Dynamic Nuclear Polarization of Biomembrane Assemblies
Abstract
:1. Introduction
2. DNP-Enhanced Solid-State NMR Spectroscopy
2.1. Basic Requirements
2.1.1. Instrumentation
2.1.2. Stable Radical Polarizing Agents
2.1.3. Gains Demonstrated
2.2. Sample Preparation Considerations
2.2.1. Biradical Polarizing Agents and Lipid Membranes
2.2.2. Freezing of Samples
2.2.3. Glassing Agents and Buffer Optimization
2.3. Evaluation of Samples and DNP Optimization
2.3.1. Measurements of Biradical Concentration and Distribution
2.3.2. Evaluating DNP Enhancement
2.3.3. Maximizing DNP Enhancements
3. DNP-Enhanced NMR Characterization of the Membrane Active Peptide KL4: A Case Study
3.1. Sample Preparation
3.2. Evaluation of Samples by X-Band EPR Spectroscopy
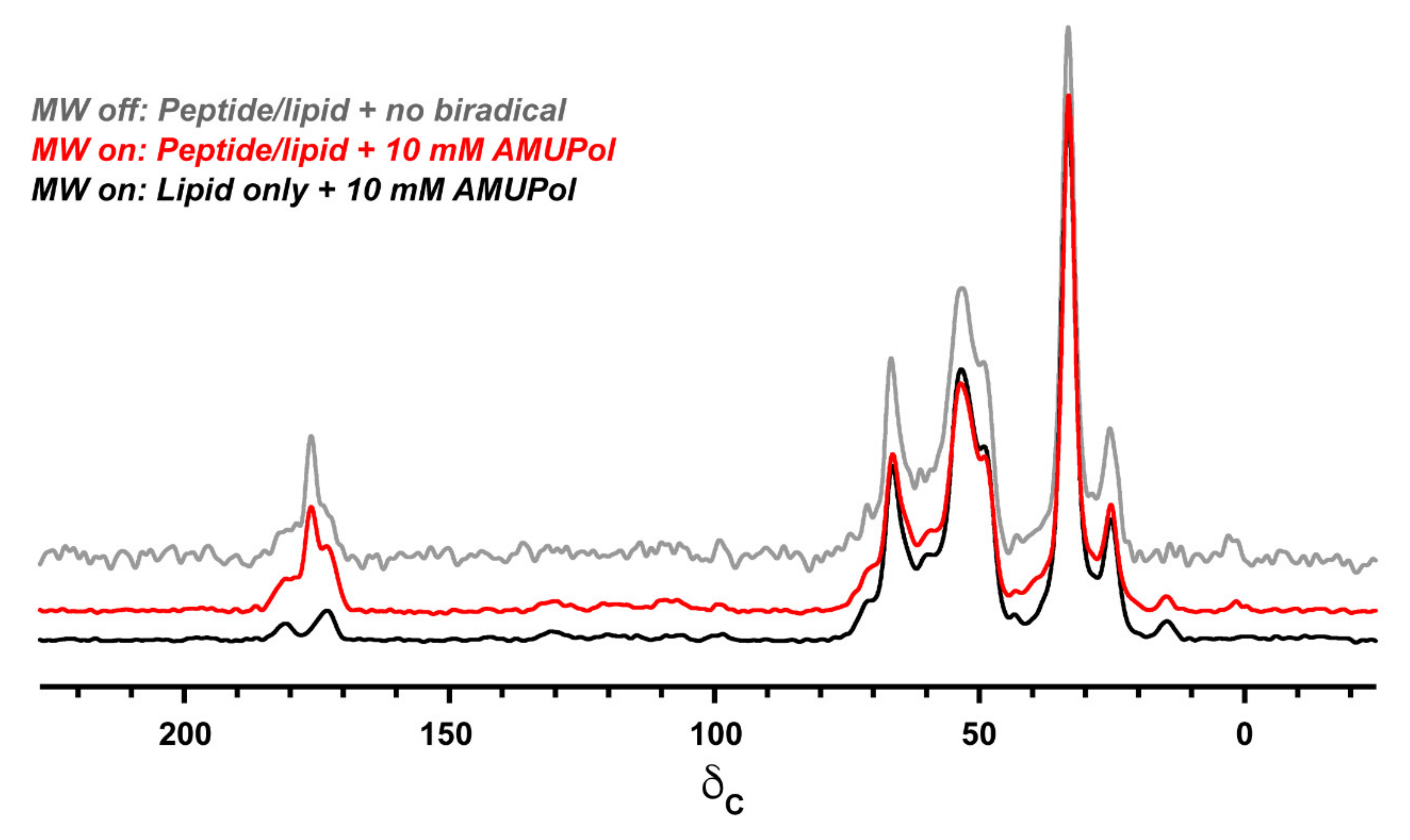
3.3. Evaluation of DNP Enhancement
3.4. Evaluation of Nuclear Spin Coherence Times
3.5. NMR Evaluation of Sample Integrity
4. Conclusions and Future Prospects for DNP of Membrane Proteins
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Amani, R.; Borcik, C.G.; Khan, N.H.; Versteeg, D.B.; Yekefallah, M.; Do, H.Q.; Coats, H.R.; Wylie, B.J. Conformational Changes upon Gating of KirBac1.1 into an Open-Activated State Revealed by Solid-State NMR and Functional Assays. Proc. Natl. Acad. Sci. USA 2020, 117, 2938–2947. [Google Scholar] [CrossRef] [Green Version]
- Shukla, R.; Medeiros-Silva, J.; Parmar, A.; Vermeulen, B.J.A.; Das, S.; Paioni, A.L.; Jekhmane, S.; Lorent, J.; Bonvin, A.M.J.J.; Baldus, M.; et al. Mode of Action of Teixobactins in Cellular Membranes. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Xiao, P.; Bolton, D.; Munro, R.A.; Brown, L.S.; Ladizhansky, V. Solid-State NMR Spectroscopy Based Atomistic View of a Membrane Protein Unfolding Pathway. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandala, V.S.; Loftis, A.R.; Shcherbakov, A.A.; Pentelute, B.L.; Hong, M. Atomic Structures of Closed and Open Influenza B M2 Proton Channel Reveal the Conduction Mechanism. Nat. Struct. Mol. Biol. 2020, 27, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Farver, R.S.; Mills, F.D.; Antharam, V.C.; Chebukati, J.N.; Fanucci, G.E.; Long, J.R. Lipid Polymorphism Induced by Surfactant Peptide SP-B(1-25). Biophys. J. 2010, 99, 1773–1782. [Google Scholar] [CrossRef] [Green Version]
- Tran, N.; Kurian, J.; Bhatt, A.; McKenna, R.; Long, J.R. Entropic Anomaly Observed in Lipid Polymorphisms Induced by Surfactant Peptide SP-B(1-25). J. Phys. Chem. B 2017, 121, 9102–9112. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Thurber, K.R.; Ghirlando, R.; Yau, W.M.; Tycko, R. Application of Millisecond Time-Resolved Solid State NMR to the Kinetics and Mechanism of Melittin Self-Assembly. Proc. Natl. Acad. Sci. USA 2019, 116, 16717–16722. [Google Scholar] [CrossRef] [Green Version]
- Maciejko, J.; Kaur, J.; Becker-Baldus, J.; Glaubitz, C. Photocycle-Dependent Conformational Changes in the Proteorhodopsin Cross-Protomer Asp–His–Trp Triad Revealed by DNP-Enhanced MAS-NMR. Proc. Natl. Acad. Sci. USA 2019, 116, 8342–8349. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, K.; Caporini, M.A.; Im, S.C.; Waskell, L.; Ramamoorthy, A. Transmembrane Interactions of Full-Length Mammalian Bitopic Cytochrome-P450-Cytochrome-b 5 Complex in Lipid Bilayers Revealed by Sensitivity-Enhanced Dynamic Nuclear Polarization Solid-State NMR Spectroscopy. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Sergeyev, I.V.; Itin, B.; Rogawski, R.; Day, L.A.; McDermott, A.E. Efficient Assignment and NMR Analysis of an Intact Virus Using Sequential Side-Chain Correlations and DNP Sensitization. Proc. Natl. Acad. Sci. USA 2017, 114, 5171–5176. [Google Scholar] [CrossRef] [Green Version]
