Enzymatic Regulation and Biological Functions of Reactive Cysteine Persulfides and Polysulfides
Abstract
:1. Introduction
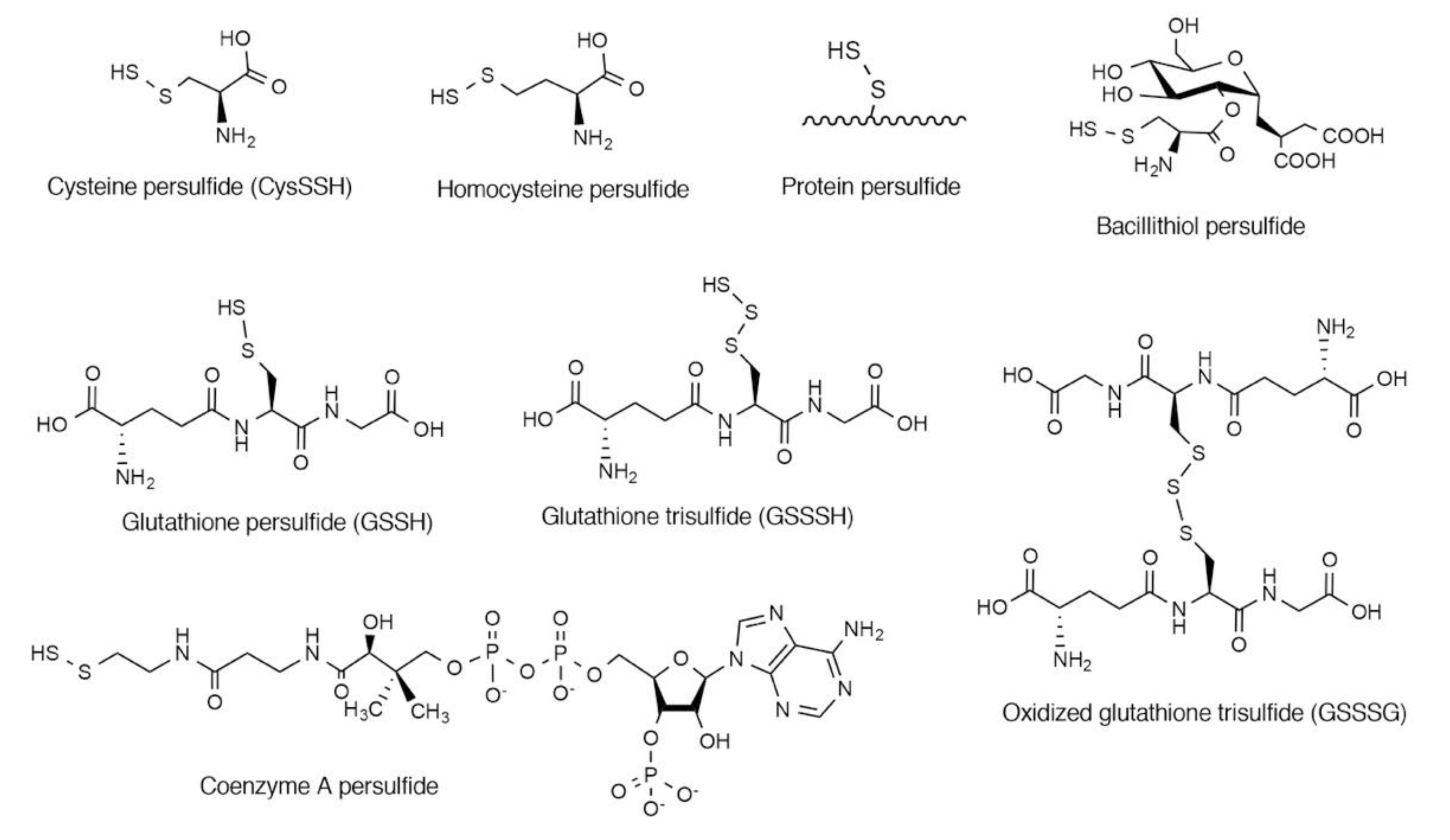
2. Endogenous Occurrence of CysSSH and Related Molecules
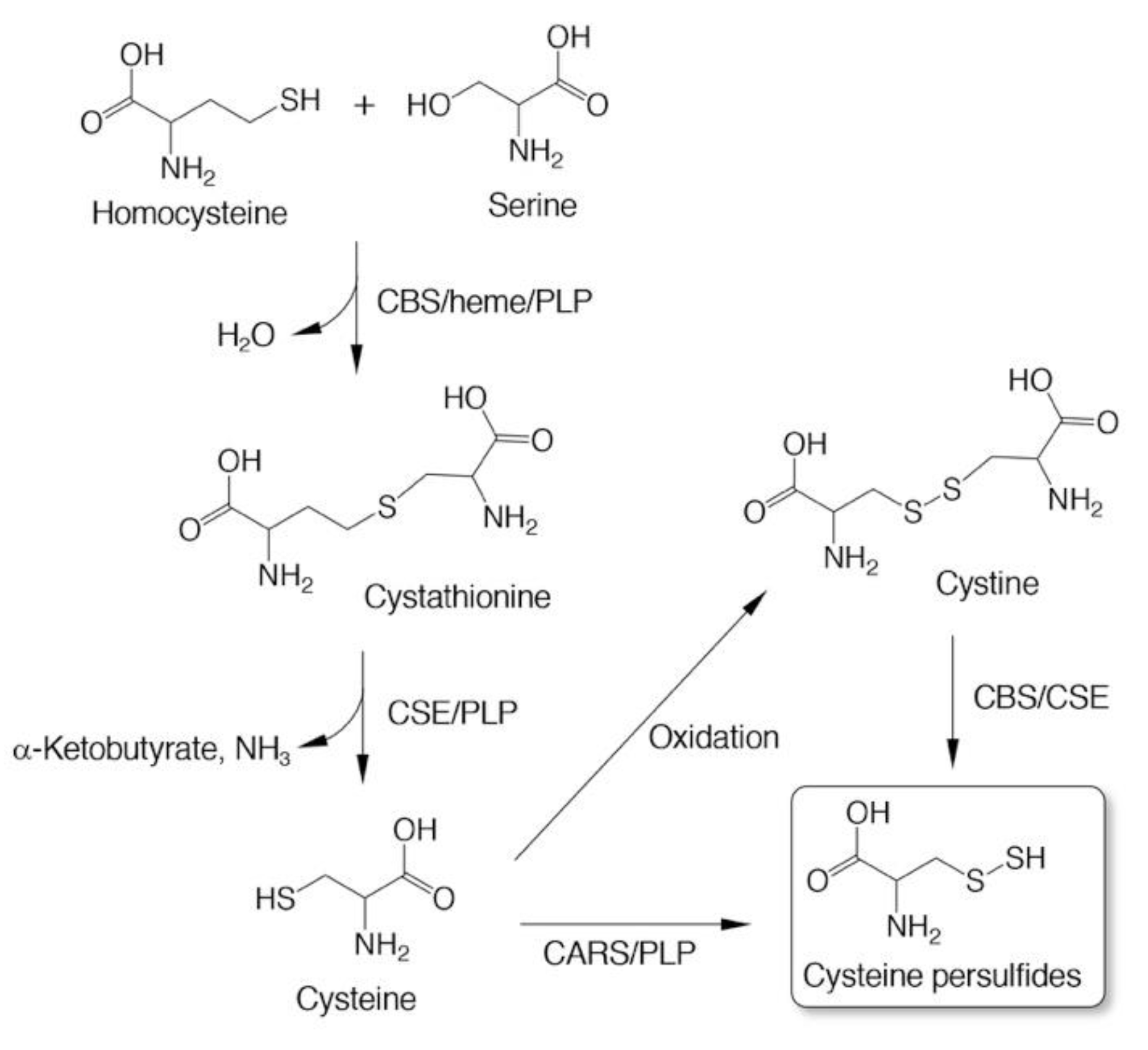
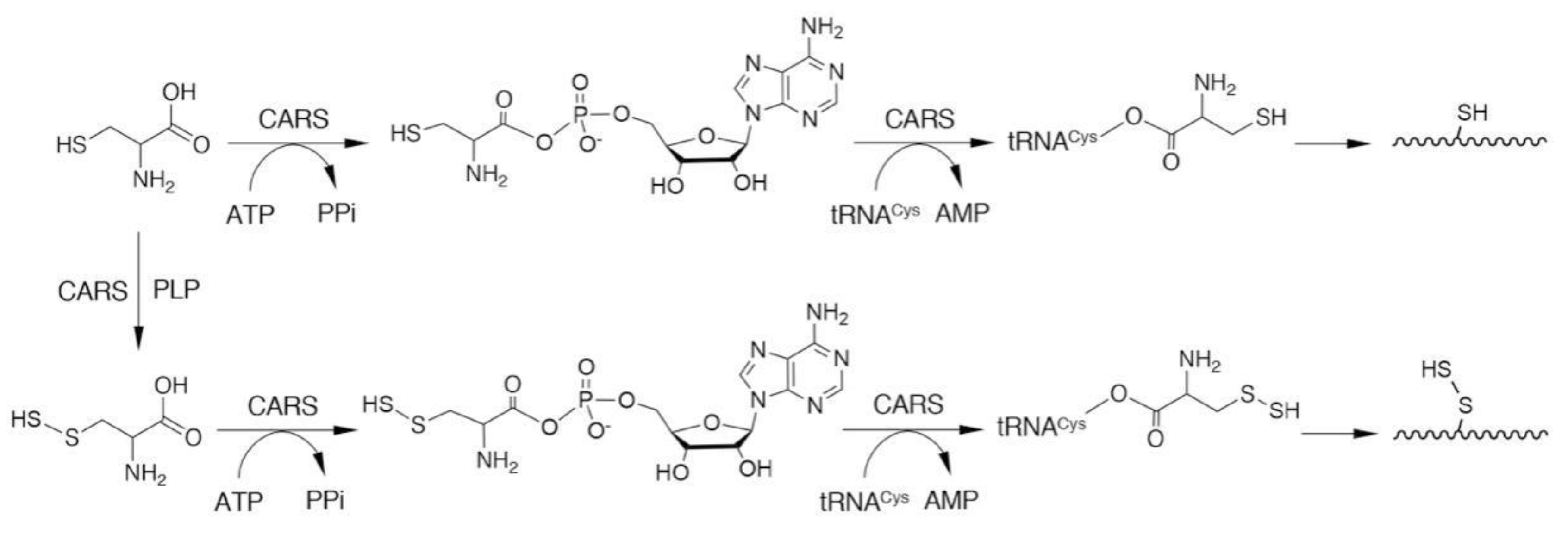
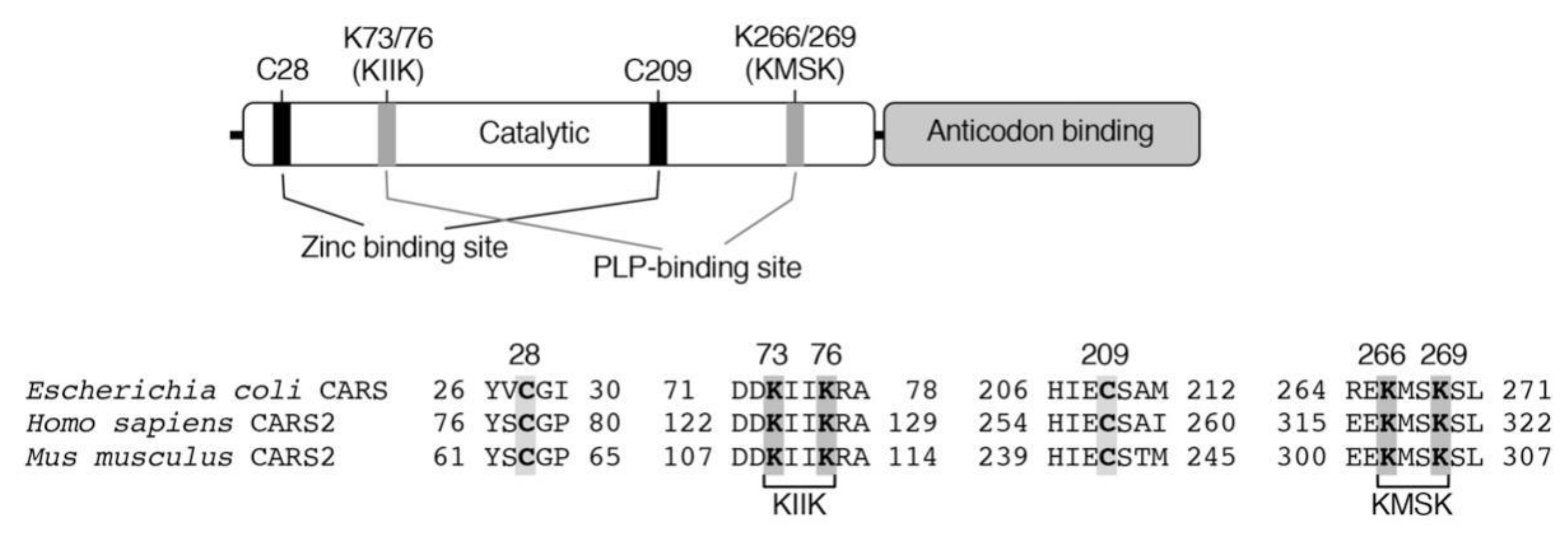
3. Biosynthesis of CysSSH and Related Molecules
4. Antioxidative and Nucleophilic Properties of Persulfides/Polysulfides
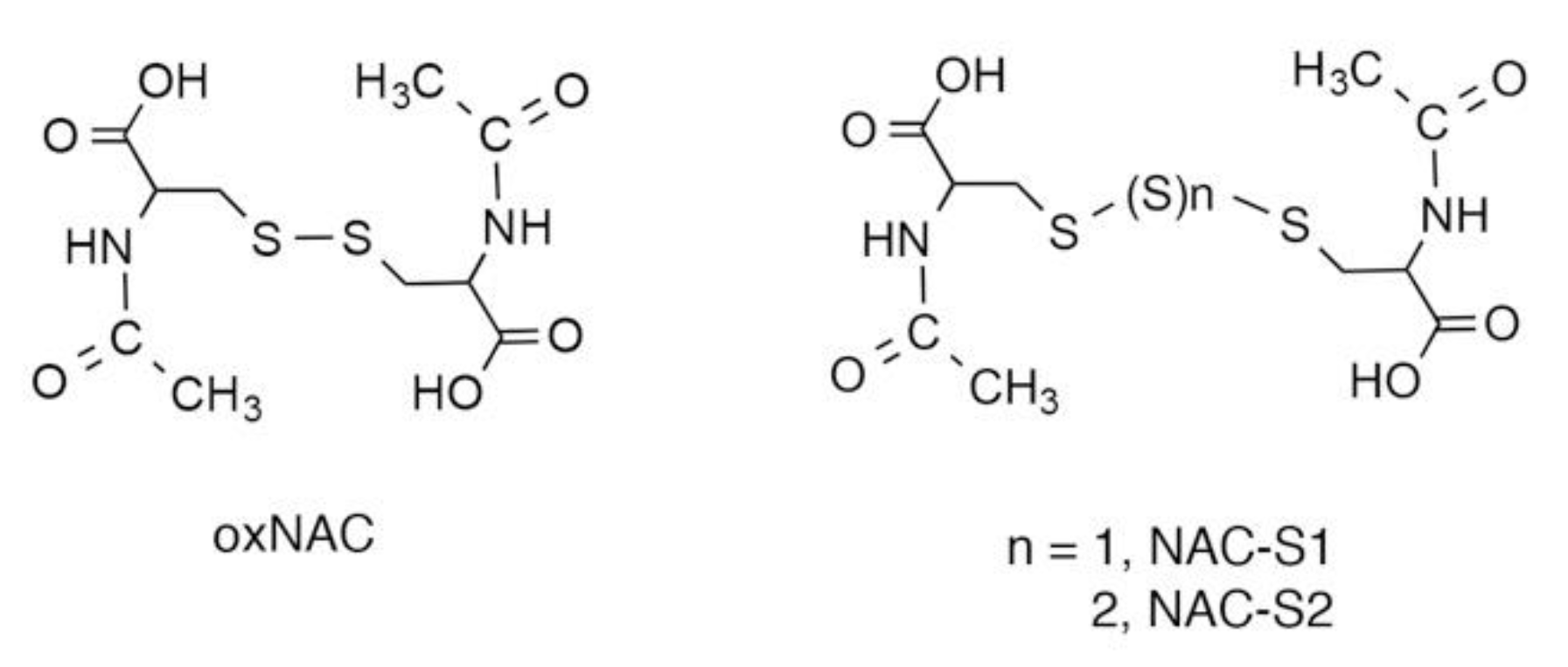
5. N-Acetyl-l-Cysteine (NAC) Polysulfides as Useful Chemical Tools to Investigate Biological Functions of Reactive Sulfur Species
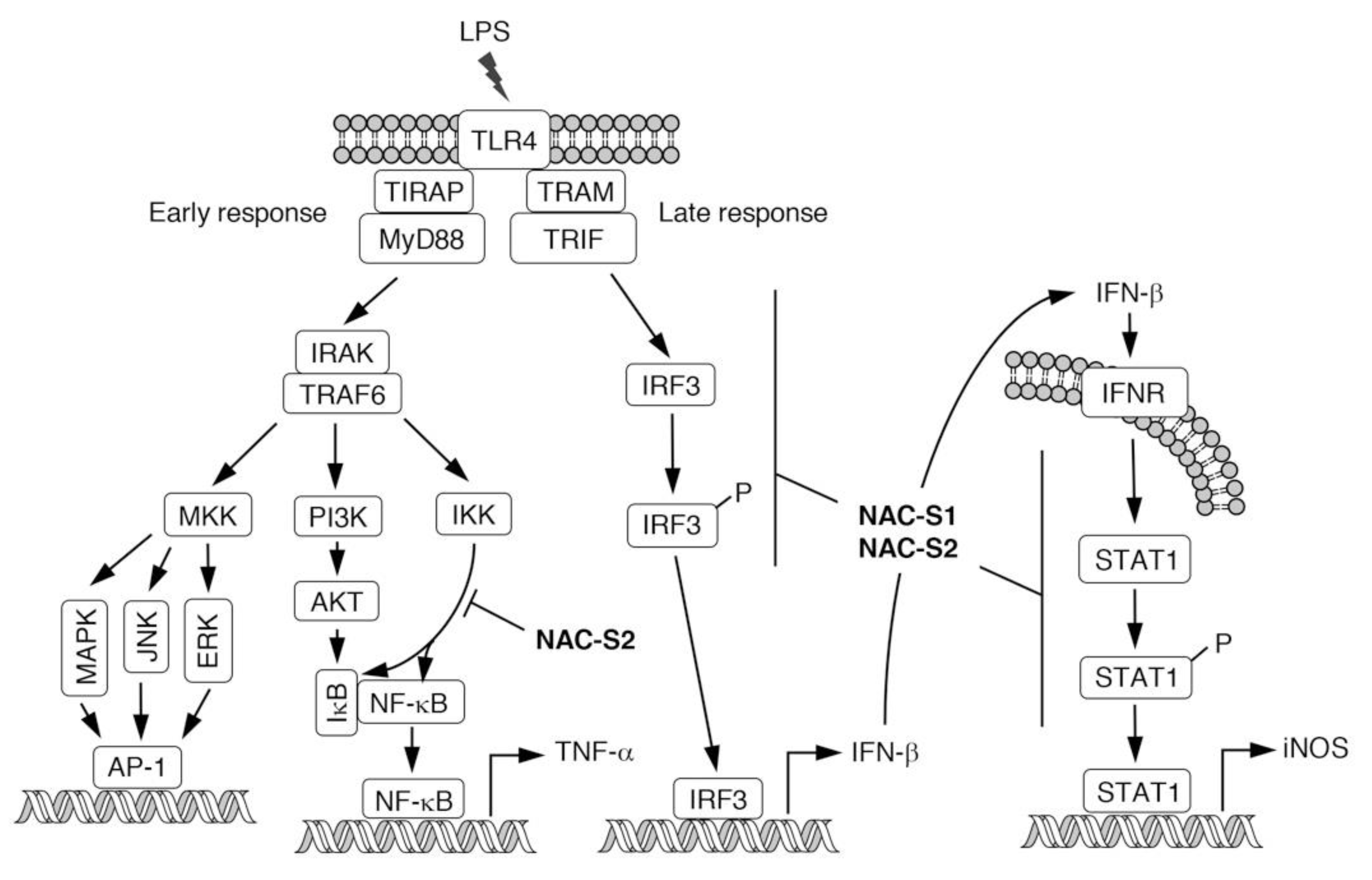
6. Anti-inflammatory Actions of NAC Polysulfides
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sawa, T.; Ono, K.; Tsutsuki, H.; Zhang, T.; Ida, T.; Nishida, M.; Akaike, T. Reactive cysteine persulphides: Occurrence, biosynthesis, antioxidant activity, methodologies, and bacterial persulphide signalling. Adv. Microb. Physiol. 2018, 72, 1–28. [Google Scholar]
- Fukuto, J.M.; Ignarro, L.J.; Nagy, P.; Wink, D.A.; Kevil, C.G.; Feelisch, M.; Cortese-Krott, M.M.; Bianco, C.L.; Kumagai, Y.; Hobbs, A.J.; et al. Biological hydropersulfides and related polysulfides—A new concept and perspective in redox biology. FEBS Lett. 2018, 592, 2140–2152. [Google Scholar] [CrossRef] [Green Version]
- Fujii, S.; Sawa, T.; Motohashi, H.; Akaike, T. Persulfide synthases that are functionally coupled with translation mediate sulfur respiration in mammalian cells. Br. J. Pharmacol. 2019, 176, 607–615. [Google Scholar] [CrossRef] [Green Version]
- Motohashi, H.; Akaike, T. Sulfur-utilizing cytoprotection and energy metabolism. Curr. Opin. Physiol. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Ida, T.; Sawa, T.; Ihara, H.; Tsuchiya, Y.; Watanabe, Y.; Kumagai, Y.; Suematsu, M.; Motohashi, H.; Fujii, S.; Matsunaga, T.; et al. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 7606–7611. [Google Scholar] [CrossRef] [Green Version]
- Akaike, T.; Ida, T.; Wei, F.Y.; Nishida, M.; Kumagai, Y.; Alam, M.M.; Ihara, H.; Sawa, T.; Matsunaga, T.; Kasamatsu, S.; et al. Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun. 2017, 8, 1177. [Google Scholar] [CrossRef] [Green Version]
