Bucky Ball Is a Novel Zebrafish Vasa ATPase Activator
Abstract
:1. Introduction
2. Materials and Methods
2.1. Zebrafish Handling and Maintenance
2.2. Microinjection
2.3. BiFC Assay
2.4. Live Cell Imaging
2.5. Quantification of Fluorescence Intensity
2.6. Recombinant Protein Expression of Buc-VBM
2.7. Recombinant Protein Expression of zfVasa (227–670) aa
2.8. Coomassie Staining
2.9. GST Pull-Down Assay
2.10. ATPase Assay
2.11. Circular Dichroism (CD) Spectroscopy
2.12. Bioinformatics Methods
2.13. Statistics
3. Results
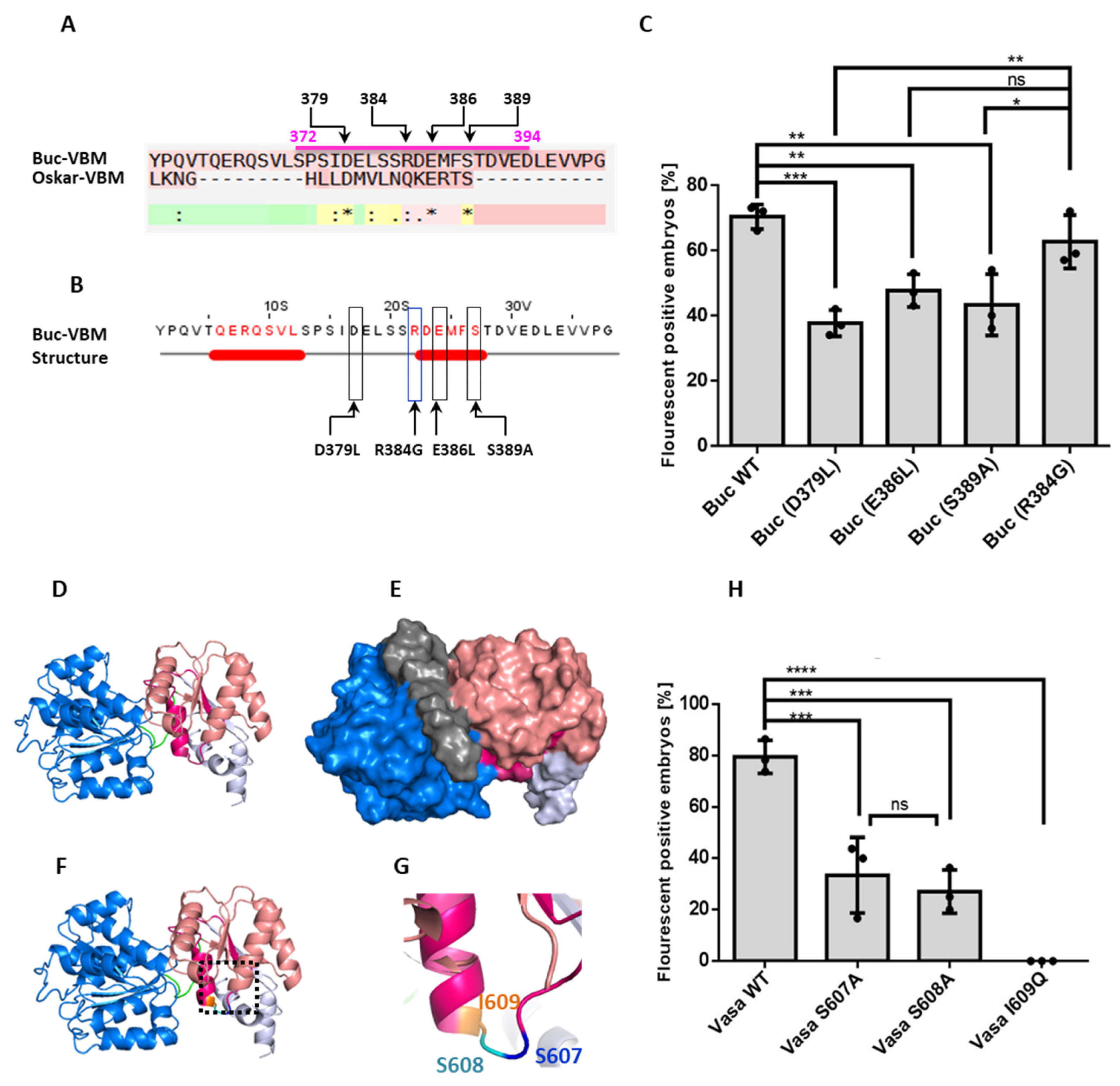
3.1. Identification of the zfVasa-Binding Motif in Buc (Buc-VBM)
3.2. Identification of the Buc Binding Motif in zfVasa (zfVasa-BBM)
3.3. Buc-VBM Adopts α-Helices from Its Disordered Nature
3.4. Homology Modeling for zfVasa
3.5. Identification of Amino Acids in the Buc-VBM, Which Are Required for zfVasa Interaction
3.6. Amino Acids in zfVasa-BBM Required for Buc Interaction
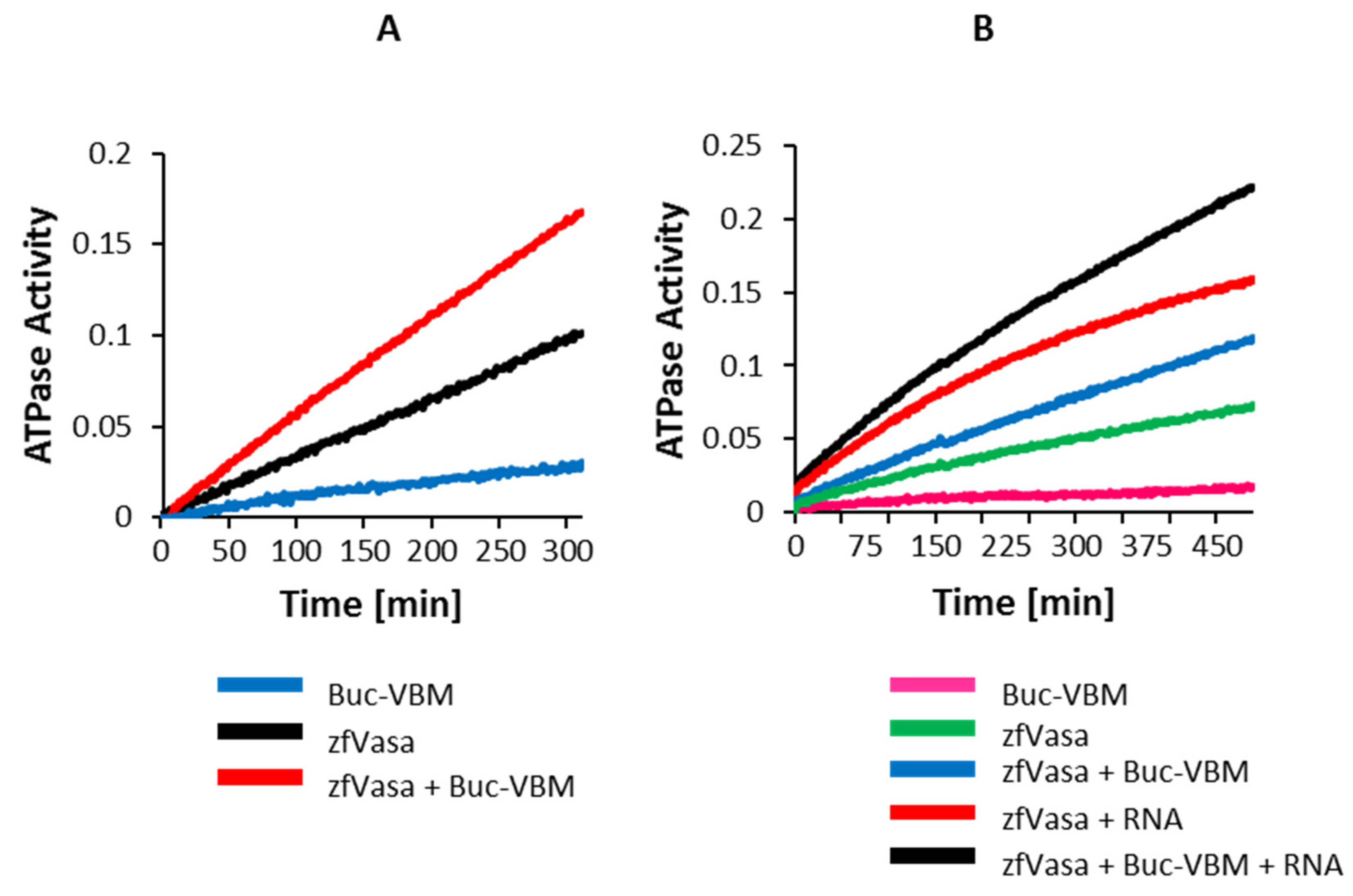
3.7. The Buc-VBM Is a Novel Activator of zfVasa ATPase Activity
3.8. The Buc-VBM and the zfVasa-BBM Inhibit PGC Formation in Zebrafish
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Santos, A.C.; Lehmann, R. Germ cell specification and migration in Drosophila and beyond. Curr. Biol. 2004, 14, 578–589. [Google Scholar] [CrossRef] [Green Version]
- Marlow, F.L. Maternal Control of Development in Vertebrates; Morgan & Claypool Life Sciences: San Rafael, CA, USA, 2010; Volume 1, ISBN 9781615040513. [Google Scholar]
- Jostes, S.; Schorle, H. Signals and transcription factors for specification of human germ cells. Stem Cell Investig. 2018, 5, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Krishnakumar, P.; Dosch, R. Germ Cell Specification: The Evolution of a Recipe to Make Germ Cells. Germ Cell 2018, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Carr, R.M.; Oranu, A.; Khungar, V. piRNA Biogenesis in Drosophila Melanogaster. Physiol. Behav. 2016, 176, 139–148. [Google Scholar] [CrossRef]
- Hartung, O.; Forbes, M.M.; Marlow, F.L. Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol. Reprod. Dev. 2015, 81, 946–961. [Google Scholar] [CrossRef] [Green Version]
