPGDVF-214 |
| P5 | 206-AVKNPGDVFQDT-217 |
| P6 | 209-NPGDVFQDTVTV-220 |
| P7 | 212-DVFQDTVTVEDL-223 |
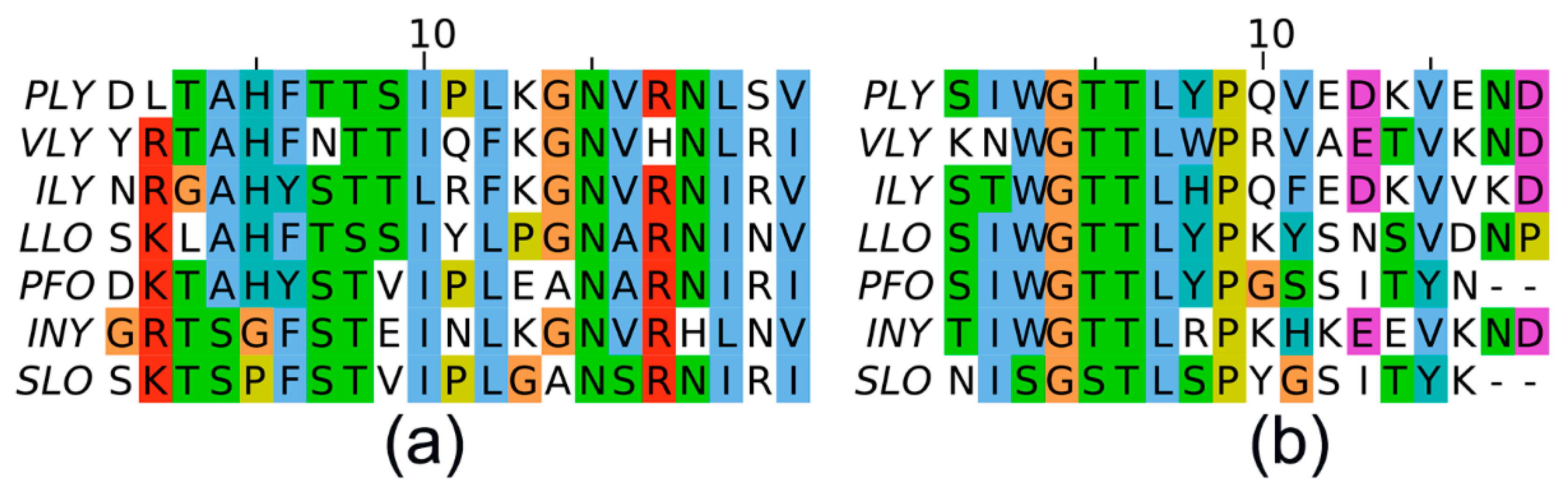
| II recognition site within PLY | 382-LSYDHQGKEVLTPKAWDRNGQDLTAHFTTSIPLKGNVRNLSV-423 |
| P8 | 378-TWDELSYDHQGK-389 |
| P9 | 381-ELSYDHQGKEVL-392 |
| P10 | 384-YDHQGKEVLTPK-395 |
| P11 | 387-QGKEVLTPKAWD-398 |
| P12 | 390-EVLTPKAWDRNG-401 |
| P13 | 393-TPKAWDRNGQDL-404 |
| P14 | 396-AWDRNGQDLTAH-407 |
| P15 | 399-RNGQDLTAHFTT-410 |
| P16 | 402-QDLTAHFTTSIP-413 |
| P17 | 405-TAHFTTSIPLKG-416 |
| P18 | 408-FTTSIPLKGNVR-419 |
| P19 | 411-SIPLKGNVRNLS-422 |
| P20 | 414-LKGNVRNLSVKI-425 |
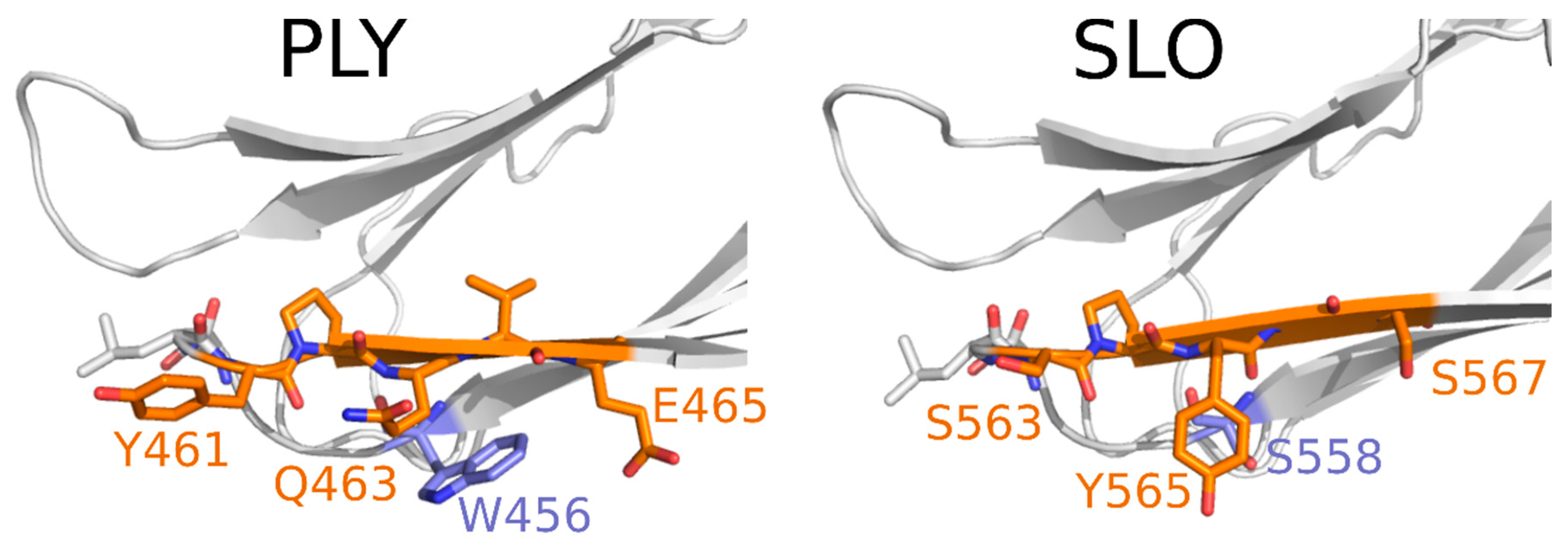
| III recognition site within PLY | 454-SIWGTTLYPQVEDKVEND-471 |
| P21 | 450-KRTISIWGTTL-460 |
| P22 | 453-ISIWGTTLYPQ-463 |
| P23 | 456-WGTTLYPQVED-466 |
| P24 | 459-TTLYPQVEDKVE-469 |
| P25 | 462-YPQVEDKVEND-471 |
| Toxin | Structure | D4 Start |
|---|---|---|
| PLY | 5AOD | 361 |
| VLY | 5IMY * | 406 |
| ILY | 1S1R | 419 |
| LLO | 4CDB * | 417 |
| PFO | 1PFO | 392 |
| INY | - | - |
| SLO | 4HSC | 463 |
| MAb Clone No. | IgG Subtype | Assay | MAb Reactivity and Cross-Reactivity with Cytolysins | ||||||
|---|---|---|---|---|---|---|---|---|---|
| PLY | ILY | VLY | PFO | LLO | INY | SLO | |||
| 6E5 | IgG2a | ELISA 1 | 0.6 | 0.2 | 0.4 | 1.7 | 67.0 | 0.8 | - |
| WB 2 | + | + | + | + | + | + | - | ||
| 3A9 | IgG1 | ELISA | 0.2 | - | - | 0.5 | 67.0 | - | - |
| WB | + | - | - | + | + | - | - | ||
| 3F3 | IgG1 | ELISA | 4.8 | - | - | - | - | - | - |
| WB | + | - | - | - | - | - | - | ||
| 12D10 | IgG1 | ELISA | 0.6 | - | - | - | - | - | - |
| WB | + | - | - | - | - | - | - | ||
| 12F11 | IgG1 | ELISA | 0.5 | - | - | - | - | - | - |
| WB | + | - | - | - | - | - | - | ||
| 6E3 | IgG2b | ELISA | 1.7 | 5.5 | 4.1 | - | - | - | - |
| WB | + | - | - | - | - | - | - | ||
| 19G1 | IgG2a | ELISA | 5.3 | 3.8 | 3.7 | - | - | - | - |
| WB | + | - | - | - | - | - | - | ||