- Kunikata, H.; Ida, T.; Sato, K.; Aizawa, N.; Sawa, T.; Tawarayama, H.; Murayama, N.; Fujii, S.; Akaike, T.; Nakazawa, T. Metabolomic profiling of reactive persulfides and polysulfides in the aqueous and vitreous humors. Sci. Rep. 2017, 7, 41984. [Google Scholar] [CrossRef]
- Numakura, T.; Sugiura, H.; Akaike, T.; Ida, T.; Fujii, S.; Koarai, A.; Yamada, M.; Onodera, K.; Hashimoto, Y.; Tanaka, R.; et al. Production of reactive persulfide species in chronic obstructive pulmonary disease. Thorax 2017, 72, 1074–1083. [Google Scholar] [CrossRef]
- Ikeda, M.; Ishima, Y.; Shibata, A.; Chuang, V.T.G.; Sawa, T.; Ihara, H.; Watanabe, H.; Xian, M.; Ouchi, Y.; Shimizu, T.; et al. Quantitative determination of polysulfide in albumins, plasma proteins and biological fluid samples using a novel combined assays approach. Anal. Chim. Acta 2017, 969, 18–25. [Google Scholar] [CrossRef]
- Peng, H.; Shen, J.; Edmonds, K.A.; Luebke, J.L.; Hickey, A.K.; Palmer, L.D.; Chang, F.J.; Bruce, K.A.; Kehl-Fie, T.E.; Skaar, E.P.; et al. Sulfide homeostasis and nitroxyl intersect via formation of reactive sulfur species in Staphylococcus aureus. mSphere 2017, 2, e00082–e17. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Fujii, S.; Matsunaga, T.; Nishimura, A.; Ono, K.; Ida, T.; Ahmed, K.A.; Okamoto, T.; Tsutsuki, H.; Sawa, T.; et al. Reactive persulfides from Salmonella typhimurium downregulate autophagy-mediated innate immunity in macrophages by inhibiting electrophilic signaling. Cell Chem. Biol. 2018, 25, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Walsh, B.J.C.; Flores-Mireles, A.L.; Peng, H.; Zhang, Y.; Zhang, Y.; Trinidad, J.C.; Hultgren, S.J.; Giedroc, D.P. Hydrogen sulfide sensing through reactive sulfur species (RSS) and nitroxyl (HNO) in Enterococcus faecalis. ACS Chem. Biol. 2018, 13, 1610–1620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Ono, K.; Tsutsuki, H.; Ihara, H.; Islam, W.; Akaike, T.; Sawa, T. Enhanced cellular polysulfides negatively regulate TLR4 signaling and mitigate lethal endotoxin shock. Cell Chem. Biol. 2019, 26, 686–698. [Google Scholar] [CrossRef]
- Hidese, R.; Mihara, H.; Esaki, N. Bacterial cysteine desulfurases: Versatile key players in biosynthetic pathways of sulfur-containing biofactors. Appl. Microbiol. Biotechnol. 