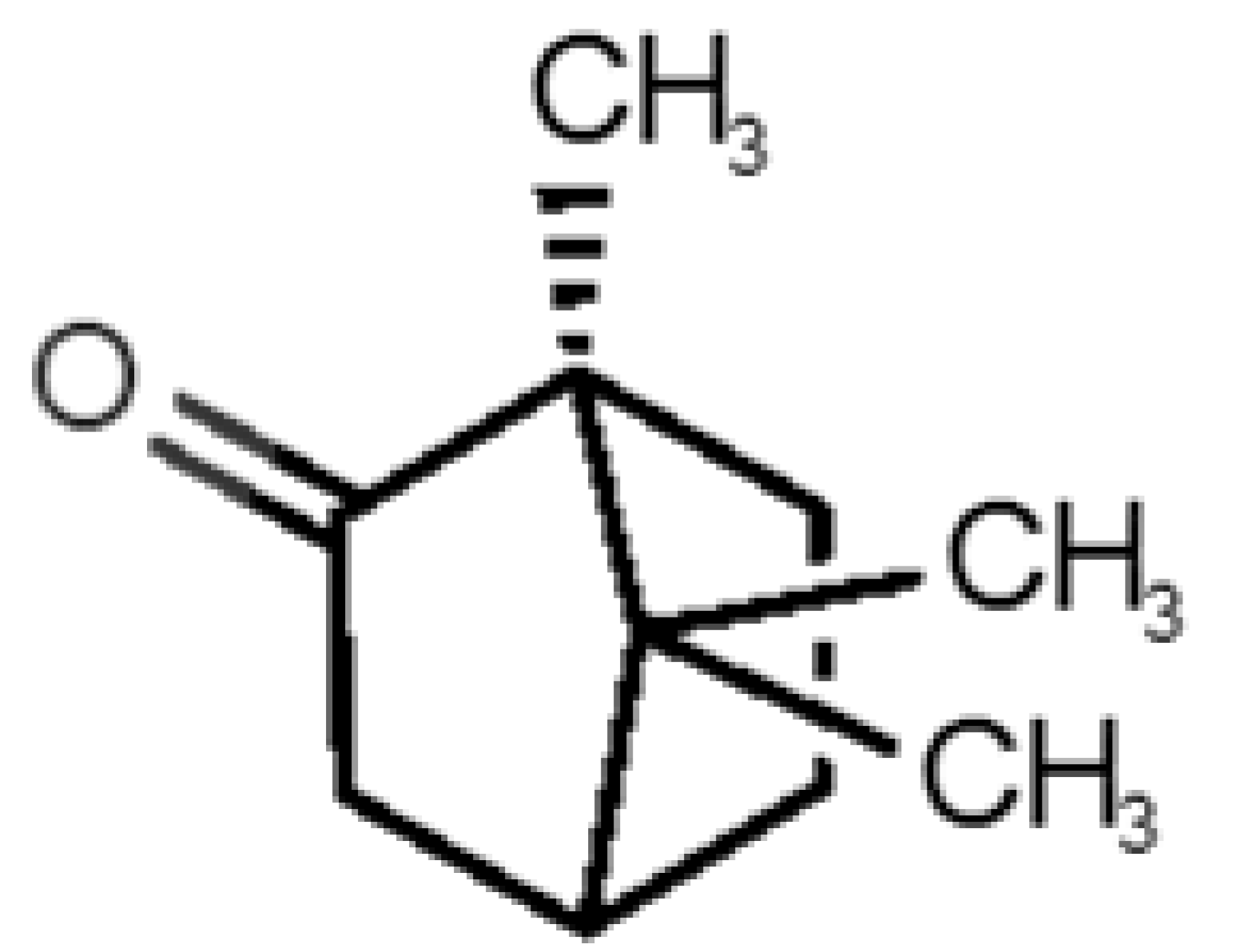
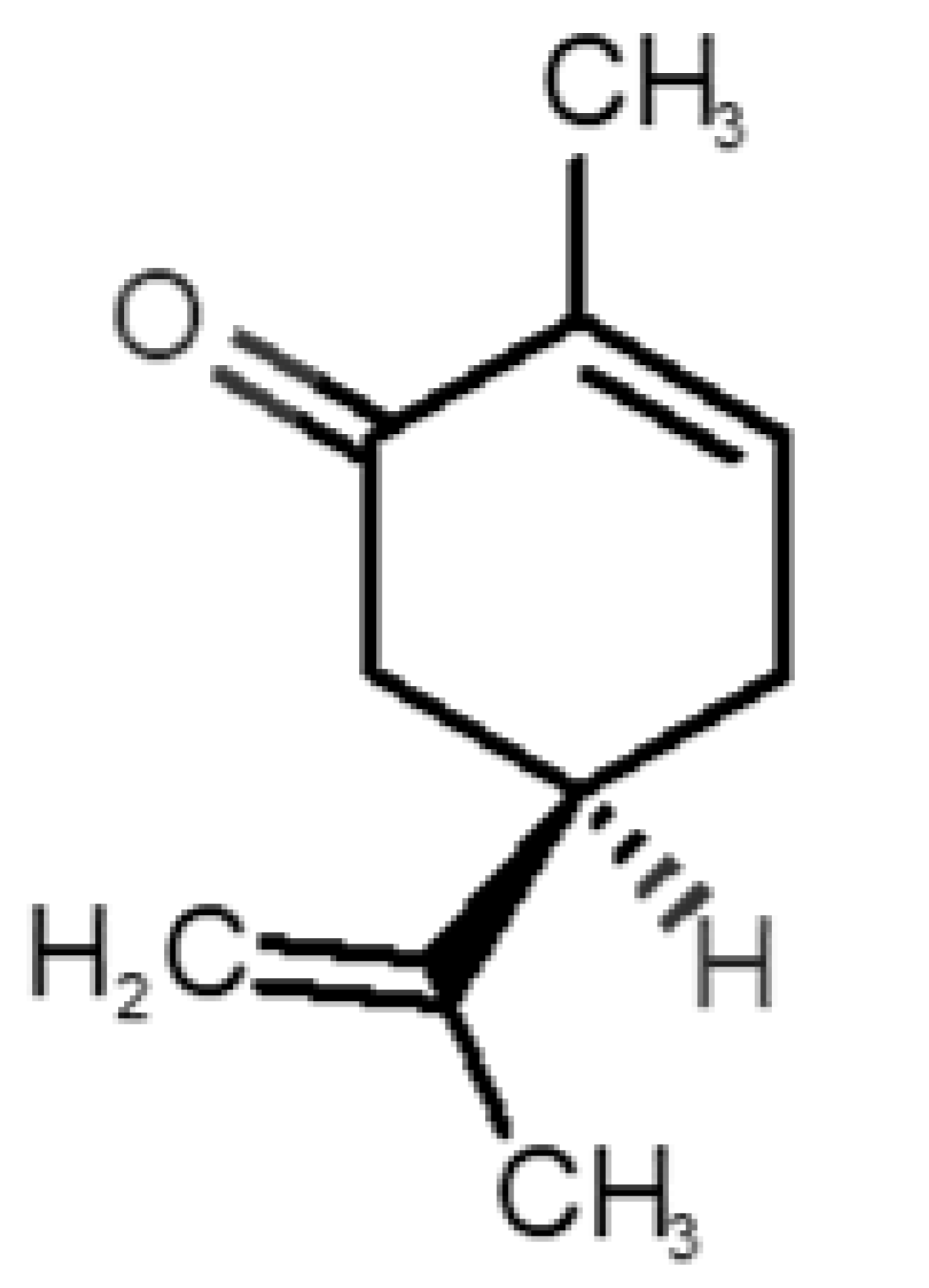
