Urm1: A Non-Canonical UBL
Abstract
:1. Introduction
2. The Urm1 Pathway
3. Cellular Phenotypes
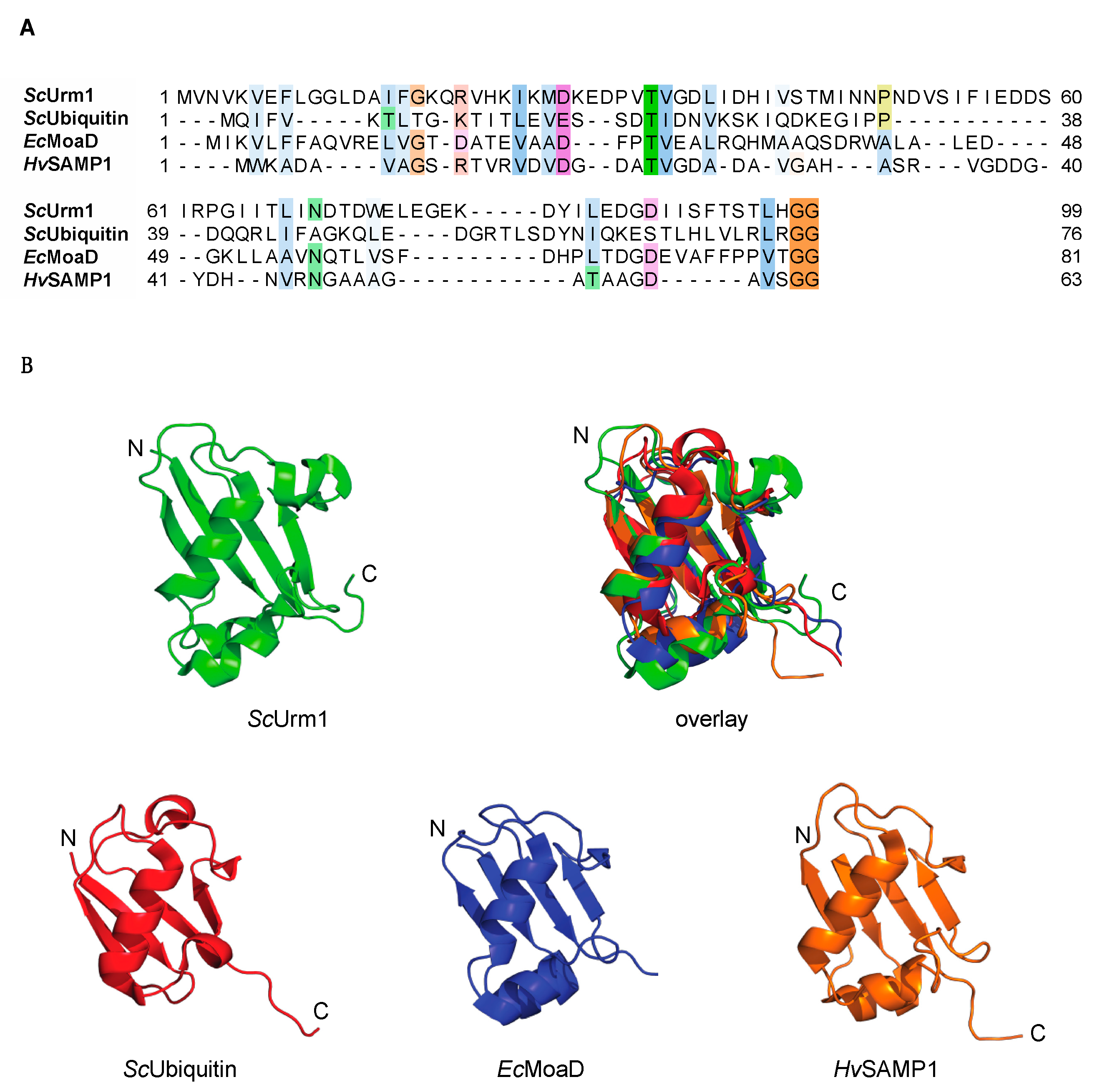
4. The Urm1-Uba4 System
5. Urm1 Conjugation
6. Molecular Phenotypes and Implications for Higher Organisms
7. Outlook
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taylor, S.V.; Kelleher, N.L.; Kinsland, C.; Chiu, H.-J.; Costello, C.A.; Backstrom, A.D.; McLafferty, F.W.; Begley, T.P. Thiamin Biosynthesis in Escherichia coli: Identification of ThiS Thiocarboxylate as the Immediate Sulfur Donor in the Thiazole Formation. J. Biol. Chem. 1998, 273, 16555–16560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furukawa, K.; Mizushima, N.; Noda, T.; Ohsumi, Y. A Protein Conjugation System in Yeast with Homology to Biosynthetic Enzyme Reaction of Prokaryotes. J. Biol. Chem. 2000, 275, 7462–7465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Zhang, J.; Wang, L.; Zhou, J.; Huang, H.; Wu, J.; Zhong, Y.; Shi, Y. Solution Structure of Urm1 and Its Implications for the Origin of Protein Modifiers. Proc. Natl. Acad. Sci. USA 2006, 103, 11625–11630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burroughs, A.M.; Iyer, L.M.; Aravind, L. Natural History of the E1-like Superfamily: Implication for Adenylation, Sulfur Transfer, and Ubiquitin Conjugation. Proteins Struct. Funct. Bioinf. 2009, 75, 895–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, J.; Kim, B.-H.; Grishin, N.V. PROMALS3D: A Tool for Multiple Protein Sequence and Structure Alignments. Nucleic Acids Res. 2008, 36, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-a Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger, LLC. The PyMOL Molecular Graphics System; Version 1.8; Schrödinger, LLC.: New York, NY, USA, 2015. [Google Scholar]
- Humbard, M.A.; Miranda, H.V.; Lim, J.-M.; Krause, D.J.; Pritz, J.R.; Zhou, G.; Chen, S.; Wells, L.; Maupin-Furlow, J.A. Ubiquitin-like Small Archaeal Modifier Proteins (SAMPs) in Haloferax Volcanii. Nature 2010, 463, 54–60. [Google Scholar] [CrossRef] [Green Version]
- Ranjan, N.; Damberger, F.F.; Sutter, M.; Allain, F.H.-T.; Weber-Ban, E. Solution Structure and Activation Mechanism of Ubiquitin-Like Small Archaeal Modifier Proteins. J. Mol. Biol. 2011, 405, 1040–1055. [Google Scholar] [CrossRef]
- Miranda, H.V.; Nembhard, N.; Su, D.; Hepowit, N.; Krause, D.J.; Pritz, J.R.; Phillips, C.; Soll, D.; Maupin-Furlow, J.A. E1- and Ubiquitin-like Proteins Provide a Direct Link between Protein Conjugation and Sulfur Transfer in Archaea. Proc. Natl. Acad. Sci. USA 2011, 108, 4417–4422. [Google Scholar] [CrossRef] [Green Version]
- Hepowit, N.L.; de Vera, I.M.S.; Cao, S.; Fu, X.; Wu, Y.; Uthandi, S.; Chavarria, N.E.; Englert, M.; Su, D.; Söll, D.; et al. Mechanistic Insight into Protein Modification and Sulfur Mobilization Activities of Noncanonical E1 and Associated Ubiquitin-like Proteins of Archaea. FEBS J. 2016, 283, 3567–3586. [Google Scholar] [CrossRef] [Green Version]
- El Yacoubi, B.; Bailly, M.; de Crécy-Lagard, V. Biosynthesis and Function of Posttranscriptional Modifications of Transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Helm, M.; Alfonzo, J.D. Posttranscriptional RNA Modifications: Playing Metabolic Games in a Cell’s Chemical Legoland. Chem. Biol. 2014, 21, 174–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansson, M.J.O.; Esberg, A.; Huang, B.; Björk, G.R.; Byström, A.S. Eukaryotic Wobble Uridine Modifications Promote a Functionally Redundant Decoding System. Mol. Cell. Biol. 2008, 28, 3301–3312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, M.; Kataoka, N.; Miyauchi, K.; Ohe, K.; Iida, K.; Yoshida, S.; Nojima, T.; Okuno, Y.; Onogi, H.; Usui, T.; et al. Rectifier of Aberrant mRNA Splicing Recovers tRNA Modification in Familial Dysautonomia. Proc. Natl. Acad. Sci. USA 2015, 112, 2764–2769. [Google Scholar] [CrossRef] [Green Version]