- Gustafson, E.A.; Wessel, G.M. Vasa genes: Emerging roles in the germ line and in multipotent cells. Bioessays 2010, 32, 626–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitou, M.; Yamaji, M. Germ cell specification in mice: Signaling, transcription regulation, and epigenetic consequences. Reproduction 2010, 139, 931–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Orazio, F.M.; Balwierz, P.J.; González, A.J.; Guo, Y.; Hernández-Rodríguez, B.; Wheatley, L.; Jasiulewicz, A.; Hadzhiev, Y.; Vaquerizas, J.M.; Cairns, B.; et al. Germ cell differentiation requires Tdrd7-dependent chromatin and transcriptome reprogramming marked by germ plasm relocalization. Dev. Cell 2021, 56, 641–656. [Google Scholar] [CrossRef] [PubMed]
- Ikenishi, K.; Kotani, M.; Tanabe, K. Ultrastructural changes associated with UV irradiation in the “germinal plasm” of Xenopus laevis. Dev. Biol. 1974, 36, 155–168. [Google Scholar] [CrossRef]
- Smith, L.D. The role of a “germinal plasm” in the formation of primordial germ cells in Rana pipiens. Dev. Biol. 1966, 14, 330–347. [Google Scholar] [CrossRef]
- Tanabe, K.; Kotani, M. Relationship between the amount of the “germinal plasm” and the number of primordial germ cells in Xenopus laevis. J. Embryol. Exp. Morphol. 1974, 31, 89–98. [Google Scholar]
- Wakahara, M. Partial characterization of “primordial germ cell forming activity” localized in vegetal pole cytoplasm in anuran eggs. J. Embryol. Exp. Morphol. 1977, 39, 221–233. [Google Scholar] [PubMed]
- Ikenishi, K.; Sakiko, N.; Okuda, T. Direct Evidence for the Presence of Germ Cell Determinant in Vegetal Pole Cytoplasm of Xenopus laevis and in a Subcellular Fraction of It: (Xenopus laevis/germ cell determinant/germ plasm/PGC induction). Dev. Growth Differ. 1986, 28, 563–568. [Google Scholar] [CrossRef]
- Illmensee, K.; Mahowald, A.P. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc. Natl. Acad. Sci. USA 1974, 71, 1016–1020. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, R.; Nüsslein-Volhard, C. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in drosophila. Cell 1986, 47, 141–152. [Google Scholar] [CrossRef]
- Ephrussi, A.; Lehmann, R. Induction of germ cell fate by oskar. Nature 1992, 358, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Wilson, J.E.; Macdonald, P.M. Overexpression of oskar directs ectopic activation of nanos and presumptive pole cell formation in Drosophila embryos. Cell 1992, 70, 849–859. [Google Scholar] [CrossRef]
- Ephrussi, A.; Dickinson, L.K.; Lehmann, R. Oskar Organizes the Germ Plasm and Directs Localization of the Posterior Determinant Nanos. Cell 1991, 66, 37–50. [Google Scholar] [CrossRef]
- Dosch, R.; Wagner, D.S.; Mintzer, K.A.; Runke, G.; Wiemelt, A.P.; Mullins, M.C. Maternal control of vertebrate development before the midblastula transition: Mutants from the zebrafish I. Dev. Cell 2004, 6, 771–780. [Google Scholar] [CrossRef] [Green Version]