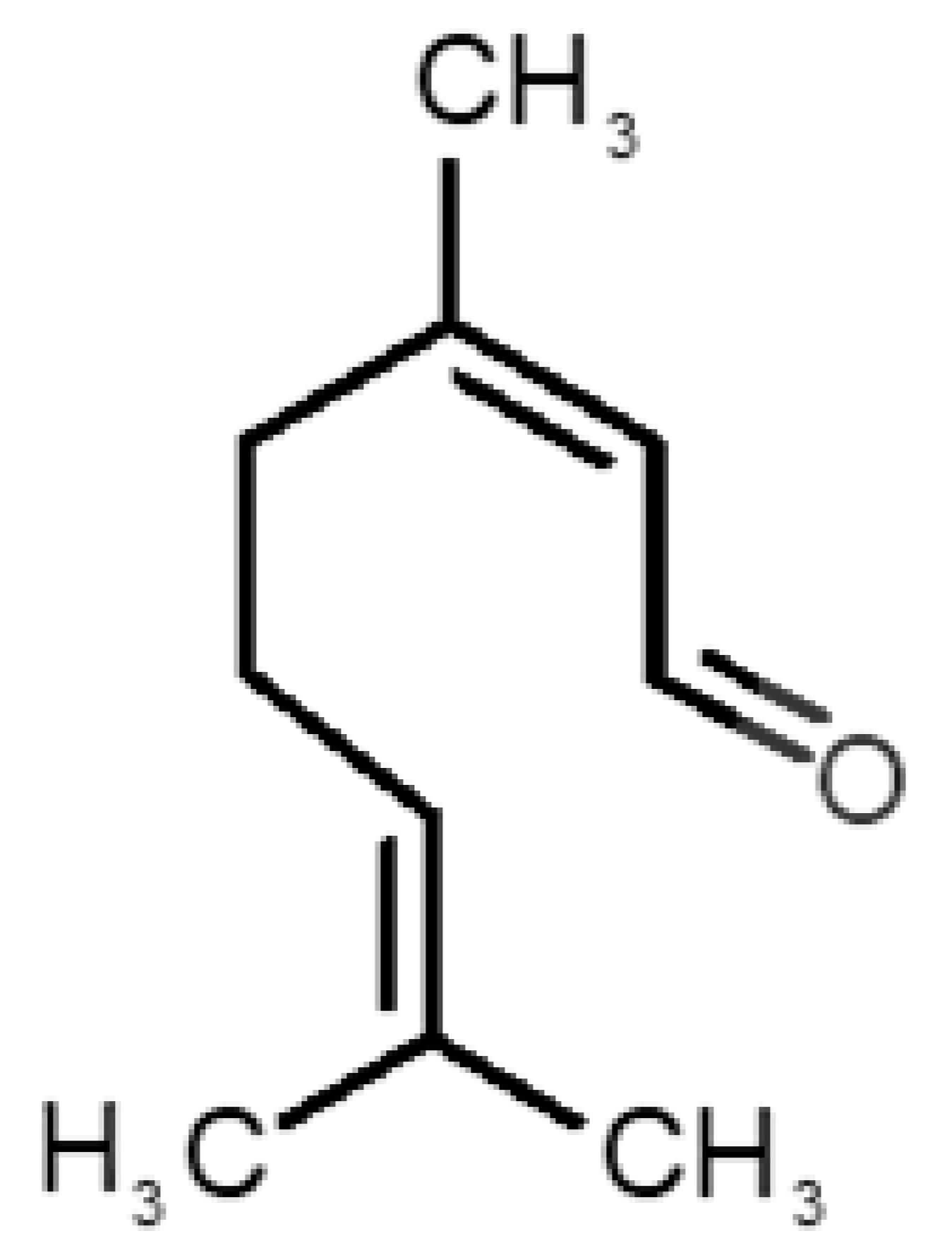
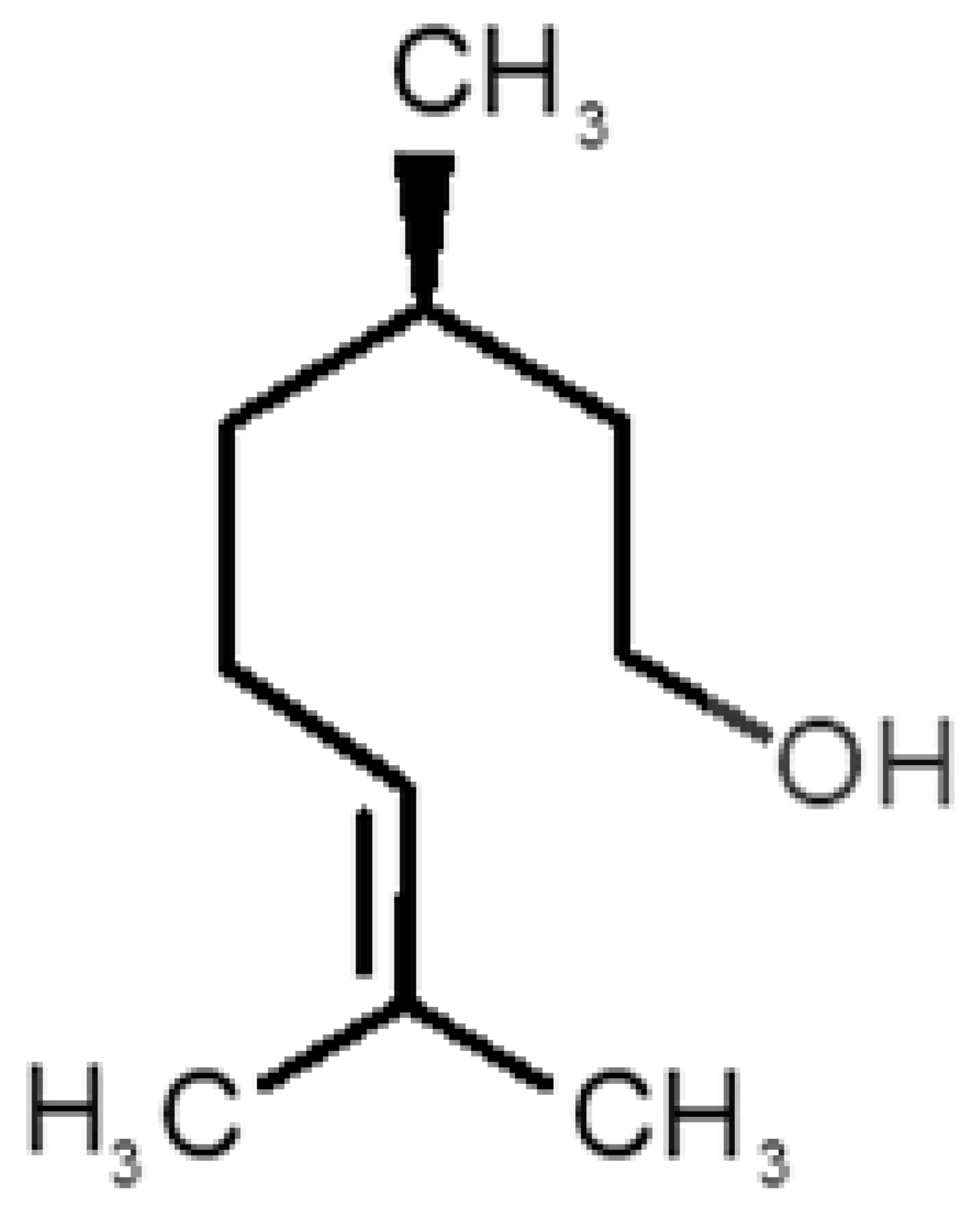