- Kalhor, H.R.; Clarke, S. Novel Methyltransferase for Modified Uridine Residues at the Wobble Position of tRNA. Mol. Cell. Biol. 2003, 23, 9283–9292. [Google Scholar] [CrossRef] [Green Version]
- Dauden, M.I.; Jaciuk, M.; Müller, C.W.; Glatt, S. Structural Asymmetry in the Eukaryotic Elongator Complex. FEBS Lett. 2018, 592, 502–515. [Google Scholar] [CrossRef] [Green Version]
- Létoquart, J.; van Tran, N.; Caroline, V.; Aleksandrov, A.; Lazar, N.; van Tilbeurgh, H.; Liger, D.; Graille, M. Insights into Molecular Plasticity in Protein Complexes from Trm9-Trm112 tRNA Modifying Enzyme Crystal Structure. Nucleic Acids Res. 2015, 43, 10989–11002. [Google Scholar] [CrossRef]
- Huang, B.; Lu, J.; Bystrom, A.S. A Genome-Wide Screen Identifies Genes Required for Formation of the Wobble Nucleoside 5-Methoxycarbonylmethyl-2-Thiouridine in Saccharomyces Cerevisiae. RNA 2008, 14, 2183–2194. [Google Scholar] [CrossRef] [Green Version]
- Adam, A.C.; Bornhövd, C.; Prokisch, H.; Neupert, W.; Hell, K. The Nfs1 Interacting Protein Isd11 Has an Essential Role in Fe/S Cluster Biogenesis in Mitochondria. EMBO J. 2006, 25, 174–183. [Google Scholar] [CrossRef] [Green Version]
- Nakai, Y.; Umeda, N.; Suzuki, T.; Nakai, M.; Hayashi, H.; Watanabe, K.; Kagamiyama, H. Yeast Nfs1p Is Involved in Thio-Modification of Both Mitochondrial and Cytoplasmic tRNAs. J. Biol. Chem. 2004, 279, 12363–12368. [Google Scholar] [CrossRef] [Green Version]
- Mühlenhoff, U.; Balk, J.; Richhardt, N.; Kaiser, J.T.; Sipos, K.; Kispal, G.; Lill, R. Functional Characterization of the Eukaryotic Cysteine Desulfurase Nfs1p from Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 36906–36915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noma, A.; Sakaguchi, Y.; Suzuki, T. Mechanistic Characterization of the Sulfur-Relay System for Eukaryotic 2-Thiouridine Biogenesis at tRNA Wobble Positions. Nucleic Acids Res. 2009, 37, 1335–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fräsdorf, B.; Radon, C.; Leimkühler, S. Characterization and Interaction Studies of Two Isoforms of the Dual Localized 3-Mercaptopyruvate Sulfurtransferase TUM1 from Humans. J. Biol. Chem. 2014, 289, 34543–34556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgenstern, M.; Stiller, S.B.; Lübbert, P.; Peikert, C.D.; Dannenmaier, S.; Drepper, F.; Weill, U.; Höß, P.; Feuerstein, R.; Gebert, M.; et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017, 19, 2836–2852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marelja, Z.; Stöcklein, W.; Nimtz, M.; Leimkühler, S. A Novel Role for Human Nfs1 in the Cytoplasm: Nfs1 Acts as a Sulfur Donor for MOCS3, a Protein Involved in Molybdenum Cofactor Biosynthesis. J. Biol. Chem. 2008, 283, 25178–25185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marelja, Z.; Mullick Chowdhury, M.; Dosche, C.; Hille, C.; Baumann, O.; Löhmannsröben, H.-G.; Leimkühler, S. The L-Cysteine Desulfurase NFS1 Is Localized in the Cytosol Where It Provides the Sulfur for Molybdenum Cofactor Biosynthesis in Humans. PLoS ONE 2013, 8, e60869. [Google Scholar] [CrossRef] [Green Version]