2011, 91, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Crack, J.C.; Subramanian, S.; Green, J.; Thomson, A.J.; Le Brun, N.E.; Johnson, M.K. Reversible cycling between cysteine persulfide-ligated [2Fe-2S] and cysteine-ligated [4Fe-4S] clusters in the FNR regulatory protein. Proc. Natl. Acad. Sci. USA 2012, 109, 15734–15739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, N.; Wei, F.Y.; Watanabe, S.; Hirayama, M.; Ohuchi, Y.; Fujimura, A.; Kaitsuka, T.; Ishii, I.; Sawa, T.; Nakayama, H.; et al. Reactive sulfur species regulate tRNA methylthiolation and contribute to insulin secretion. Nucleic Acids Res. 2017, 45, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Takata, T.; Tsukuda, A.; Tsuchiya, Y.; Akaike, T.; Watanabe, Y. The active-site cysteine residue of Ca2+/calmodulin-dependent protein kinase I is protected from irreversible modification via generation of polysulfidation. Nitric Oxide 2019, 86, 68–75. [Google Scholar] [CrossRef]
- Takata, T.; Araki, S.; Tsuchiya, Y.; Watanabe, Y. Persulfide signaling in stress-initiated CaM kinase response. Antioxid. Redox Signal. 2020, in press. [Google Scholar] [CrossRef]
- Fukuto, J.M.; Carrington, S.J.; Tantillo, D.J.; Harrison, J.G.; Ignarro, L.J.; Freeman, B.A.; Chen, A.; Wink, D.A. Small molecule signaling agents: The integrated chemistry and biochemistry of nitrogen oxides, oxides of carbon, dioxygen, hydrogen sulfide, and their derived species. Chem. Res. Toxicol. 2012, 25, 769–793. [Google Scholar] [CrossRef]
- Rudyk, O.; Rowan, A.; Prysyazhna, O.; Krasemann, S.; Hartmann, K.; Zhang, M.; Shah, A.M.; Ruppert, C.; Weiss, A.; Schermuly, R.T.; et al. Oxidation of PKGIα mediates an endogenous adaptation to pulmonary hypertension. Proc. Natl. Acad. Sci. USA 2019, 116, 13016–13025. [Google Scholar] [CrossRef] [Green Version]
- Francoleon, N.E.; Carrington, S.J.; Fukuto, J.M. The reaction of H2S with oxidized thiols: Generation of persulfides and implications to H2S biology. Arch. Biochem. Biophys. 2011, 516, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Hamid, H.A.; Tanaka, A.; Ida, T.; Nishimura, A.; Matsunaga, T.; Fujii, S.; Morita, M.; Sawa, T.; Fukuto, J.M.; Nagy, P.; et al. Polysulfide stabilization by tyrosine and hydroxyphenyl-containing derivatives that is important for a reactive sulfur metabolomics analysis. Redox Biol. 2019, 21, 101096. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Akaike, T.; Sawa, T.; Kumagai, Y.; Wink, D.A.; Tantillo, D.J.; Hobbs, A.J.; Nagy, P.; Xian, M.; Lin, J.; et al. Redox chemistry and chemical biology of H2S, hydropersulfides, and derived species: Implications of their possible biological activity and utility. Free Radic. Biol. Med. 2014, 77, 82–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, P.J. New anti-inflammatory targets for chronic obstructive pulmonary disease. Nat. Rev. Drug Discov. 2013, 12, 543–559. [Google Scholar] [CrossRef] [PubMed]