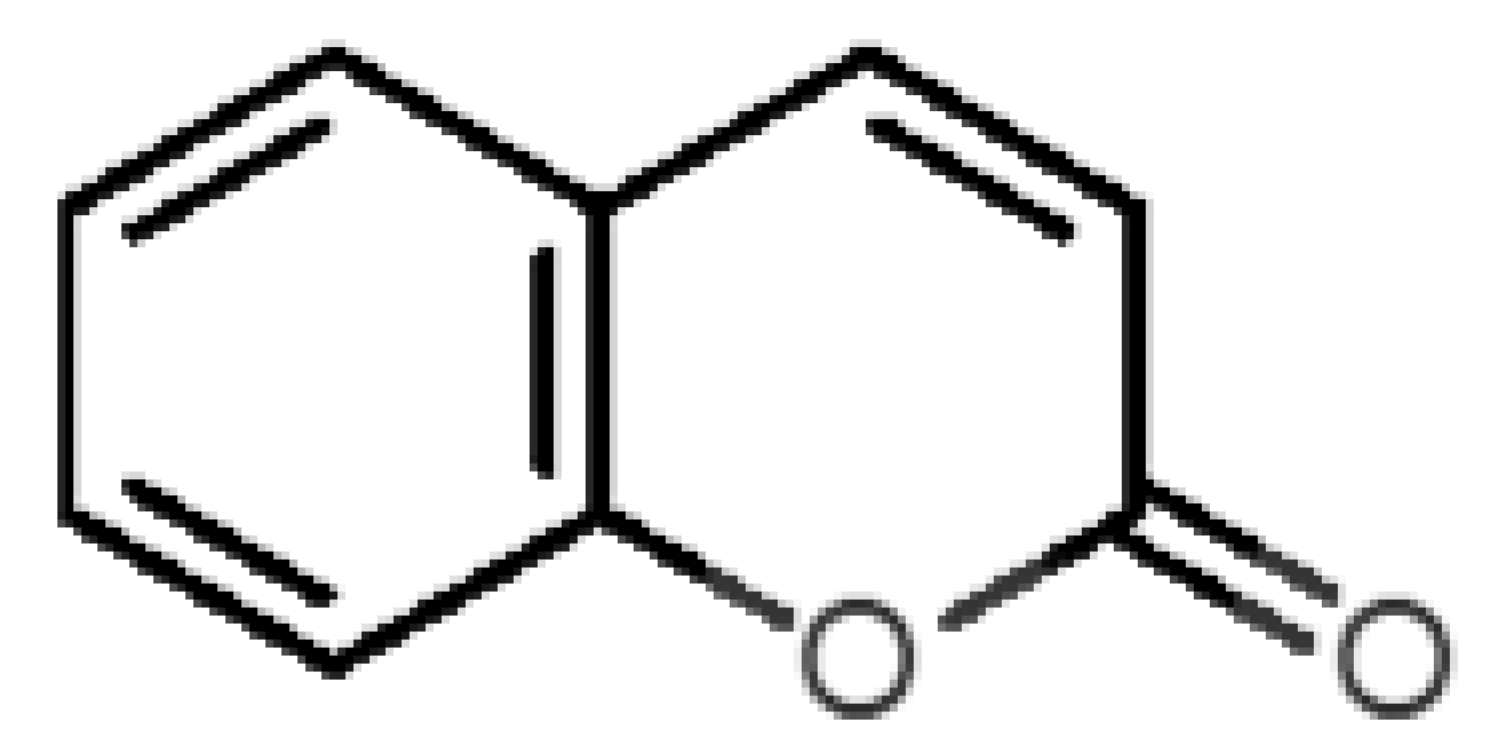