- Leidel, S.; Pedrioli, P.G.A.; Bucher, T.; Brost, R.; Costanzo, M.; Schmidt, A.; Aebersold, R.; Boone, C.; Hofmann, K.; Peter, M. Ubiquitin-Related Modifier Urm1 Acts as a Sulphur Carrier in Thiolation of Eukaryotic transferRNA. Nature 2009, 458, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, J.; Chowdhury, M.M.; Hänzelmann, P.; Nimtz, M.; Lee, E.-Y.; Schindelin, H.; Leimkühler, S. The Sulfurtransferase Activity of Uba4 Presents a Link between Ubiquitin-like Protein Conjugation and Activation of Sulfur Carrier Proteins. Biochemistry 2008, 47, 6479–6489. [Google Scholar] [CrossRef]
- Liu, Y.; Vinyard, D.J.; Reesbeck, M.E.; Suzuki, T.; Manakongtreecheep, K.; Holland, P.L.; Brudvig, G.W.; Söll, D. A [3Fe-4S] Cluster Is Required for tRNA Thiolation in Archaea and Eukaryotes. Proc. Natl. Acad. Sci. USA 2016, 113, 12703–12708. [Google Scholar] [CrossRef] [Green Version]
- Arragain, S.; Bimai, O.; Legrand, P.; Caillat, S.; Ravanat, J.-L.; Touati, N.; Binet, L.; Atta, M.; Fontecave, M.; Golinelli-Pimpaneau, B. Nonredox Thiolation in tRNA Occurring via Sulfur Activation by a [4Fe-4S] Cluster. Proc. Natl. Acad. Sci. USA 2017, 114, 7355–7360. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Asai, S.; Narai, S.; Nambu, S.; Omura, N.; Sakaguchi, Y.; Suzuki, T.; Ikeda-Saito, M.; Watanabe, K.; Yao, M.; et al. Biochemical and Structural Characterization of Oxygen-Sensitive 2-Thiouridine Synthesis Catalyzed by an Iron-Sulfur Protein TtuA. Proc. Natl. Acad. Sci. USA 2017, 114, 4954–4959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakai, Y.; Nakai, M.; Lill, R.; Suzuki, T.; Hayashi, H. Thio Modification of Yeast Cytosolic tRNA Is an Iron-Sulfur Protein-Dependent Pathway. Mol. Cell. Biol. 2007, 27, 2841–2847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, C.M.; Christman, G.D.; Snellinger, A.M.; Johnston, M.V.; Mueller, E.G. Direct Evidence for Enzyme Persulfide and Disulfide Intermediates during 4-Thiouridine Biosynthesis. Chem. Commun. 2006, 3104. [Google Scholar] [CrossRef] [PubMed]
- Kambampati, R.; Lauhon, C.T. Evidence for the Transfer of Sulfane Sulfur from IscS to ThiI during the in Vitro Biosynthesis of 4-Thiouridine in Escherichia coli tRNA. J. Biol. Chem. 2000, 275, 10727–10730. [Google Scholar] [CrossRef] [Green Version]
- Palenchar, P.M.; Buck, C.J.; Cheng, H.; Larson, T.J.; Mueller, E.G. Evidence That ThiI, an Enzyme Shared between Thiamin and 4-Thiouridine Biosynthesis, May Be a Sulfurtransferase That Proceeds through a Persulfide Intermediate. J. Biol. Chem. 2000, 275, 8283–8286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterman, D.G.; Ortiz-Lombardía, M.; Fogg, M.J.; Koonin, E.V.; Antson, A.A. Crystal Structure of Bacillus Anthracis ThiI, a tRNA-Modifying Enzyme Containing the Predicted RNA-Binding THUMP Domain. J. Mol. Biol. 2006, 356, 97–110. [Google Scholar] [CrossRef]
- Damon, J.R.; Pincus, D.; Ploegh, H.L. tRNA Thiolation Links Translation to Stress Responses in Saccharomyces cerevisiae. Mol. Biol. Cell 2015, 26, 270–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyagi, K.; Pedrioli, P.G.A. Protein Degradation and Dynamic tRNA Thiolation Fine-Tune Translation at Elevated Temperatures. Nucleic Acids Res. 2015, 43, 4701–4712. [Google Scholar] [CrossRef] [Green Version]
- Alings, F.; Sarin, L.P.; Fufezan, C.; Drexler, H.C.A.; Leidel, S.A. An Evolutionary Approach Uncovers a Diverse Response of tRNA 2-Thiolation to Elevated Temperatures in Yeast. RNA 2015, 21, 202–212. [Google Scholar] [CrossRef] [Green Version]