- Kyogoku, Y.; Sugiura, H.; Ichikawa, T.; Numakura, T.; Koarai, A.; Yamada, M.; Fujino, N.; Tojo, Y.; Onodera, K.; Tanaka, R.; et al. Nitrosative stress in patients with asthma-chronic obstructive pulmonary disease overlap. J. Allergy Clin. Immunol. 2019, 144, 972–983 e14. [Google Scholar] [CrossRef] [Green Version]
- Postma, D.S.; Rabe, K.F. The Asthma-COPD Overlap Syndrome. N. Engl. J. Med. 2015, 373, 1241–1249. [Google Scholar] [CrossRef] [Green Version]
- Conrady, D.G.; Brescia, C.C.; Horii, K.; Weiss, A.A.; Hassett, D.J.; Herr, A.B. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc. Natl. Acad. Sci. USA 2008, 105, 19456–19461. [Google Scholar] [CrossRef] [Green Version]
- Takagi, H.; Ohtsu, I. L-Cysteine Metabolism and Fermentation in Microorganisms. Adv. Biochem. Eng. Biotechnol. 2017, 159, 129–151. [Google Scholar]
- Miles, E.W.; Kraus, J.P. Cystathionine β-synthase: Structure, function, regulation, and location of homocystinuria-causing mutations. J. Biol. Chem. 2004, 279, 29871–29874. [Google Scholar] [CrossRef] [Green Version]
- Stipanuk, M.H.; Ueki, I. Dealing with methionine/homocysteine sulfur: Cysteine metabolism to taurine and inorganic sulfur. J. Inherit. Metab. Dis. 2011, 34, 17–32. [Google Scholar] [CrossRef] [Green Version]
- Carter, C.W., Jr. Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem. 1993, 62, 715–748. [Google Scholar] [CrossRef]
- Woese, C.R.; Olsen, G.J.; Ibba, M.; Soll, D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000, 64, 202–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, M.; Yang, X.L.; Schimmel, P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 2010, 11, 668–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.M.; Christian, T.; Newberry, K.J.; Perona, J.J.; Hou, Y.M. Zinc-mediated amino acid discrimination in cysteinyl-tRNA synthetase. J. Mol. Biol. 2003, 327, 911–917. [Google Scholar] [CrossRef]