- Wagner, D.S.; Dosch, R.; Mintzer, K.A.; Wiemelt, A.P.; Mullins, M.C. Maternal control of development at the midblastula transition and beyond: Mutants from the zebrafish II. Dev. Cell 2004, 6, 781–790. [Google Scholar] [CrossRef] [Green Version]
- Bontems, F.; Stein, A.; Marlow, F.; Lyautey, J.; Gupta, T.; Mullins, M.C.; Dosch, R. Bucky Ball Organizes Germ Plasm Assembly in Zebrafish. Curr. Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef] [Green Version]
- Krishnakumar, P.; Riemer, S.; Perera, R.; Lingner, T.; Goloborodko, A.; Khalifa, H.; Bontems, F.; Kaufholz, F.; El-Brolosy, M.A.; Dosch, R. Functional equivalence of germ plasm organizers. PLoS Genet. 2018, 14, e1007696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeske, M.; Müller, C.W.; Ephrussi, A. The LOTUS domain is a conserved DEAD-box RNA helicase regulator essential for the recruitment of Vasa to the germ plasm and nuage. Genes Dev. 2017, 31, 939–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeske, M.; Bordi, M.; Glatt, S.; Müller, S.; Rybin, V.; Müller, C.W.; Ephrussi, A. The crystal structure of the Drosophila germline inducer Oskar identifies two domains with distinct Vasa Helicase- and RNA-binding activities. Cell Rep. 2015, 12, 587–598. [Google Scholar] [CrossRef] [Green Version]
- Heim, A.E.; Hartung, O.; Rothhämel, S.; Ferreira, E.; Jenny, A.; Marlow, F.L. Oocyte polarity requires a Bucky ball-dependent feedback amplification loop. Development 2014, 141, 842–854. [Google Scholar] [CrossRef] [Green Version]
- Roovers, E.F.; Kaaij, L.J.T.; Redl, S.; Bronkhorst, A.W.; Wiebrands, K.; de Jesus Domingues, A.M.; Huang, H.Y.; Han, C.T.; Riemer, S.; Dosch, R.; et al. Tdrd6a Regulates the Aggregation of Buc into Functional Subcellular Compartments that Drive Germ Cell Specification. Dev. Cell 2018, 46, 285–301.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, F.; Miao, R.; Xiao, R.; Mei, J. m6A reader Igf2bp3 enables germ plasm assembly by m6A-dependent regulation of gene expression in zebrafish. Sci. Bull. 2021, 66, 1119–1128. [Google Scholar] [CrossRef]
- Perera, R.P.; Dosch, R. Chapter 24 with Bimolecular Fluorescent Complementation; Springer: New York, NY, USA; Humana: New York, NY, USA, 2021; Volume 2218, pp. 303–317. [Google Scholar] [CrossRef]
- Riemer, S.; Bontems, F.; Krishnakumar, P.; Gömann, J.; Dosch, R. A functional Bucky ball-GFP transgene visualizes germ plasm in living zebrafish. Gene Expr. Patterns 2015, 18, 44–52. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio Rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–41. [Google Scholar] [CrossRef]