- Goehring, A.S.; Rivers, D.M.; Sprague, G.F. Urmylation: A Ubiquitin-like Pathway That Functions during Invasive Growth and Budding in Yeast. Mol. Biol. Cell 2003, 14, 4329–4341. [Google Scholar] [CrossRef] [Green Version]
- Goehring, A.S.; Rivers, D.M.; Sprague, G.F. Attachment of the Ubiquitin-Related Protein Urm1p to the Antioxidant Protein Ahp1p. Eukaryot. Cell 2003, 2, 930–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthies, A.; Nimtz, M.; Leimkühler, S. Molybdenum Cofactor Biosynthesis in Humans: Identification of a Persulfide Group in the Rhodanese-like Domain of MOCS3 by Mass Spectrometry. Biochemistry 2005, 44, 7912–7920. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.M.; Dosche, C.; Löhmannsröben, H.-G.; Leimkühler, S. Dual Role of the Molybdenum Cofactor Biosynthesis Protein MOCS3 in tRNA Thiolation and Molybdenum Cofactor Biosynthesis in Humans. J. Biol. Chem. 2012, 287, 17297–17307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendel, R.R. The Molybdenum Cofactor. J. Biol. Chem. 2013, 288, 13165–13172. [Google Scholar] [CrossRef] [Green Version]
- Leimkühler, S.; Bühning, M.; Beilschmidt, L. Shared Sulfur Mobilization Routes for tRNA Thiolation and Molybdenum Cofactor Biosynthesis in Prokaryotes and Eukaryotes. Biomolecules 2017, 7, 5. [Google Scholar] [CrossRef] [Green Version]
- Esberg, A.; Huang, B.; Johansson, M.J.O.; Byström, A.S. Elevated Levels of Two tRNA Species Bypass the Requirement for Elongator Complex in Transcription and Exocytosis. Mol. Cell 2006, 24, 139–148. [Google Scholar] [CrossRef]
- Bjork, G.R.; Huang, B.; Persson, O.P.; Bystrom, A.S. A Conserved Modified Wobble Nucleoside (Mcm5s2U) in Lysyl-tRNA Is Required for Viability in Yeast. RNA 2007, 13, 1245–1255. [Google Scholar] [CrossRef] [Green Version]
- Termathe, M.; Leidel, S.A. The Uba4 Domain Interplay Is Mediated via a Thioester That Is Critical for tRNA Thiolation through Urm1 Thiocarboxylation. Nucleic Acids Res. 2018, 46, 5171–5181. [Google Scholar] [CrossRef] [Green Version]
- Clayden, J.; Greeves, N.; Warren, S.; Wothers, P. Organic Chemistry, 1st ed.; Oxford University Press: New York, NY, USA, 2001; ISBN 978-0-19-850346-0. [Google Scholar]
- Pickart, C.M.; Kim, A. Substrate Properties of Site-Specific Mutant Ubiquitin Protein (G76A) Reveal Unexpected Mechanistic Features of Ubiquitin-Activating Enzyme (E1). J. Biol. Chem. 1994, 269, 7115–7123. [Google Scholar] [CrossRef]
- Huang, D.T.; Hunt, H.W.; Zhuang, M.; Ohi, M.D.; Holton, J.M.; Schulman, B.A. Basis for a Ubiquitin-like Protein Thioester Switch Toggling E1–E2 Affinity. Nature 2007, 445, 394–398. [Google Scholar] [CrossRef]
- Olsen, S.K.; Capili, A.D.; Lu, X.; Tan, D.S.; Lima, C.D. Active Site Remodelling Accompanies Thioester Bond Formation in the SUMO E1. Nature 2010, 463, 906–912. [Google Scholar] [CrossRef] [Green Version]
- Schulman, B.A.; Wade Harper, J. Ubiquitin-like Protein Activation by E1 Enzymes: The Apex for Downstream Signalling Pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.; Schindelin, H. Structural Insights into E1-Catalyzed Ubiquitin Activation and Transfer to Conjugating Enzymes. Cell 2008, 134, 268–278. [Google Scholar] [CrossRef] [Green Version]
- Leimkühler, S.; Wuebbens, M.M.; Rajagopalan, K.V. Characterization of Escherichia coli MoeB and Its Involvement in the Activation of Molybdopterin Synthase for the Biosynthesis of the Molybdenum Cofactor. J. Biol. Chem. 2001, 276, 34695–34701. [Google Scholar] [CrossRef] [Green Version]
- Hochstrasser, M. Evolution and Function of Ubiquitin-like Protein-Conjugation Systems. Nat. Cell Biol. 2000, 2, E153–E157. [Google Scholar] [CrossRef]