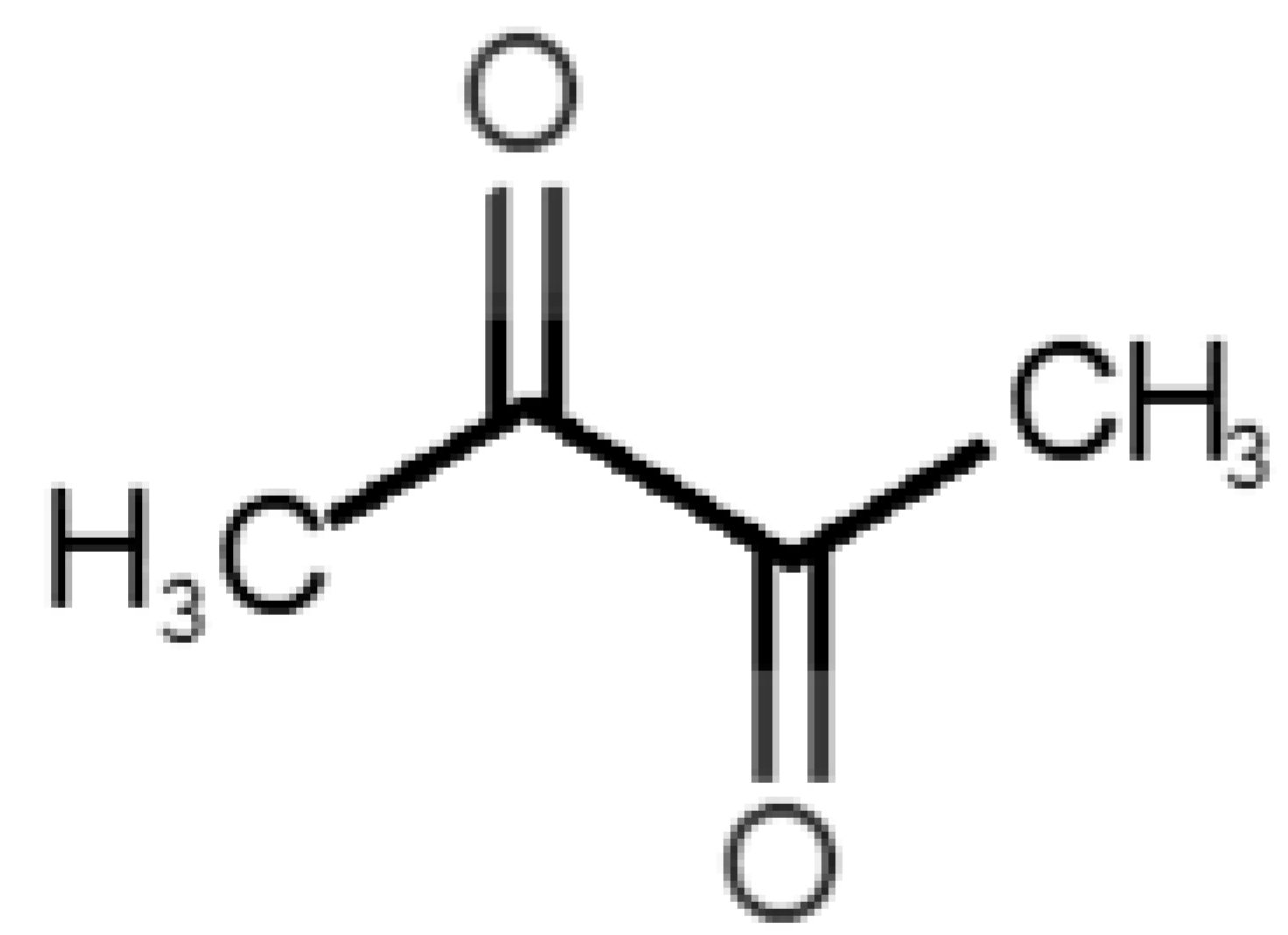
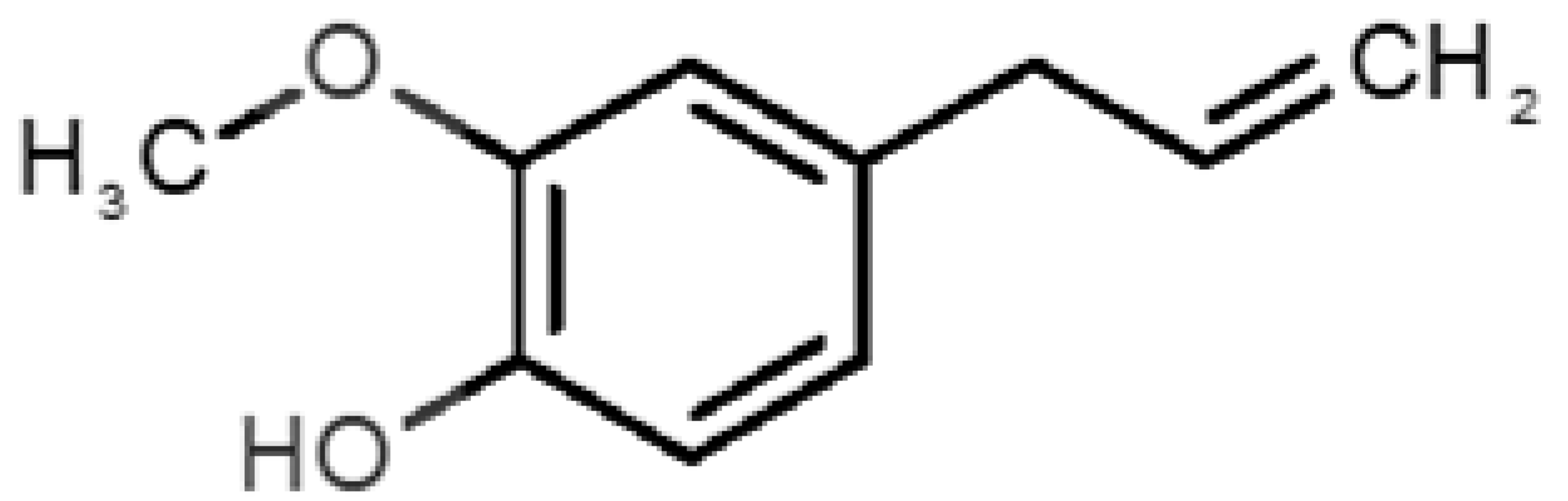