- Hallmann, K.; Zsurka, G.; Moskau-Hartmann, S.; Kirschner, J.; Korinthenberg, R.; Ruppert, A.K.; Ozdemir, O.; Weber, Y.; Becker, F.; Lerche, H.; et al. A homozygous splice-site mutation in CARS2 is associated with progressive myoclonic epilepsy. Neurology 2014, 83, 2183–2187. [Google Scholar] [CrossRef]
- Coughlin, C.R., 2nd; Scharer, G.H.; Friederich, M.W.; Yu, H.C.; Geiger, E.A.; Creadon-Swindell, G.; Collins, A.E.; Vanlander, A.V.; Coster, R.V.; Powell, C.A.; et al. Mutations in the mitochondrial cysteinyl-tRNA synthase gene, CARS2, lead to a severe epileptic encephalopathy and complex movement disorder. J. Med. Genet. 2015, 52, 532–540. [Google Scholar] [CrossRef] [Green Version]
- Doka, E.; Pader, I.; Biro, A.; Johansson, K.; Cheng, Q.; Ballago, K.; Prigge, J.R.; Pastor-Flores, D.; Dick, T.P.; Schmidt, E.E.; et al. A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci. Adv. 2016, 2, e1500968. [Google Scholar] [CrossRef] [Green Version]
- Jung, M.; Kasamatsu, S.; Matsunaga, T.; Akashi, S.; Ono, K.; Nishimura, A.; Morita, M.; Abdul Hamid, H.; Fujii, S.; Kitamura, H.; et al. Protein polysulfidation-dependent persulfide dioxygenase activity of ethylmalonic encephalopathy protein 1. Biochem. Biophys. Res. Commun. 2016, 480, 180–186. [Google Scholar] [CrossRef] [Green Version]
- Zivanovic, J.; Kouroussis, E.; Kohl, J.B.; Adhikari, B.; Bursac, B.; Schott-Roux, S.; Petrovic, D.; Miljkovic, J.L.; Thomas-Lopez, D.; Jung, Y.; et al. Selective persulfide detection reveals evolutionarily conserved antiaging effects of S-sulfhydration. Cell Metab. 2019, 30, 1152–1170. [Google Scholar] [CrossRef]
- Heppner, D.E.; Hristova, M.; Ida, T.; Mijuskovic, A.; Dustin, C.M.; Bogdandi, V.; Fukuto, J.M.; Dick, T.P.; Nagy, P.; Li, J.; et al. Cysteine perthiosulfenic acid (Cys-SSOH): A novel intermediate in thiol-based redox signaling? Redox Biol. 2018, 14, 379–385. [Google Scholar] [CrossRef]
- Nishimura, A.; Shimoda, K.; Tanaka, T.; Toyama, T.; Nishiyama, K.; Shinkai, Y.; Numaga-Tomita, T.; Yamazaki, D.; Kanda, Y.; Akaike, T.; et al. Depolysulfidation of Drp1 induced by low-dose methylmercury exposure increases cardiac vulnerability to hemodynamic overload. Sci. Signal. 2019, 12, eaaw1920. [Google Scholar] [CrossRef] [PubMed]