- Zhang, W.; Urban, A.; Mihara, H.; Leimkühler, S.; Kurihara, T.; Esaki, N. IscS Functions as a Primary Sulfur-Donating Enzyme by Interacting Specifically with MoeB and MoaD in the Biosynthesis of Molybdopterin in Escherichia coli. J. Biol. Chem. 2010, 285, 2302–2308. [Google Scholar] [CrossRef] [Green Version]
- Begley, T.P.; Ealick, S.E.; McLafferty, F.W. Thiamin Biosynthesis: Still Yielding Fascinating Biological Chemistry. Biochem. Soc. Trans. 2012, 40, 555–560. [Google Scholar] [CrossRef] [Green Version]
- Xi, J.; Ge, Y.; Kinsland, C.; McLafferty, F.W.; Begley, T.P. Biosynthesis of the Thiazole Moiety of Thiamin in Escherichia coli: Identification of an Acyldisulfide-Linked Protein-Protein Conjugate That Is Functionally Analogous to the Ubiquitin/E1 Complex. Proc. Natl. Acad. Sci. USA 2001, 98, 8513–8518. [Google Scholar] [CrossRef] [Green Version]
- Buetow, L.; Huang, D.T. Structural Insights into the Catalysis and Regulation of E3 Ubiquitin Ligases. Nat. Rev. Mol. Cell Biol. 2016, 17, 626–642. [Google Scholar] [CrossRef] [Green Version]
- Kerscher, O.; Felberbaum, R.; Hochstrasser, M. Modification of Proteins by Ubiquitin and Ubiquitin-Like Proteins. Annu. Rev. Cell Dev. Biol. 2006, 22, 159–180. [Google Scholar] [CrossRef] [Green Version]
- Hochstrasser, M. Biochemical Functions of Ubiquitin and Ubiquitin-like Protein Conjugation. In Protein Degradation Series; Mayer, R.J., Ciechanover, A.J., Rechsteiner, M., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2007; pp. 249–278. ISBN 978-3-527-61932-0. [Google Scholar]
- Pabis, M.; Termathe, M.; Ravichandran, K.E.; Kienast, S.D.; Krutyhołowa, R.; Sokołowski, M.; Jankowska, U.; Grudnik, P.; Leidel, S.A.; Glatt, S. Molecular Basis for the Bifunctional Uba4–Urm1 Sulfur-relay System in tRNA Thiolation and Ubiquitin-like Conjugation. EMBO J. 2020, 39. [Google Scholar] [CrossRef]
- Wang, J.; Chen, Y. Role of the Zn2+ Motif of E1 in SUMO Adenylation. J. Biol. Chem. 2010, 285, 23732–23738. [Google Scholar] [CrossRef] [Green Version]
- Park, S.G.; Cha, M.-K.; Jeong, W.; Kim, I.-H. Distinct Physiological Functions of Thiol Peroxidase Isoenzymes in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 5723–5732. [Google Scholar] [CrossRef] [Green Version]
- Van der Veen, A.G.; Schorpp, K.; Schlieker, C.; Buti, L.; Damon, J.R.; Spooner, E.; Ploegh, H.L.; Jentsch, S. Role of the Ubiquitin-like Protein Urm1 as a Noncanonical Lysine-Directed Protein Modifier. Proc. Natl. Acad. Sci. USA 2011, 108, 1763–1770. [Google Scholar] [CrossRef] [Green Version]
- Khoshnood, B.; Dacklin, I.; Grabbe, C. Urm1: An Essential Regulator of JNK Signaling and Oxidative Stress in Drosophila melanogaster. Cell. Mol. Life Sci. 2016, 73, 1939–1954. [Google Scholar] [CrossRef]
- Khoshnood, B.; Dacklin, I.; Grabbe, C. A Proteomics Approach to Identify Targets of the Ubiquitin-like Molecule Urm1 in Drosophila melanogaster. PLoS ONE 2017, 12, e0185611. [Google Scholar] [CrossRef] [Green Version]
- Anjum, R.S.; Bray, S.M.; Blackwood, J.K.; Kilkenny, M.L.; Coelho, M.A.; Foster, B.M.; Li, S.; Howard, J.A.; Pellegrini, L.; Albers, S.-V.; et al. Involvement of a Eukaryotic-like Ubiquitin-Related Modifier in the Proteasome Pathway of the Archaeon Sulfolobus acidocaldarius. Nat. Commun. 2015, 6, 8163. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Orgel, L.E. Oxidative Acylation Using Thioacids. Nature 1997, 389, 52–54. [Google Scholar] [CrossRef]