- Frederick, K.K.; Michaelis, V.K.; Caporini, M.A.; Andreas, L.B.; Debelouchina, G.T.; Griffin, R.G.; Lindquist, S. Combining DNP NMR with Segmental and Specific Labeling to Study a Yeast Prion Protein Strain That Is Not Parallel In-Register. Proc. Natl. Acad. Sci. USA 2017, 114, 3642–3647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mak-Jurkauskas, M.L.; Bajaj, V.S.; Hornstein, M.K.; Belenky, M.; Griffin, R.G.; Herzfeld, J. Energy Transformations Early in the Bacteriorhodopsin Photocycle Revealed by DNP-Enhanced Solid-State NMR. Proc. Natl. Acad. Sci. USA 2008, 105, 883–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajaj, V.S.; Mak-Jurkauskas, M.L.; Belenky, M.; Herzfeld, J.; Griffin, R.G. Functional and Shunt States of Bacteriorhodopsin Resolved by 250 GHz Dynamic Nuclear Polarization-Enhanced Solid-State NMR. Proc. Natl. Acad. Sci. USA 2009, 106, 9244–9249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mentink-Vigier, F.; Mathies, G.; Liu, Y.; Barra, A.L.; Caporini, M.A.; Lee, D.; Hediger, S.; Griffin, R.; De Paëpe, G. Efficient Cross-Effect Dynamic Nuclear Polarization without Depolarization in High-Resolution MAS NMR. Chem. Sci. 2017, 8, 8150–8163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mentink-Vigier, F.; Marin-Montesinos, I.; Jagtap, A.P.; Halbritter, T.; van Tol, J.; Hediger, S.; Lee, D.; Sigurdsson, S.T.; De Paëpe, G. Computationally Assisted Design of Polarizing Agents for Dynamic Nuclear Polarization Enhanced NMR: The AsymPol Family. J. Am. Chem. Soc. 2018, 140, 11013–11019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lund, A.; Casano, G.; Menzildjian, G.; Kaushik, M.; Stevanato, G.; Yulikov, M.; Jabbour, R.; Wisser, D.; Renom-Carrasco, M.; Thieuleux, C.; et al. TinyPols: A Family of Water-Soluble Binitroxides Tailored for Dynamic Nuclear Polarization Enhanced NMR Spectroscopy at 18.8 and 21.1 T. Chem. Sci. 2020, 11, 2810–2818. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Hu, K.N.; Joo, C.G.; Swager, T.M.; Griffin, R.G. TOTAPOL: A Biradical Polarizing Agent for Dynamic Nuclear Polarization Experiments in Aqueous Media. J. Am. Chem. Soc. 2006, 128, 11385–11390. [Google Scholar] [CrossRef]
- Sauvée, C.; Rosay, M.; Casano, G.; Aussenac, F.; Weber, R.T.; Ouari, O.; Tordo, P. Highly Efficient, Water-Soluble Polarizing Agents for Dynamic Nuclear Polarization at High Frequency. Angew. Chem. Int. Ed. 2013, 52, 10858–10861. [Google Scholar] [CrossRef]
- Kiesewetter, M.K.; Corzilius, B.; Smith, A.A.; Griffin, R.G.; Swager, T.M. Dynamic Nuclear Polarization with a Water-Soluble Rigid Biradical. J. Am. Chem. Soc. 2012, 134, 4537–4540. [Google Scholar] [CrossRef] [Green Version]
- Dubroca, T.; Smith, A.N.; Pike, K.J.; Froud, S.; Wylde, R.; Trociewitz, B.; McKay, J.; Mentink-Vigier, F.; van Tol, J.; Wi, S.; et al. A Quasi-Optical and Corrugated Waveguide Microwave Transmission System for Simultaneous Dynamic Nuclear Polarization NMR on Two Separate 14.1 T Spectrometers. J. Magn. Reson. 2018, 289, 35–44. [Google Scholar] [CrossRef]
- Rosay, M.; Tometich, L.; Pawsey, S.; Bader, R.; Schauwecker, R.; Blank, M.; Borchard, P.M.; Cauffman, S.R.; Felch, K.L.; Weber, R.T.; et al. Solid-State Dynamic Nuclear Polarization at 263 GHz: Spectrometer Design and Experimental Results. Phys. Chem. Chem. Phys. 2010, 12, 5850–5860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sergeyev, I.V.; Aussenac, F.; Purea, A.; Reiter, C.; Bryerton, E.; Retzloff, S.; Hesler, J.; Tometich, L.; Rosay, M. Efficient 263 GHz Magic Angle Spinning DNP at 100 K Using Solid-State Diode Sources. Solid State Nucl. Magn. Reson. 2019, 100, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Thurber, K.R.; Yau, W.M.; Tycko, R. Low-Temperature Dynamic Nuclear Polarization at 9.4 T with a 30 MW Microwave Source. J. Magn. Reson. 2010, 204, 303–313. [Google Scholar] [CrossRef] [Green Version]
- Purea, A.; Reiter, C.; Dimitriadis, A.I.; de Rijk, E.; Aussenac, F.; Sergeyev, I.; Rosay, M.; Engelke, F. Improved Waveguide Coupling for 1.3 mm MAS DNP Probes at 263 GHz. J. Magn. Reson. 2019, 302, 43–49. [Google Scholar] [CrossRef]