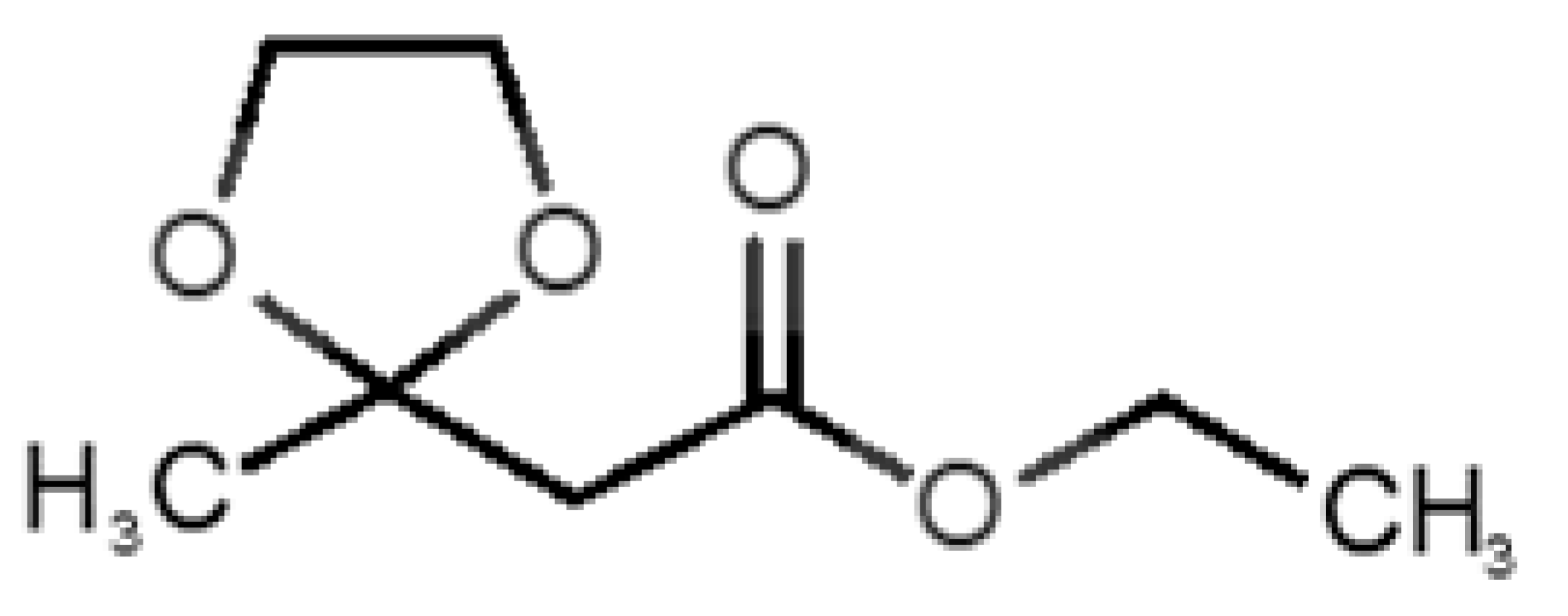
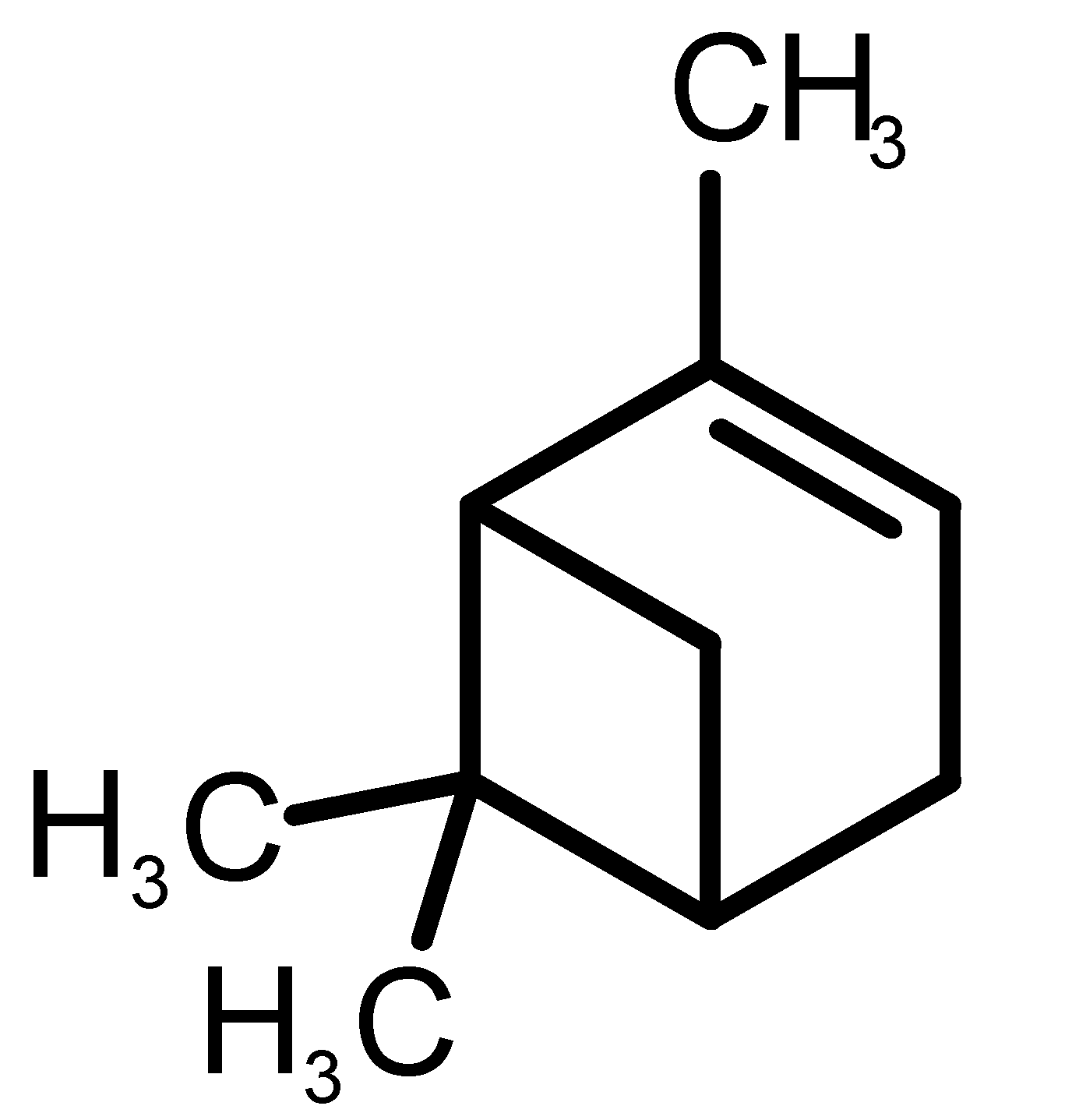