- Doka, E.; Ida, T.; Dagnell, M.; Abiko, Y.; Luong, N.C.; Balog, N.; Takata, T.; Espinosa, B.; Nishimura, A.; Cheng, Q.; et al. Control of protein function through oxidation and reduction of persulfidated states. Sci. Adv. 2020, 6, eaax8358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, B.D.; Snyder, S.H. H2S: A novel gasotransmitter that signals by sulfhydration. Trends Biochem. Sci. 2015, 40, 687–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Du, J.; Tang, C.; Huang, Y.; Jin, H. H2S-induced sulfhydration: Biological function and detection methodology. Front. Pharmacol. 2017, 8, 608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, G.; Zhao, S.; Xie, L.; Han, Y.; Ji, Y. Protein S-sulfhydration by hydrogen sulfide in cardiovascular system. Br. J. Pharmacol. 2018, 175, 1146–1156. [Google Scholar] [CrossRef]
- Li, H.; Liu, H.; Chen, Z.; Zhao, R.; Wang, Q.; Ran, M.; Xia, Y.; Hu, X.; Liu, J.; Xian, M.; et al. Using resonance synchronous spectroscopy to characterize the reactivity and electrophilicity of biologically relevant sulfane sulfur. Redox Biol. 2019, 24, 101179. [Google Scholar] [CrossRef]
- Everett, S.A.; Wardman, P. Perthiols as antioxidants: Radical-scavenging and prooxidative mechanisms. Methods Enzymol. 1995, 251, 55–69. [Google Scholar]
- Ihara, H.; Kasamatsu, S.; Kitamura, A.; Nishimura, A.; Tsutsuki, H.; Ida, T.; Ishizaki, K.; Toyama, T.; Yoshida, E.; Abdul Hamid, H.; et al. Exposure to electrophiles impairs reactive persulfide-dependent redox signaling in neuronal cells. Chem. Res. Toxicol. 2017, 30, 1673–1684. [Google Scholar] [CrossRef]
- Sawa, T.; Zaki, M.H.; Okamoto, T.; Akuta, T.; Tokutomi, Y.; Kim-Mitsuyama, S.; Ihara, H.; Kobayashi, A.; Yamamoto, M.; Fujii, S.; et al. Protein S-guanylation by the biological signal 8-nitroguanosine 3′,5′-cyclic monophosphate. Nat. Chem. Biol. 2007, 3, 727–735. [Google Scholar] [CrossRef] [Green Version]
- Nishida, M.; Sawa, T.; Kitajima, N.; Ono, K.; Inoue, H.; Ihara, H.; Motohashi, H.; Yamamoto, M.; Suematsu, M.; Kurose, H.; et al. Hydrogen sulfide anion regulates redox signaling via electrophile sulfhydration. Nat. Chem. Biol. 2012, 8, 714–724. [Google Scholar] [CrossRef]
- Powell, C.R.; Dillon, K.M.; Wang, Y.; Carrazzone, R.J.; Matson, J.B. A persulfide donor responsive to reactive oxygen species: Insights into reactivity and therapeutic potential. Angew. Chem. Int. Ed. 2018, 57, 6324–6328. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Yu, B.; Li, Z.; Yuan, Z.; Organ, C.L.; Trivedi, R.K.; Wang, S.; Lefer, D.J.; Wang, B. An esterase-sensitive prodrug approach for controllable delivery of persulfide species. Angew. Chem. Int. Ed. Engl. 2017, 56, 11749–11753. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Xu, S.; Radford, M.N.; Zhang, W.; Kelly, S.S.; Day, J.J.; Xian, M. O→S relay deprotection: A general approach to controllable donors of reactive sulfur species. Angew. Chem. Int. Ed. 2018, 57, 5893–5897. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.; Venkatesh, Y.; Das, J.; Gangopadhyay, M.; Maiti, T.K.; Singh, N.D.P. One- and two-photon-activated cysteine persulfide donors for biological targeting. J. Org. Chem. 2019, 84, 11441–11449. [Google Scholar] [CrossRef] [PubMed]
- Collin, M.; Anuar, F.B.; Murch, O.; Bhatia, M.; Moore, P.K.; Thiemermann, C. Inhibition of endogenous hydrogen sulfide formation reduces the organ injury caused by endotoxemia. Br. J. Pharmacol. 2005, 146, 498–505. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.L.; Pan, L.L.; Long, F.; Wu, W.J.; Yan, D.; Xu, P.; Liu, S.Y.; Qin, M.; Jia, W.W.; Liu, X.H.; et al. Endogenous hydrogen sulfide ameliorates NOX4 induced oxidative stress in LPS-stimulated macrophages and mice. Cell Physiol. Biochem. 2018, 47, 458–474. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Signaling to NF-κB by Toll-like receptors. Trends Mol. Med. 2007, 13, 460–469. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011, 34, 637–650. [Google Scholar] [CrossRef] [Green Version]
- Young, S.H.; Ye, J.; Frazer, D.G.; Shi, X.; Castranova, V. Molecular mechanism of tumor necrosis factor-α production in 1→3-β-glucan (zymosan)-activated macrophages. J. Biol. Chem. 2001, 276, 20781–20787. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Sun, R.; Wei, H.; Tian, Z. Toll-like receptor 3 ligand attenuates LPS-induced liver injury by down-regulation of toll-like receptor 4 expression on macrophages. Proc. Natl. Acad. Sci. USA 2005, 102, 17077–17082. [Google Scholar] [CrossRef] [Green Version]
- Cavaillon, J.M. Exotoxins and endotoxins: Inducers of inflammatory cytokines. Toxicon 2018, 149, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Kuzmich, N.N.; Sivak, K.V.; Chubarev, V.N.; Porozov, Y.B.; Savateeva-Lyubimova, T.N.; Peri, F. TLR4 signaling pathway modulators as potential therapeutics in inflammation and sepsis. Vaccines 2017, 5, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.; Xiong, Y.; Li, Q.; Yang, H. Inhibition of toll-like receptor signaling as a promising therapy for inflammatory diseases: A journey from molecular to nano therapeutics. Front. Physiol. 2017, 8, 508. [Google Scholar] [CrossRef] [PubMed]