| 3C10 | IgG1 | ELISA | 0.5 | - | - | - | - | - | - |
| WB | + | - | - | - | - | - | - | ||
| 9F2 | IgG1 | ELISA | 0.2 | - | - | - | - | - | - |
| WB | + | - | - | - | - | - | - | ||
| 14E4 | IgG1 | ELISA | 0.3 | - | - | - | - | - | - |
| WB | + | - | - | - | - | - | - | ||
| MAb Clone | Identified MAb Recognition Sites, aa | Protein Structure Elements | PLY Fragments | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |||
| 14E4 | 196–220 | D1 | + | + | + | |||||
| 9F2 | 381–402 | D4 | + | + | + | + | + | |||
| 3C10 | 403–423 | L3 in D4 | + | + | + | + | ||||
| 12D10 | + | + | + | + | ||||||
| 3A9 | 454–471 | L1 in D4 | + | |||||||
| 6E5 | + | |||||||||
| 12F11 | + | |||||||||
| 3F3 | not determined | + | ||||||||
| 6E3 | + | |||||||||
| 19G1 | + | |||||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kucinskaite-Kodze, I.; Simanavicius, M.; Dapkunas, J.; Pleckaityte, M.; Zvirbliene, A. Mapping of Recognition Sites of Monoclonal Antibodies Responsible for the Inhibition of Pneumolysin Functional Activity. Biomolecules 2020, 10, 1009. https://doi.org/10.3390/biom10071009
Kucinskaite-Kodze I, Simanavicius M, Dapkunas J, Pleckaityte M, Zvirbliene A. Mapping of Recognition Sites of Monoclonal Antibodies Responsible for the Inhibition of Pneumolysin Functional Activity. Biomolecules. 2020; 10(7):1009. https://doi.org/10.3390/biom10071009
Chicago/Turabian StyleKucinskaite-Kodze, Indre, Martynas Simanavicius, Justas Dapkunas, Milda Pleckaityte, and Aurelija Zvirbliene. 2020. "Mapping of Recognition Sites of Monoclonal Antibodies Responsible for the Inhibition of Pneumolysin Functional Activity" Biomolecules 10, no. 7: 1009. https://doi.org/10.3390/biom10071009
APA StyleKucinskaite-Kodze, I., Simanavicius, M., Dapkunas, J., Pleckaityte, M., & Zvirbliene, A. (2020). Mapping of Recognition Sites of Monoclonal Antibodies Responsible for the Inhibition of Pneumolysin Functional Activity. Biomolecules, 10(7), 1009. https://doi.org/10.3390/biom10071009