- Mentink-Vigier, F.; Barra, A.-L.; van Tol, J.; Hediger, S.; Lee, D.; De Paëpe, G. De Novo Prediction of Cross-Effect Efficiency for Magic Angle Spinning Dynamic Nuclear Polarization. Phys. Chem. Chem. Phys. 2019, 21, 2166–2176. [Google Scholar] [CrossRef] [PubMed]
- Mentink-Vigier, F.; Vega, S.; De Paëpe, G. Fast and Accurate MAS–DNP Simulations of Large Spin Ensembles. Phys. Chem. Chem. Phys. 2017, 19, 3506–3522. [Google Scholar] [CrossRef]
- Yamamoto, K.; Caporini, M.A.; Im, S.-C.C.; Waskell, L.; Ramamoorthy, A. Cellular Solid-State NMR Investigation of a Membrane Protein Using Dynamic Nuclear Polarization. Biochim. Biophys. Acta Biomembr. 2015, 1848, 342–349. [Google Scholar] [CrossRef] [Green Version]
- Jantschke, A.; Koers, E.; Mance, D.; Weingarth, M.; Brunner, E.; Baldus, M. Insight into the Supramolecular Architecture of Intact Diatom Biosilica from DNP-Supported Solid-State NMR Spectroscopy. Angew. Chem. Int. Ed. 2015, 54, 15069–15073. [Google Scholar] [CrossRef]
- Takahashi, H.; Ayala, I.; Bardet, M.; De Paëpe, G.; Simorre, J.-P.P.; Hediger, S. Solid-State NMR on Bacterial Cells: Selective Cell Wall Signal Enhancement and Resolution Improvement Using Dynamic Nuclear Polarization. J. Am. Chem. Soc. 2013, 135, 5105–5110. [Google Scholar] [CrossRef]
- Narasimhan, S.; Scherpe, S.; Lucini Paioni, A.; van der Zwan, J.; Folkers, G.E.; Ovaa, H.; Baldus, M. DNP-Supported Solid-State NMR Spectroscopy of Proteins Inside Mammalian Cells. Angew. Chem. 2019, 131, 13103–13107. [Google Scholar] [CrossRef] [Green Version]
- Brownbill, N.J.; Gajan, D.; Lesage, A.; Emsley, L.; Blanc, F. Oxygen-17 Dynamic Nuclear Polarisation Enhanced Solid-State NMR Spectroscopy at 18.8 T. Chem. Commun. 2017, 53, 2563–2566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurencin, D.; Smith, M.E. Development of 43Ca Solid State NMR Spectroscopy as a Probe of Local Structure in Inorganic and Molecular Materials. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 68, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Leroy, C.; Crevant, C.; Bonhomme-Coury, L.; Babonneau, F.; Laurencin, D.; Bonhomme, C.; De Paëpe, G. Interfacial Ca2+ Environments in Nanocrystalline Apatites Revealed by Dynamic Nuclear Polarization Enhanced 43Ca NMR Spectroscopy. Nat. Commun. 2017, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Thurber, K.R.; Tycko, R. Theory for Cross Effect Dynamic Nuclear Polarization under Magic-Angle Spinning in Solid State Nuclear Magnetic Resonance: The Importance of Level Crossings. J. Chem. Phys. 2012, 137, 084508. [Google Scholar] [CrossRef]
- Mentink-Vigier, F.; Akbey, Ü.; Hovav, Y.; Vega, S.; Oschkinat, H.; Feintuch, A. Fast Passage Dynamic Nuclear Polarization on Rotating Solids. J. Magn. Reson. 2012, 224, 13–21. [Google Scholar] [CrossRef]
- Hovav, Y.; Feintuch, A.; Vega, S. Theoretical Aspects of Dynamic Nuclear Polarization in the Solid State—The Cross Effect. J. Magn. Reson. 2012, 214, 29–41. [Google Scholar] [CrossRef]
- Hu, K.N.; Debelouchina, G.T.; Smith, A.A.; Griffin, R.G. Quantum Mechanical Theory of Dynamic Nuclear Polarization in Solid Dielectrics. J. Chem. Phys. 2011, 134, 125105. [Google Scholar] [CrossRef] [Green Version]
- Mentink-Vigier, F.; Akbey, Ü.; Oschkinat, H.; Vega, S.; Feintuch, A. Theoretical Aspects of Magic Angle Spinning—Dynamic Nuclear Polarization. J. Magn. Reson. 2015, 258, 102–120. [Google Scholar] [CrossRef]
- Mentink-Vigier, F.; Paul, S.; Lee, D.; Feintuch, A.; Hediger, S.; Vega, S.; De Paëpe, G. Nuclear Depolarization and Absolute Sensitivity in Magic-Angle Spinning Cross Effect Dynamic Nuclear Polarization. Phys. Chem. Chem. Phys. 2015, 17, 21824–21836. [Google Scholar] [CrossRef]
- Matsuki, Y.; Nakamura, S.; Fukui, S.; Suematsu, H.; Fujiwara, T. Closed-Cycle Cold Helium Magic-Angle Spinning for Sensitivity-Enhanced Multi-Dimensional Solid-State NMR. J. Magn. Reson. 2015, 259, 76–81. [Google Scholar] [CrossRef] [Green Version]