- Park, C.-M.; Weerasinghe, L.; Day, J.J.; Fukuto, J.M.; Xian, M. Persulfides: Current Knowledge and Challenges in Chemistry and Chemical Biology. Mol. Biosyst. 2015, 11, 1775–1785. [Google Scholar] [CrossRef] [Green Version]
- Paulsen, C.E.; Carroll, K.S. Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem. Rev. 2013, 113, 4633–4679. [Google Scholar] [CrossRef]
- Brachmann, C.; Kaduhr, L.; Jüdes, A.; Ravichandran, K.E.; West, J.D.; Glatt, S.; Schaffrath, R. Redox Requirements for Ubiquitin-like Urmylation of Ahp1, a 2-Cys Peroxiredoxin from Yeast. Redox Biol. 2020, 30, 101438. [Google Scholar] [CrossRef]
- Schlieker, C.D.; Van der Veen, A.G.; Damon, J.R.; Spooner, E.; Ploegh, H.L. A Functional Proteomics Approach Links the Ubiquitin-Related Modifier Urm1 to a tRNA Modification Pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 18255–18260. [Google Scholar] [CrossRef] [Green Version]
- Borodovsky, A.; Ovaa, H.; Kolli, N.; Gan-Erdene, T.; Wilkinson, K.D.; Ploegh, H.L.; Kessler, B.M. Chemistry-Based Functional Proteomics Reveals Novel Members of the Deubiquitinating Enzyme Family. Chem. Biol. 2002, 9, 1149–1159. [Google Scholar] [CrossRef] [Green Version]
- Nedialkova, D.D.; Leidel, S.A. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015, 161, 1606–1618. [Google Scholar] [CrossRef] [Green Version]
- Tükenmez, H.; Xu, H.; Esberg, A.; Byström, A.S. The Role of Wobble Uridine Modifications in +1 Translational Frameshifting in Eukaryotes. Nucleic Acids Res. 2015, 43, 9489–9499. [Google Scholar] [CrossRef] [Green Version]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.S.; Weissman, J.S. Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef] [Green Version]
- Zinshteyn, B.; Gilbert, W.V. Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling. PLoS Genet. 2013, 9, e1003675. [Google Scholar] [CrossRef] [Green Version]
- Chou, H.-J.; Donnard, E.; Gustafsson, H.T.; Garber, M.; Rando, O.J. Transcriptome-Wide Analysis of Roles for tRNA Modifications in Translational Regulation. Mol. Cell 2017, 68, 978–992.e4. [Google Scholar] [CrossRef] [Green Version]
- Klassen, R.; Ciftci, A.; Funk, J.; Bruch, A.; Butter, F.; Schaffrath, R. tRNA Anticodon Loop Modifications Ensure Protein Homeostasis and Cell Morphogenesis in Yeast. Nucleic Acids Res. 2016, 44, 10946–10959. [Google Scholar] [CrossRef] [Green Version]
- Tavares, J.F.; Davis, N.K.; Poim, A.; Reis, A.; Kellner, S.; Sousa, I.; Soares, A.R.; Moura, G.M.R.; Dedon, P.C.; Santos, M. tRNA-Modifying Enzyme Mutations Induce Codon-Specific Mistranslation and Protein Aggregation in Yeast. RNA Biol. 2020, 1–13. [Google Scholar] [CrossRef]
- Rezgui, V.A.N.; Tyagi, K.; Ranjan, N.; Konevega, A.L.; Mittelstaet, J.; Rodnina, M.V.; Peter, M.; Pedrioli, P.G.A. tRNA tKUUU, tQUUG, and tEUUC Wobble Position Modifications Fine-Tune Protein Translation by Promoting Ribosome A-Site Binding. Proc. Natl. Acad. Sci. USA 2013, 110, 12289–12294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranjan, N.; Rodnina, M.V. Thio-Modification of tRNA at the Wobble Position as Regulator of the Kinetics of Decoding and Translocation on the Ribosome. J. Am. Chem. Soc. 2017, 139, 5857–5864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirchner, S.; Ignatova, Z. Emerging Roles of tRNA in Adaptive Translation, Signalling Dynamics and Disease. Nat. Rev. Genet. 2015, 16, 98–112. [Google Scholar] [CrossRef]
- Schaffrath, R.; Leidel, S.A. Wobble Uridine Modifications—A Reason to Live, a Reason to Die?! RNA Biol. 2017, 14, 1209–1222. [Google Scholar] [CrossRef]