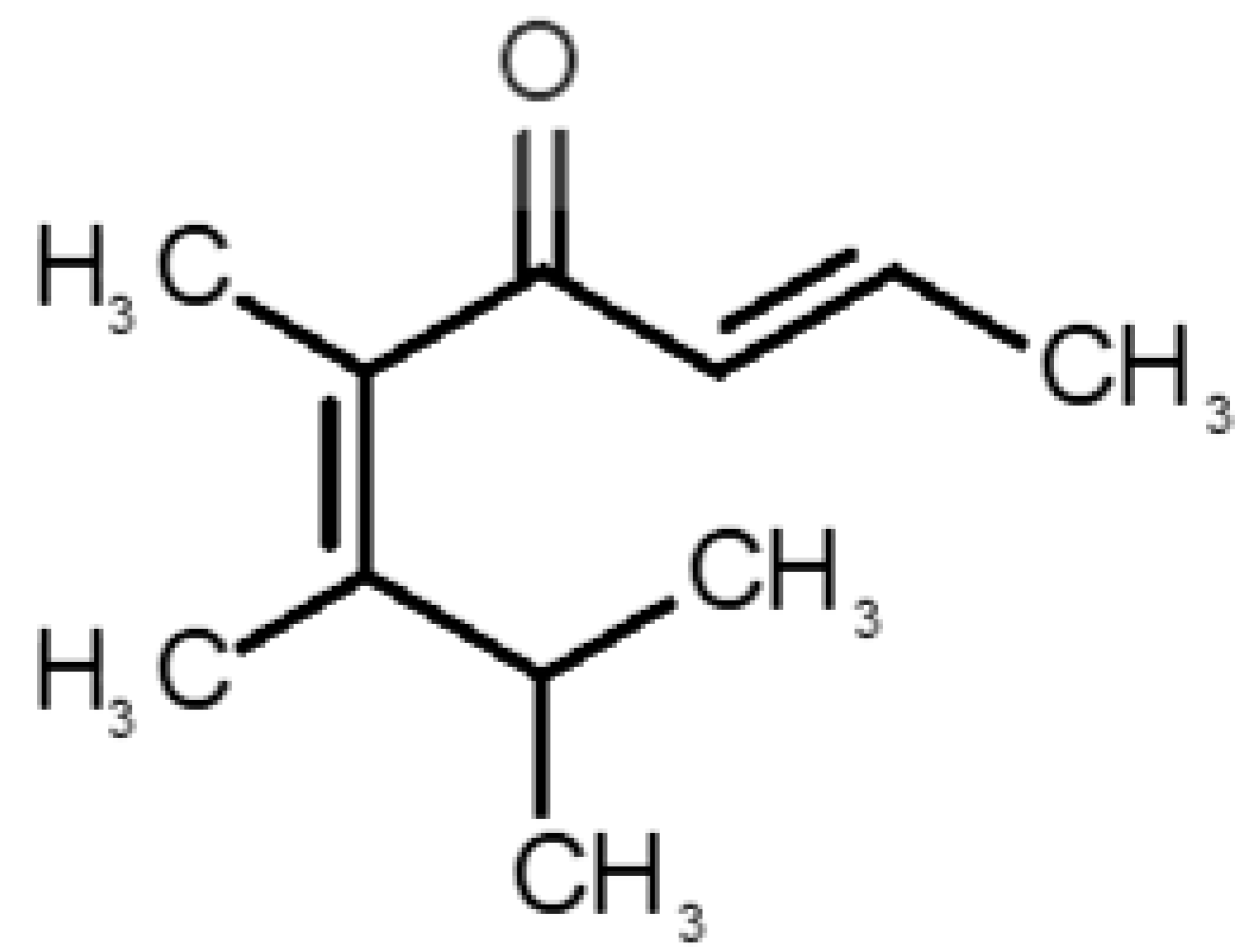