- Matsuki, Y.; Ueda, K.; Idehara, T.; Ikeda, R.; Ogawa, I.; Nakamura, S.; Toda, M.; Anai, T.; Fujiwara, T. Helium-Cooling and -Spinning Dynamic Nuclear Polarization for Sensitivity-Enhanced Solid-State NMR at 14 T and 30 K. J. Magn. Reson. 2012, 225, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Bouleau, E.; Saint-Bonnet, P.; Hediger, S.; De Paëpe, G. Ultra-Low Temperature MAS-DNP. J. Magn. Reson. 2016, 264, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Bouleau, E.; Saint-Bonnet, P.; Mentink-Vigier, F.; Takahashi, H.; Jacquot, J.F.; Bardet, M.; Aussenac, F.; Purea, A.; Engelke, F.; Hediger, S.; et al. Pushing NMR Sensitivity Limits Using Dynamic Nuclear Polarization with Closed-Loop Cryogenic Helium Sample Spinning. Chem. Sci. 2015, 6, 6806–6812. [Google Scholar] [CrossRef] [Green Version]
- Corzilius, B. Theory of Solid Effect and Cross Effect Dynamic Nuclear Polarization with Half-Integer High-Spin Metal Polarizing Agents in Rotating Solids. Phys. Chem. Chem. Phys. 2016, 18, 27190–27204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, K.O.; Jawla, S.; Temkin, R.J.; Griffin, R.G. Pulsed Dynamic Nuclear Polarization. eMagRes 2019, 8, 339–352. [Google Scholar]
- Berruyer, P.; Emsley, L.; Lesage, A. DNP in Materials Science: Touching the Surface. eMagRes 2018, 7, 93–104. [Google Scholar]
- Becerra, L.R.; Gerfen, G.J.; Temkin, R.J.; Singel, D.J.; Griffin, R.G. Dynamic Nuclear Polarization with a Cyclotron Resonance Maser at 5 T. Phys. Rev. Lett. 1993, 71, 3561–3564. [Google Scholar] [CrossRef] [PubMed]
- Blank, M.; Felch, K.L. Millimeter-Wave Sources for DNP-NMR. eMagRes 2018, 7, 155–166. [Google Scholar]
- Thurber, K.; Tycko, R. Low-Temperature Dynamic Nuclear Polarization with Helium-Cooled Samples and Nitrogen-Driven Magic-Angle Spinning. J. Magn. Reson. 2016, 264, 99–106. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, M.; Narasimhan, S.; de Heus, C.; Mance, D.; van Doorn, S.; Houben, K.; Popov-Čeleketić, D.; Damman, R.; Katrukha, E.A.; Jain, P.; et al. EGFR Dynamics Change during Activation in Native Membranes as Revealed by NMR. Cell 2016, 167, 1241–1251. [Google Scholar] [CrossRef] [Green Version]
- Wisser, D.; Karthikeyan, G.; Lund, A.; Casano, G.; Karoui, H.; Yulikov, M.; Menzildjian, G.; Pinon, A.C.; Purea, A.; Engelke, F.; et al. BDPA-Nitroxide Biradicals Tailored for Efficient Dynamic Nuclear Polarization Enhanced Solid-State NMR at Magnetic Fields up to 21.1 T. J. Am. Chem. Soc. 2018, 140, 13340–13349. [Google Scholar] [CrossRef] [Green Version]
- Hu, K.N.; Song, C.; Yu, H.H.; Swager, T.M.; Griffin, R.G. High-Frequency Dynamic Nuclear Polarization Using Biradicals: A Multifrequency EPR Lineshape Analysis. J. Chem. Phys. 2008, 128. [Google Scholar] [CrossRef]
- Hu, K.N.; Yu, H.H.; Swager, T.M.; Griffin, R.G. Dynamic Nuclear Polarization with Biradicals. J. Am. Chem. Soc. 2004, 126, 10844–10845. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, Y.; Maly, T.; Ouari, O.; Karoui, H.; Le Moigne, F.; Rizzato, E.; Lyubenova, S.; Herzfeld, J.; Prisner, T.; Tordo, P.; et al. Dynamic Nuclear Polarization with a Rigid Biradical. Angew. Chem. Int. Ed. 2009, 48, 4996–5000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mentink-Vigier, F. Optimizing Nitroxide Biradicals for Cross-Effect MAS-DNP: The Role of: G -Tensors’ Distance. Phys. Chem. Chem. Phys. 2020, 22, 3643–3652. [Google Scholar] [CrossRef] [PubMed]
- Sauvée, C.; Casano, G.; Abel, S.; Rockenbauer, A.; Akhmetzyanov, D.; Karoui, H.; Siri, D.; Aussenac, F.; Maas, W.; Weber, R.T.; et al. Tailoring of Polarizing Agents in the BTurea Series for Cross-Effect Dynamic Nuclear Polarization in Aqueous Media. Chem. Eur. J. 2016, 22, 5598–5606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagtap, A.P.; Geiger, M.A.; Stöppler, D.; Orwick-Rydmark, M.; Oschkinat, H.; Sigurdsson, S.T. BcTol: A Highly Water-Soluble Biradical for Efficient Dynamic Nuclear Polarization of Biomolecules. Chem. Commun. 2016, 52, 7020–7023. [Google Scholar] [CrossRef]