- Kiianitsa, K.; Solinger, J.A.; Heyer, W.D. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal. Biochem. 2003, 321, 266–271. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Tommaso, P.; Moretti, S.; Xenarios, I.; Orobitg, M.; Montanyola, A.; Chang, J.M.; Taly, J.F.; Notredame, C. T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011, 39, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Kerppola, T.K. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 2006, 7, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Kerppola, T.K. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev. Biophys. 2008, 37, 465–487. [Google Scholar] [CrossRef] [Green Version]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, D61–D65. Available online: http://academic.oup.com/nar/article/35/suppl_1/D61/1099759?login=true (accessed on 20 July 2019). [CrossRef] [Green Version]
- Karimi, K.; Fortriede, J.D.; Lotay, V.S.; Burns, K.A.; Wang, D.Z.; Fisher, M.E.; Pells, T.J.; James-Zorn, C.; Wang, Y.; Ponferrada, V.G.; et al. Xenbase: A genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res. 2018, 46, D861–D868. Available online: http://academic.oup.com/nar/article/46/D1/D861/4559118 (accessed on 15 August 2019). [CrossRef]
- Ashok Kumar, T. CFSSP: Chou and Fasman Secondary Structure Prediction server. Wide Spectr. 2013, 1, 15–19. [Google Scholar] [CrossRef]
- Singh, H.; Singh, S.; Singh Raghava, G.P. Peptide Secondary Structure Prediction using Evolutionary Information. bioRxiv 2019, 558791. [Google Scholar] [CrossRef] [Green Version]
- Cole, C.; Barber, J.D.; Barton, G.J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008, 36, 197–201. [Google Scholar] [CrossRef] [Green Version]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef]
- Sengoku, T.; Nureki, O.; Nakamura, A.; Kobayashi, S.; Yokoyama, S. Structural Basis for RNA Unwinding by the DEAD-Box Protein Drosophila Vasa. Cell 2006, 125, 287–300. [Google Scholar] [CrossRef] [Green Version]
- Norma, J. Greenfield Using circular dichroism spectra to estimate protein secondary structure. ProQuest Diss. Theses 2012, 1, 218. [Google Scholar] [CrossRef]
- Laureto, P.P.; De Donadi, M.; Scaramella, E.; Frare, E.; Fontana, A. Trifluoroethanol-assisted protein folding: Fragment 53-103 of bovine α-lactalbumin. Biosystems 2001, 1548, 29–37. [Google Scholar]