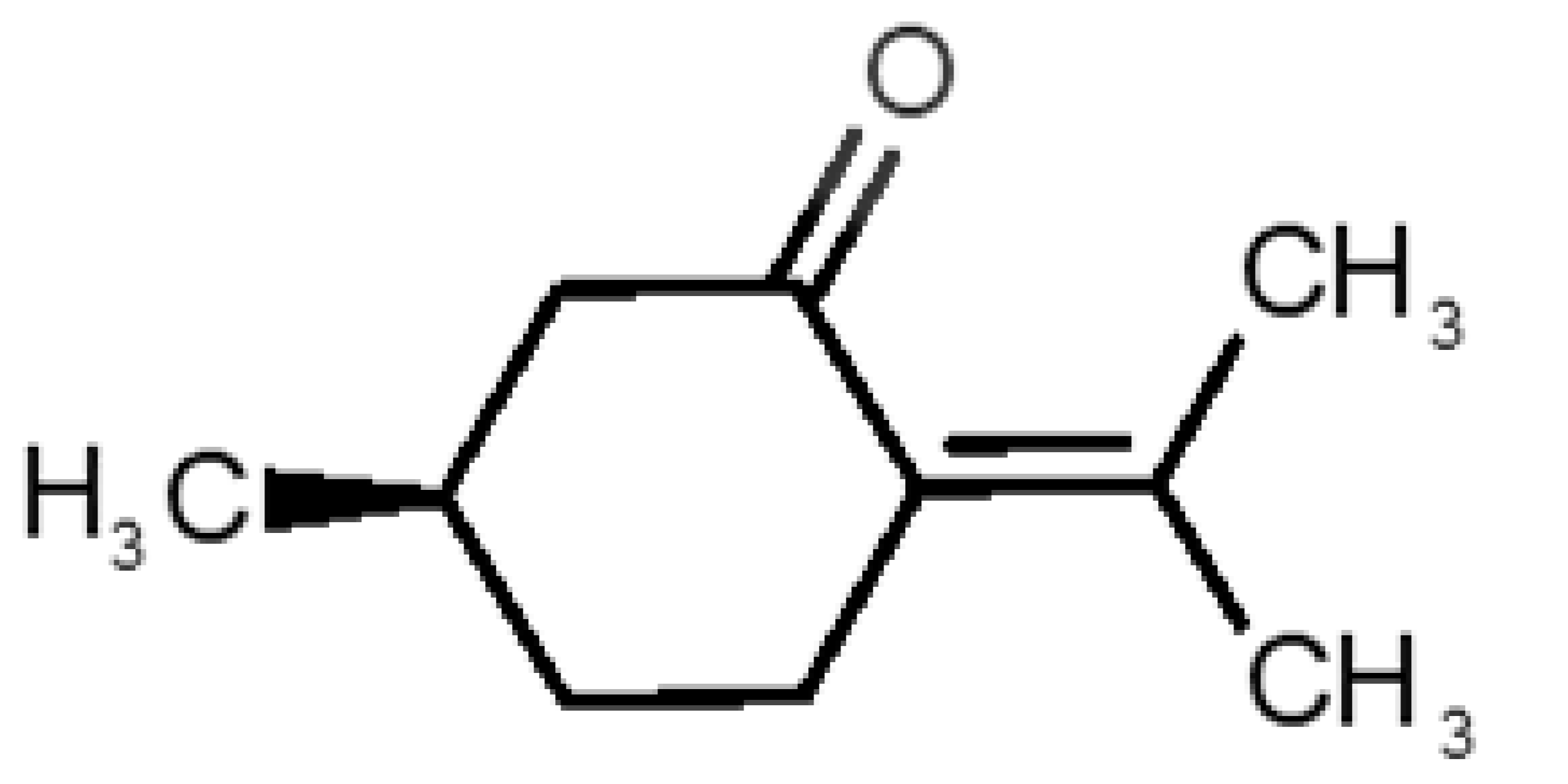
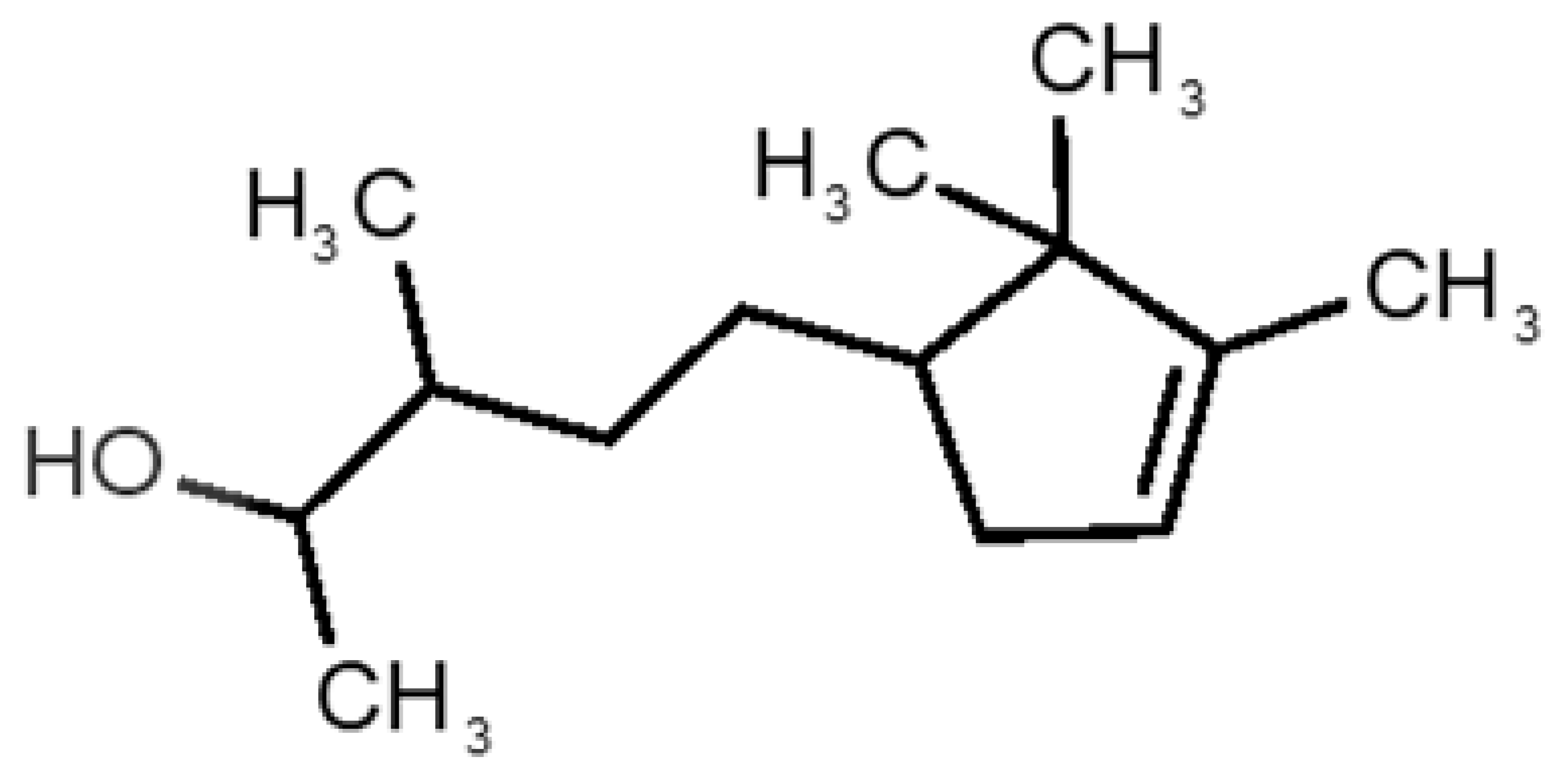