- Geiger, M.-A.; Jagtap, A.P.; Kaushik, M.; Sun, H.; Stöppler, D.; Sigurdsson, S.T.; Corzilius, B.; Oschkinat, H. Efficiency of Water-Soluble Nitroxide Biradicals for Dynamic Nuclear Polarization in Rotating Solids at 9.4 T: BcTol-M and Cyolyl-TOTAPOL as New Polarizing Agents. Chem. Eur. J. 2018, 24, 13485–13494. [Google Scholar] [CrossRef]
- Mathies, G.; Caporini, M.A.; Michaelis, V.K.; Liu, Y.; Hu, K.-N.; Mance, D.; Zweier, J.L.; Rosay, M.; Baldus, M.; Griffin, R.G. Efficient Dynamic Nuclear Polarization at 800 MHz/527 GHz with Trityl-Nitroxide Biradicals. Angew. Chem. Int. Ed. 2015, 54, 11770–11774. [Google Scholar] [CrossRef] [Green Version]
- Rossini, A.J.; Zagdoun, A.; Lelli, M.; Gajan, D.; Rascón, F.; Rosay, M.; Maas, W.E.; Copéret, C.; Lesage, A.; Emsley, L. One Hundred Fold Overall Sensitivity Enhancements for Silicon-29 NMR Spectroscopy of Surfaces by Dynamic Nuclear Polarization with CPMG Acquisition. Chem. Sci. 2012, 3, 108–115. [Google Scholar] [CrossRef]
- Kobayashi, T.; Lafon, O.; Lilly Thankamony, A.S.; Slowing, I.I.; Kandel, K.; Carnevale, D.; Vitzthum, V.; Vezin, H.; Amoureux, J.P.; Bodenhausen, G.; et al. Analysis of Sensitivity Enhancement by Dynamic Nuclear Polarization in Solid-State NMR: A Case Study of Functionalized Mesoporous Materials. Phys. Chem. Chem. Phys. 2013, 15, 5553–5562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debelouchina, G.T.; Bayro, M.J.; Van Der Wel, P.C.A.; Caporini, M.A.; Barnes, A.B.; Rosay, M.; Maas, W.E.; Griffin, R.G. Dynamic Nuclear Polarization-Enhanced Solid-State NMR Spectroscopy of GNNQQNY Nanocrystals and Amyloid Fibrils. Phys. Chem. Chem. Phys. 2010, 12, 5911–5919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lafon, O.; Thankamony, A.S.L.; Kobayashi, T.; Carnevale, D.; Vitzthum, V.; Slowing, I.I.; Kandel, K.; Vezin, H.; Amoureux, J.P.; Bodenhausen, G.; et al. Mesoporous Silica Nanoparticles Loaded with Surfactant: Low Temperature Magic Angle Spinning 13C and 29Si NMR Enhanced by Dynamic Nuclear Polarization. J. Phys. Chem. C 2013, 117, 1375–1382. [Google Scholar] [CrossRef] [Green Version]
- Lesage, A.; Lelli, M.; Gajan, D.; Caporini, M.A.; Vitzthum, V.; Miéville, P.; Alauzun, J.; Roussey, A.; Thieuleux, C.; Mehdi, A.; et al. Surface Enhanced NMR Spectroscopy by Dynamic Nuclear Polarization. J. Am. Chem. Soc. 2010, 132, 15459–15461. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.N.; Caporini, M.A.; Fanucci, G.E.; Long, J.R. A Method for Dynamic Nuclear Polarization Enhancement of Membrane Proteins. Angew. Chem. Int. Ed. 2015, 54, 1542–1546. [Google Scholar] [CrossRef]
- Smith, A.N.; Twahir, U.T.; Dubroca, T.; Fanucci, G.E.; Long, J.R. Molecular Rationale for Improved Dynamic Nuclear Polarization of Biomembranes. J. Phys. Chem. B 2016, 120, 7880–7888. [Google Scholar] [CrossRef] [Green Version]
- Maly, T.; Cui, D.; Griffin, R.G.; Miller, A.-F. 1 H Dynamic Nuclear Polarization Based on an Endogenous Radical. J. Phys. Chem. B 2012, 116, 7055–7065. [Google Scholar] [CrossRef]
- Kirilina, E.P.; Grigoriev, I.A.; Dzuba, S.A. Orientational Motion of Nitroxides in Molecular Glasses: Dependence on the Chemical Structure, on the Molecular Size of the Probe, and on the Type of the Matrix. J. Chem. Phys. 2004, 121, 12465. [Google Scholar] [CrossRef]
- Takahashi, H.; Fernández-De-Alba, C.; Lee, D.; Maurel, V.; Gambarelli, S.; Bardet, M.; Hediger, S.; Barra, A.L.; De Paëpe, G. Optimization of an Absolute Sensitivity in a Glassy Matrix during DNP-Enhanced Multidimensional Solid-State NMR Experiments. J. Magn. Reson. 2014, 239, 91–99. [Google Scholar] [CrossRef]
- Rubinstein, P.; Dobrila, L.; Rosenfield, R.E.; Adamson, J.W.; Migliaccio, G.; Migliaccio, A.R.; Taylor, P.E.; Stevens, C.E. Processing and Cryopreservation of Placental/Umbilical Cord Blood for Unrelated Bone Marrow Reconstitution. Proc. Natl. Acad. Sci. USA 1995, 92, 10119–10122. [Google Scholar] [CrossRef] [Green Version]