- Rubio-Texeira, M. Urmylation Controls Nil1p and Gln3p-Dependent Expression of Nitrogen-Catabolite Repressed Genes in Saccharomyces cerevisiae. FEBS Lett. 2007, 581, 541–550. [Google Scholar] [CrossRef] [Green Version]
- Laxman, S.; Sutter, B.M.; Wu, X.; Kumar, S.; Guo, X.; Trudgian, D.C.; Mirzaei, H.; Tu, B.P. Sulfur Amino Acids Regulate Translational Capacity and Metabolic Homeostasis through Modulation of tRNA Thiolation. Cell 2013, 154, 416–429. [Google Scholar] [CrossRef] [Green Version]
- Karlsborn, T.; Mahmud, A.K.M.F.; Tükenmez, H.; Byström, A.S. Loss of ncm5 and mcm5 Wobble Uridine Side Chains Results in an Altered Metabolic Profile. Metabolomics 2016, 12, 177. [Google Scholar] [CrossRef] [Green Version]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic Expression Programs in the Response of Yeast Cells to Environmental Changes. Mol. Biol. Cell 2000, 11, 17. [Google Scholar] [CrossRef]
- Bauer, F.; Matsuyama, A.; Candiracci, J.; Dieu, M.; Scheliga, J.; Wolf, D.A.; Yoshida, M.; Hermand, D. Translational Control of Cell Division by Elongator. Cell Rep. 2012, 1, 424–433. [Google Scholar] [CrossRef] [Green Version]
- Neukranz, Y.; Kotter, A.; Beilschmidt, L.; Marelja, Z.; Helm, M.; Gräf, R.; Leimkühler, S. Analysis of the Cellular Roles of MOCS3 Identifies a MOCS3-Independent Localization of NFS1 at the Tips of the Centrosome. Biochemistry 2019, 58, 1786–1798. [Google Scholar] [CrossRef]
- Sarin, L.P.; Leidel, S.A. Modify or Die? —RNA Modification Defects in Metazoans. RNA Biol. 2014, 11, 1555–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres, A.G.; Batlle, E.; Ribas de Pouplana, L. Role of tRNA Modifications in Human Diseases. Trends Mol. Med. 2014, 20, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-T.; Hims, M.M.; Shetty, R.S.; Mull, J.; Liu, L.; Leyne, M.; Slaugenhaupt, S.A. Loss of Mouse Ikbkap, a Subunit of Elongator, Leads to Transcriptional Deficits and Embryonic Lethality That Can Be Rescued by Human IKBKAP. Mol. Cell. Biol. 2009, 29, 736–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laguesse, S.; Creppe, C.; Nedialkova, D.D.; Prévot, P.-P.; Borgs, L.; Huysseune, S.; Franco, B.; Duysens, G.; Krusy, N.; Lee, G.; et al. A Dynamic Unfolded Protein Response Contributes to the Control of Cortical Neurogenesis. Dev. Cell 2015, 35, 553–567. [Google Scholar] [CrossRef] [Green Version]
- Slaugenhaupt, S.A.; Blumenfeld, A.; Gill, S.P.; Leyne, M.; Mull, J.; Cuajungco, M.P.; Liebert, C.B.; Chadwick, B.; Idelson, M.; Reznik, L.; et al. Tissue-Specific Expression of a Splicing Mutation in the IKBKAP Gene Causes Familial Dysautonomia. Am. J. Hum. Genet. 2001, 8. [Google Scholar] [CrossRef] [Green Version]
- Anderson, S.L.; Coli, R.; Daly, I.W.; Kichula, E.A.; Rork, M.J.; Volpi, S.A.; Ekstein, J.; Rubin, B.Y. Familial Dysautonomia Is Caused by Mutations of the IKAP Gene. Am. J. Hum. Genet. 2001, 68, 753–758. [Google Scholar] [CrossRef] [Green Version]
- Karlsborn, T.; Tükenmez, H.; Chen, C.; Byström, A.S. Familial Dysautonomia (FD) Patients Have Reduced Levels of the Modified Wobble Nucleoside mcm5s2U in tRNA. Biochem. Biophys. Res. Commun. 2014, 454, 441–445. [Google Scholar] [CrossRef] [Green Version]
- Kojic, M.; Wainwright, B. The Many Faces of Elongator in Neurodevelopment and Disease. Front. Mol. Neurosci. 2016, 9. [Google Scholar] [CrossRef]
- Shaheen, R.; Mark, P.; Prevost, C.T.; AlKindi, A.; Alhag, A.; Estwani, F.; Al-Sheddi, T.; Alobeid, E.; Alenazi, M.M.; Ewida, N.; et al. Biallelic Variants in CTU2 Cause DREAM-PL Syndrome and Impair Thiolation of tRNA Wobble U34. Hum. Mutat. 2019, 40, 2108–2120. [Google Scholar] [CrossRef]