- Walgers, R.; Lee, T.C.; Cammers-Goodwin, A. An indirect chaotropic mechanism for the stabilization of helix conformation of peptides in aqueous trifluoroethanol and hexafluoro-2- propanol. J. Am. Chem. Soc. 1998, 120, 5073–5079. [Google Scholar] [CrossRef]
- Kaczka, P.; Winiewska, M.; Zhukov, I.; Rempoła, B.; Bolewska, K.; Łozinski, T.; Ejchart, A.; Poznańska, A.; Wierzchowski, K.L.; Poznański, J. The TFE-induced transient native-like structure of the intrinsically disordered σ_4^704 domain of Escherichia coli RNA polymerase. Eur. Biophys. J. 2014, 43, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Kohji Ikenishi, T.S.T. Involvement of the protein of Xenopus vasa homolog (Xenopus vasa-like gene 1, XVLG1) in the differentiation of primordial germ cells. Dev. Growth Differ. 1997, 39, 625–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimaoka, K.; Mukumoto, Y.; Tanigawa, Y.; Komiya, T. Xenopus Vasa Homolog XVLG1 is Essential for Migration and Survival of Primordial Germ Cells. Zoolog. Sci. 2017, 34, 93–104. [Google Scholar] [CrossRef]
- Cipriani, P.G.; Bay, O.; Zinno, J.; Gutwein, M.; Gan, H.H.; Mayya, K.; Chung, G.; Chen, J.; Fahs, H.; Guan, Y.; et al. Novel LOTUS-domain proteins are organizational hubs that recruit C. elegans Vasa to germ granules. eLife 2021, 10, 1–41. [Google Scholar] [CrossRef]
- Perera R and RD. Unpublished work. 2021.
- Marnik, E.A.; Fuqua, J.H.; Sharp, C.S.; Rochester, J.D.; Xu, E.L.; Holbrook, S.E.; Updike, D.L. Germline maintenance through the multifaceted activities of GLH/Vasa in caenorhabditis elegans P Granules. Genetics 2019, 213, 923–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boke, E.; Ruer, M.; Wühr, M.; Coughlin, M.; Lemaitre, R.; Gygi, S.P.; Alberti, S.; Drechsel, D.; Hyman, A.A.; Mitchison, T.J. Amyloid-like Self-Assembly of a Cellular Compartment. Cell 2016, 166, 637–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perera, R.P.; Shaikhqasem, A.; Rostam, N.; Dickmanns, A.; Ficner, R.; Tittmann, K.; Dosch, R. Bucky Ball Is a Novel Zebrafish Vasa ATPase Activator. Biomolecules 2021, 11, 1507. https://doi.org/10.3390/biom11101507
Perera RP, Shaikhqasem A, Rostam N, Dickmanns A, Ficner R, Tittmann K, Dosch R. Bucky Ball Is a Novel Zebrafish Vasa ATPase Activator. Biomolecules. 2021; 11(10):1507. https://doi.org/10.3390/biom11101507
Chicago/Turabian StylePerera, Roshan Priyarangana, Alaa Shaikhqasem, Nadia Rostam, Achim Dickmanns, Ralf Ficner, Kai Tittmann, and Roland Dosch. 2021. "Bucky Ball Is a Novel Zebrafish Vasa ATPase Activator" Biomolecules 11, no. 10: 1507. https://doi.org/10.3390/biom11101507