- Leavesley, A.; Wilson, C.B.; Sherwin, M.; Han, S. Effect of Water/Glycerol Polymorphism on Dynamic Nuclear Polarization. Phys. Chem. Chem. Phys. 2018, 20, 9897–9903. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, D.; Bhat, S.V. Vitrification, Relaxation and Free Volume in Glycerol–Water Binary Liquid Mixture: Spin Probe ESR Studies. J. Non-Cryst. Solids 2009, 355, 2433–2438. [Google Scholar] [CrossRef] [Green Version]
- Metz, G.; Wu, X.; Smith, S.O. Ramped-Amplitude Cross Polarization in Magic-Angle-Spinning NMR. J. Magn. Reson. Ser. A 1994, 110, 219–227. [Google Scholar] [CrossRef]
- Ni, Q.Z.; Daviso, E.; Can, T.V.; Markhasin, E.; Jawla, S.K.; Swager, T.M.; Temkin, R.J.; Herzfeld, J.; Griffin, R.G. High Frequency Dynamic Nuclear Polarization. Acc. Chem. Res. 2013, 46, 1933–1941. [Google Scholar] [CrossRef]
- McDaniel, R.V.; McIntosh, T.J.; Simon, S.A. Nonelectrolyte Substitution for Water in Phosphatidylcholine Bilayers. Biochim. Biophys. Acta Biomembr. 1983, 731, 97–108. [Google Scholar] [CrossRef]
- Schrader, A.M.; Cheng, C.-Y.; Israelachvili, J.N.; Han, S. Communication: Contrasting Effects of Glycerol and DMSO on Lipid Membrane Surface Hydration Dynamics and Forces. J. Chem. Phys. 2016, 145, 041101. [Google Scholar] [CrossRef] [Green Version]
- Viennet, T.; Viegas, A.; Kuepper, A.; Arens, S.; Gelev, V.; Petrov, O.; Grossmann, T.N.; Heise, H.; Etzkorn, M. Selective Protein Hyperpolarization in Cell Lysates Using Targeted Dynamic Nuclear Polarization. Angew. Chem. Int. Ed. 2016, 55, 10746–10750. [Google Scholar] [CrossRef]
- Good, D.B.; Voinov, M.A.; Bolton, D.; Ward, M.E.; Sergeyev, I.V.; Caporini, M.; Scheffer, P.; Lo, A.; Rosay, M.; Marek, A.; et al. A Biradical-Tagged Phospholipid as a Polarizing Agent for Solid-State MAS Dynamic Nuclear Polarization NMR of Membrane Proteins. Solid State Nucl. Magn. Reson. 2019, 100, 92–101. [Google Scholar] [CrossRef]
- Fernández-de-Alba, C.; Takahashi, H.; Richard, A.; Chenavier, Y.; Dubois, L.; Maurel, V.; Lee, D.; Hediger, S.; De Paëpe, G.; De Paëpe, G. Matrix-Free DNP-Enhanced NMR Spectroscopy of Liposomes Using a Lipid-Anchored Biradical. Chem. Eur. J. 2015, 21, 4512–4517. [Google Scholar] [CrossRef]
- Sani, M.-A.; Zhu, S.; Hofferek, V.; Separovic, F. Nitroxide Spin–Labeled Peptides for DNP-NMR in-Cell Studies. FASEB J. 2019, 33, 11021–11027. [Google Scholar] [CrossRef] [Green Version]
- Voinov, M.A.; Good, D.B.; Ward, M.E.; Milikisiyants, S.; Marek, A.; Caporini, M.A.; Rosay, M.; Munro, R.A.; Ljumovic, M.; Brown, L.S.; et al. Cysteine-Specific Labeling of Proteins with a Nitroxide Biradical for Dynamic Nuclear Polarization NMR. J. Phys. Chem. B 2015, 119, 10180–10190. [Google Scholar] [CrossRef] [PubMed]
- Rogawski, R.; Sergeyev, I.V.; Li, Y.; Ottaviani, M.F.; Cornish, V.; McDermott, A.E. Dynamic Nuclear Polarization Signal Enhancement with High-Affinity Biradical Tags. J. Phys. Chem. B 2017, 121, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- van der Cruijsen, E.A.W.; Koers, E.J.; Sauvée, C.; Hulse, R.E.; Weingarth, M.; Ouari, O.; Perozo, E.; Tordo, P.; Baldus, M. Biomolecular DNP-Supported NMR Spectroscopy Using Site-Directed Spin Labeling. Chem. Eur. J. 2015, 21, 12971–12977. [Google Scholar] [CrossRef] [PubMed]
- Wylie, B.J.; Dzikovski, B.G.; Pawsey, S.; Caporini, M.; Rosay, M.; Freed, J.H.; McDermott, A.E. Dynamic Nuclear Polarization of Membrane Proteins: Covalently Bound Spin-Labels at Protein-Protein Interfaces. J. Biomol. NMR 2015, 61, 361–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.; Gerfen, G.J.; Rousseau, D.L.; Yeh, S.R. Ultrafast Microfluidic Mixer and Freeze-Quenching Device. Anal. Chem. 2003, 75, 5381–5386. [Google Scholar] [CrossRef]