- Ain, Q.U.; Batool, M.; Choi, S. TLR4-targeting therapeutics: Structural basis and computer-aided drug discovery approaches. Molecules 2020, 25, 627. [Google Scholar] [CrossRef] [Green Version]
- Bachtell, R.; Hutchinson, M.R.; Wang, X.; Rice, K.C.; Maier, S.F.; Watkins, L.R. Targeting the toll of drug abuse: The translational potential of toll-like receptor 4. CNS Neurol. Disord. Drug Targets 2015, 14, 692–699. [Google Scholar] [CrossRef]
- Liu, J.; Buisman-Pijlman, F.; Hutchinson, M.R. Toll-like receptor 4: Innate immune regulator of neuroimmune and neuroendocrine interactions in stress and major depressive disorder. Front. Neurosci. 2014, 8, 309. [Google Scholar] [CrossRef] [Green Version]
- De Paola, M.; Sestito, S.E.; Mariani, A.; Memo, C.; Fanelli, R.; Freschi, M.; Bendotti, C.; Calabrese, V.; Peri, F. Synthetic and natural small molecule TLR4 antagonists inhibit motoneuron death in cultures from ALS mouse model. Pharmacol. Res. 2016, 103, 180–187. [Google Scholar] [CrossRef]
- Thakur, K.K.; Saini, J.; Mahajan, K.; Singh, D.; Jayswal, D.P.; Mishra, S.; Bishayee, A.; Sethi, G.; Kunnumakkara, A.B. Therapeutic implications of toll-like receptors in peripheral neuropathic pain. Pharmacol. Res. 2017, 115, 224–232. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawa, T.; Motohashi, H.; Ihara, H.; Akaike, T. Enzymatic Regulation and Biological Functions of Reactive Cysteine Persulfides and Polysulfides. Biomolecules 2020, 10, 1245. https://doi.org/10.3390/biom10091245
Sawa T, Motohashi H, Ihara H, Akaike T. Enzymatic Regulation and Biological Functions of Reactive Cysteine Persulfides and Polysulfides. Biomolecules. 2020; 10(9):1245. https://doi.org/10.3390/biom10091245
Chicago/Turabian StyleSawa, Tomohiro, Hozumi Motohashi, Hideshi Ihara, and Takaaki Akaike. 2020. "Enzymatic Regulation and Biological Functions of Reactive Cysteine Persulfides and Polysulfides" Biomolecules 10, no. 9: 1245. https://doi.org/10.3390/biom10091245
APA StyleSawa, T., Motohashi, H., Ihara, H., & Akaike, T. (2020). Enzymatic Regulation and Biological Functions of Reactive Cysteine Persulfides and Polysulfides. Biomolecules, 10(9), 1245. https://doi.org/10.3390/biom10091245