- Delaunay, S.; Rapino, F.; Tharun, L.; Zhou, Z.; Heukamp, L.; Termathe, M.; Shostak, K.; Klevernic, I.; Florin, A.; Desmecht, H.; et al. Elp3 Links tRNA Modification to IRES-Dependent Translation of LEF1 to Sustain Metastasis in Breast Cancer. J. Exp. Med. 2016, 213, 2503–2523. [Google Scholar] [CrossRef] [Green Version]
- Rapino, F.; Delaunay, S.; Rambow, F.; Zhou, Z.; Tharun, L.; De Tullio, P.; Sin, O.; Shostak, K.; Schmitz, S.; Piepers, J.; et al. Codon-Specific Translation Reprogramming Promotes Resistance to Targeted Therapy. Nature 2018, 558, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Silvera, D.; Formenti, S.C.; Schneider, R.J. Translational Control in Cancer. Nat. Rev. Cancer 2010, 10, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Hleihel, R.; Khoshnood, B.; Dacklin, I.; Omran, H.; Mouawad, C.; Dassouki, Z.; El-Sabban, M.; Shirinian, M.; Grabbe, C.; Bazarbachi, A. The HTLV-1 Oncoprotein Tax Is Modified by the Ubiquitin Related Modifier 1 (Urm1). Retrovirology 2018, 15, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhat, M.; Robichaud, N.; Hulea, L.; Sonenberg, N.; Pelletier, J.; Topisirovic, I. Targeting the Translation Machinery in Cancer. Nat. Rev. Drug Discov. 2015, 14, 261–278. [Google Scholar] [CrossRef]
- Boriack-Sjodin, P.A.; Ribich, S.; Copeland, R.A. RNA-Modifying Proteins as Anticancer Drug Targets. Nat. Rev. Drug Discov. 2018, 17, 435–453. [Google Scholar] [CrossRef]
- Soucy, T.A.; Smith, P.G.; Milhollen, M.A.; Berger, A.J.; Gavin, J.M.; Adhikari, S.; Brownell, J.E.; Burke, K.E.; Cardin, D.P.; Critchley, S.; et al. An Inhibitor of NEDD8-Activating Enzyme as a New Approach to Treat Cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef]
- He, X.; Riceberg, J.; Soucy, T.; Koenig, E.; Minissale, J.; Gallery, M.; Bernard, H.; Yang, X.; Liao, H.; Rabino, C.; et al. Probing the Roles of SUMOylation in Cancer Cell Biology by Using a Selective SAE Inhibitor. Nat. Chem. Biol. 2017, 13, 1164–1171. [Google Scholar] [CrossRef]
- Misra, M.; Kuhn, M.; Löbel, M.; An, H.; Statsyuk, A.V.; Sotriffer, C.; Schindelin, H. Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme. Structure 2017, 25, 1120–1129.e3. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Walvekar, A.S.; Liang, S.; Rashida, Z.; Shah, P.; Laxman, S. A tRNA Modification Balances Carbon and Nitrogen Metabolism by Regulating Phosphate Homeostasis. eLife 2019, 8, e44795. [Google Scholar] [CrossRef]
- Gupta, R.; Laxman, S. tRNA Wobble-Uridine Modifications as Amino Acid Sensors and Regulators of Cellular Metabolic State. Curr. Genet. 2020, 66, 475–480. [Google Scholar] [CrossRef]
- Chan, C.; Pham, P.; Dedon, P.C.; Begley, T.J. Lifestyle modifications: Coordinating the tRNA epitranscriptome with codon bias to adapt translation during stress responses. Genome Biol. 2018, 19, 228. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Termathe, M.; Leidel, S.A. Urm1: A Non-Canonical UBL. Biomolecules 2021, 11, 139. https://doi.org/10.3390/biom11020139
Termathe M, Leidel SA. Urm1: A Non-Canonical UBL. Biomolecules. 2021; 11(2):139. https://doi.org/10.3390/biom11020139
Chicago/Turabian StyleTermathe, Martin, and Sebastian A. Leidel. 2021. "Urm1: A Non-Canonical UBL" Biomolecules 11, no. 2: 139. https://doi.org/10.3390/biom11020139
APA StyleTermathe, M., & Leidel, S. A. (2021). Urm1: A Non-Canonical UBL. Biomolecules, 11(2), 139. https://doi.org/10.3390/biom11020139