- Hu, K.N.; Yau, W.M.; Tycko, R. Detection of a Transient Intermediate in a Rapid Protein Folding Process by Solid-State Nuclear Magnetic Resonance. J. Am. Chem. Soc. 2010, 132, 24–25. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, T.; Jeon, J.; Okuno, Y.; Chiliveri, S.C.; Clore, G.M. Submillisecond Freezing Permits Cryoprotectant-Free EPR Double Electron−Electron Resonance Spectroscopy. ChemPhysChem 2020, 21, 1224–1229. [Google Scholar] [CrossRef]
- Kubicki, D.J.; Rossini, A.J.; Purea, A.; Zagdoun, A.; Ouari, O.; Tordo, P.; Engelke, F.; Lesage, A.; Emsley, L. Amplifying Dynamic Nuclear Polarization of Frozen Solutions by Incorporating Dielectric Particles. J. Am. Chem. Soc. 2014, 136, 15711–15718. [Google Scholar] [CrossRef]
- Le, D.; Ziarelli, F.; Phan, T.N.T.; Mollica, G.; Thureau, P.; Aussenac, F.; Ouari, O.; Gigmes, D.; Tordo, P.; Viel, S. Up to 100% Improvement in Dynamic Nuclear Polarization Solid-State NMR Sensitivity Enhancement of Polymers by Removing Oxygen. Macromol. Rapid Commun. 2015, 36, 1416–1421. [Google Scholar] [CrossRef]
- Fahy, G.M.; MacFarlane, D.R.; Angell, C.A.; Meryman, H.T. Vitrification as an Approach to Cryopreservation. Cryobiology 1984, 21, 407–426. [Google Scholar] [CrossRef]
- Ringe, D.; Petsko, G.A. The “glass Transition” in Protein Dynamics: What It Is, Why It Occurs, and How to Exploit It. Biophys. Chem. 2003, 105, 667–680. [Google Scholar] [CrossRef]
- Copié, V.; McDermott, A.E.; Beshah, K.; Griffin, R.G.; Williams, J.C.; Spijker-Assink, M.; Gebhard, R.; Lugtenburg, J.; Herzfeld, J. Deuterium Solid-State Nuclear Magnetic Resonance Studies of Methyl Group Dynamics in Bacteriorhodopsin and Retinal Model Compounds: Evidence for a 6-s-Trans Chromophore in the Protein. Biochemistry 1994, 33, 3280–3286. [Google Scholar] [CrossRef] [PubMed]
- Beshah, K.; Olejniczak, E.T.; Griffin, R.G. Deuterium NMR Study of Methyl Group Dynamics in L-Alanine. J. Chem. Phys. 1987, 86, 4730–4736. [Google Scholar] [CrossRef]
- Gupta, R.; Lu, M.; Hou, G.; Caporini, M.A.; Rosay, M.; Maas, W.; Struppe, J.; Suiter, C.; Ahn, J.; Byeon, I.J.L.; et al. Dynamic Nuclear Polarization Enhanced MAS NMR Spectroscopy for Structural Analysis of HIV-1 Protein Assemblies. J. Phys. Chem. B 2016, 120, 329–339. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Zhang, H.; Lu, M.; Hou, G.; Caporini, M.; Rosay, M.; Maas, W.; Struppe, J.; Ahn, J.; Byeon, I.J.L.; et al. Dynamic Nuclear Polarization Magic-Angle Spinning Nuclear Magnetic Resonance Combined with Molecular Dynamics Simulations Permits Detection of Order and Disorder in Viral Assemblies. J. Phys. Chem. B 2019, 123, 5048–5058. [Google Scholar] [CrossRef] [PubMed]
- Kratochvílová, I.; Kopečná, O.; Bačíková, A.; Pagáčová, E.; Falková, I.; Follett, S.E.; Elliott, K.W.; Varga, K.; Golan, M.; Falk, M. Changes in Cryopreserved Cell Nuclei Serve as Indicators of Processes during Freezing and Thawing. Langmuir 2019, 35, 7496–7508. [Google Scholar] [CrossRef] [PubMed]
- Mazur, P.; Seki, S. Survival of Mouse Oocytes after Being Cooled in a Vitrification Solution to −196 °C at 95° to 70,000 °C/Min and Warmed at 610° to 118,000 °C/Min: A New Paradigm for Cryopreservation by Vitrification. Cryobiology 2011, 62, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.-Y.; Song, J.; Pas, J.; Meijer, L.H.H.; Han, S. DMSO Induces Dehydration near Lipid Membrane Surfaces. Biophys. J. 2015, 109, 330–339. [Google Scholar] [CrossRef] [Green Version]
- Williams-Smith, D.L.; Bray, R.C.; Barber, M.J.; Tsopanakis, A.D.; Vincent, S.P. Changes in Apparent PH on Freezing Aqueous Buffer Solutions and Their Relevance to Biochemical Electron-Paramagnetic-Resonance Spectroscopy. Biochem. J. 1977, 167, 593–600. [Google Scholar] [CrossRef]
- Brooke, D.; Movahed, N.; Bothner, B. Universal Buffers for Use in Biochemistry and Biophysical Experiments. AIMS Biophys. 2015, 2, 336–342. [Google Scholar] [CrossRef]
- Liao, S.Y.; Lee, M.; Wang, T.; Sergeyev, I.V.; Hong, M. Efficient DNP NMR of Membrane Proteins: Sample Preparation Protocols, Sensitivity, and Radical Location. J. Biomol. NMR 2016, 64, 223–237. [Google Scholar] [CrossRef]
- Hediger, S.; Lee, D.; Mentink-Vigier, F.; De Paëpe, G. MAS-DNP Enhancements: Hyperpolarization, Depolarization, and Absolute Sensitivity. eMagRes 2007, 105–116. [Google Scholar]
- Nagaraj, M.; Franks, T.W.; Saeidpour, S.; Schubeis, T.; Oschkinat, H.; Ritter, C.; van Rossum, B.-J. Surface Binding of TOTAPOL Assists Structural Investigations of Amyloid Fibrils by Dynamic Nuclear Polarization NMR Spectroscopy. ChemBioChem 2016, 17, 1308–1311. [Google Scholar] [CrossRef] [PubMed]
- Braide-Moncoeur, O.; Tran, N.T.; Long, J.R. Peptide-Based Synthetic Pulmonary Surfactant for the Treatment of Respiratory Distress Disorders. Curr. Opin. Chem. Biol. 2016, 32, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Antharam, V.C.; Elliott, D.W.; Mills, F.D.; Suzanne Farver, R.; Sternin, E.; Long, J.R. Penetration Depth of Surfactant Peptide KL4 into Membranes Is Determined by Fatty Acid Saturation. Biophys. J. 2009, 96, 4085–4098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, A.L.; Braide, O.; Mills, F.D.; Fanucci, G.E.; Long, J.R. Residue Specific Partitioning of KL4 into Phospholipid Bilayers. Biochim. Biophys. Acta Biomembr. 2014, 1838, 3212–3219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, J.R.; Mills, F.D.; Ganesh, O.K.; Antharam, V.C.; Farver, R.S. Partitioning, Dynamics, and Orientation of Lung Surfactant Peptide KL4 in Phospholipid Bilayers. Biochim. Biophys. Acta Biomembr. 2010, 1798, 216–222. [Google Scholar] [CrossRef] [Green Version]
- Mills, F.D.; Antharam, V.C.; Ganesh, O.K.; Elliott, D.W.; McNeill, S.A.; Long, J.R. The Helical Structure of Surfactant Peptide KL 4 When Bound to POPC: POPG Lipid Vesicles†. Biochemistry 2008, 47, 8292–8300. [Google Scholar] [CrossRef] [Green Version]
- Samuel-Landtiser, M.; Zachariah, C.; Williams, C.R.; Edison, A.S.; Long, J.R. Incorporation of Isotopically Enriched Amino Acids. Curr. Protoc. Protein Sci. 2007, 47, 26. [Google Scholar] [CrossRef]
- Salnikov, E.S.; Abel, S.; Karthikeyan, G.; Karoui, H.; Aussenac, F.; Tordo, P.; Bechinger, B.; Ouari, O. Dynamic Nuclear Polarization/Solid-State NMR Spectroscopy of Membrane Polypeptides: Free-Radical Optimization for Matrix-Free Lipid Bilayer Samples. ChemPhysChem 2017, 18, 2103–2113. [Google Scholar] [CrossRef]






| Sample | Buffer | Lipid Glycerol | Lipid C=O | Lipid CH2 | Lipid C=C | Peptide C=O |
|---|---|---|---|---|---|---|
| KL4/Lipids-1 | 36 | 22 | 13 | 13 | 14 | 13 |
| KL4/Lipids-2 | 23 | 15 | 8 | 8 | N.D. | 8 |
| Lipids only | 34 | 58 | 48 | 45 | 34 | -- |
| Sample | Buffer | Lipid Glycerol | Lipid C=O | Lipid CH2 | Lipid C=C | Peptide C=O |
|---|---|---|---|---|---|---|
| KL4/Lipids-1 | 2.7 ± 0.7 s | 2.2 ± 0.2 s | 3.5 ± 0.3 s | 3.3 ± 0.2 s | 4.2 ± 0.7 s | 3.6 ± 0.1 s |
| KL4/Lipids-2 | 1.4 ± 0.1 | 1.6 ± 0.1 | 2.3 ± 0.2 | 2.2 ± 0.1 | 2.0 ± 0.6 | 1.8 ± 0.1 |
| Lipids only | 3.2 ± 0.6 | 3.5 ± 0.4 | 8.8 ± 0.5 | 7.7 ± 0.2 | 8.1 ± 1.2 | -- |
| Sample | Buffer | Lipid Glycerol | Lipid C=O | Lipid CH2 | Peptide C=O |
|---|---|---|---|---|---|
| KL4/Lipids | 15 ± 4 ms | 4.9 ± 0.3 ms | 33 ± 3 ms | 8.0 ± 0.7 ms | 37.1 ± 1.5 ms |
| Lipids only | 9.9 ± 0.9 | 4.3 ± 0.2 | 35 ± 2 | 6.9 ± 0.5 | -- |
| Control | 82 ± 20 | 3.2 ± 0.9 | 73 ± 5 | 11 ± 4 | 64 ± 18 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, N.T.; Mentink-Vigier, F.; Long, J.R. Dynamic Nuclear Polarization of Biomembrane Assemblies. Biomolecules 2020, 10, 1246. https://doi.org/10.3390/biom10091246
Tran NT, Mentink-Vigier F, Long JR. Dynamic Nuclear Polarization of Biomembrane Assemblies. Biomolecules. 2020; 10(9):1246. https://doi.org/10.3390/biom10091246
Chicago/Turabian StyleTran, Nhi T., Frédéric Mentink-Vigier, and Joanna R. Long. 2020. "Dynamic Nuclear Polarization of Biomembrane Assemblies" Biomolecules 10, no. 9: 1246. https://doi.org/10.3390/biom10091246