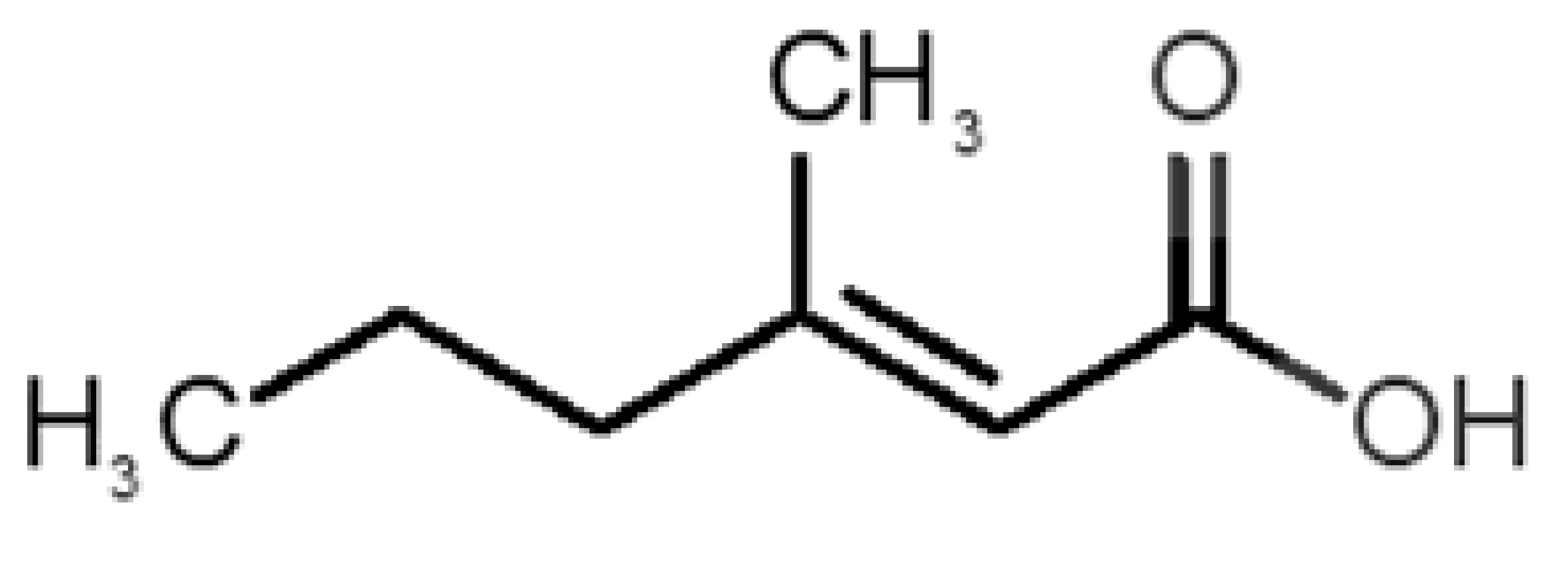
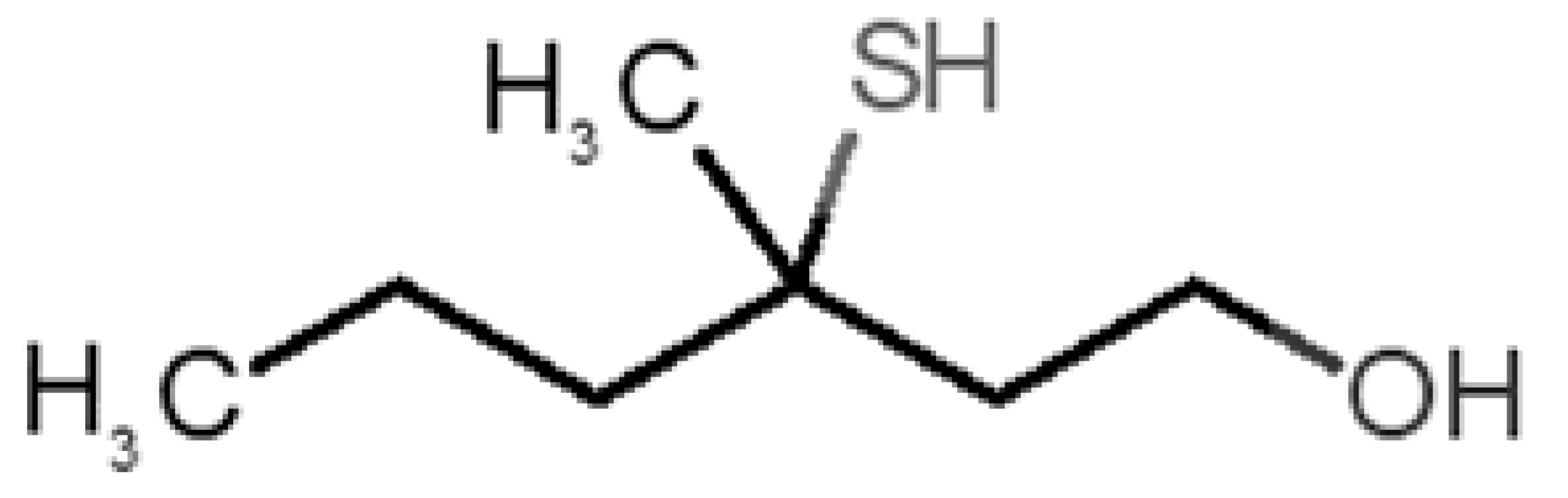
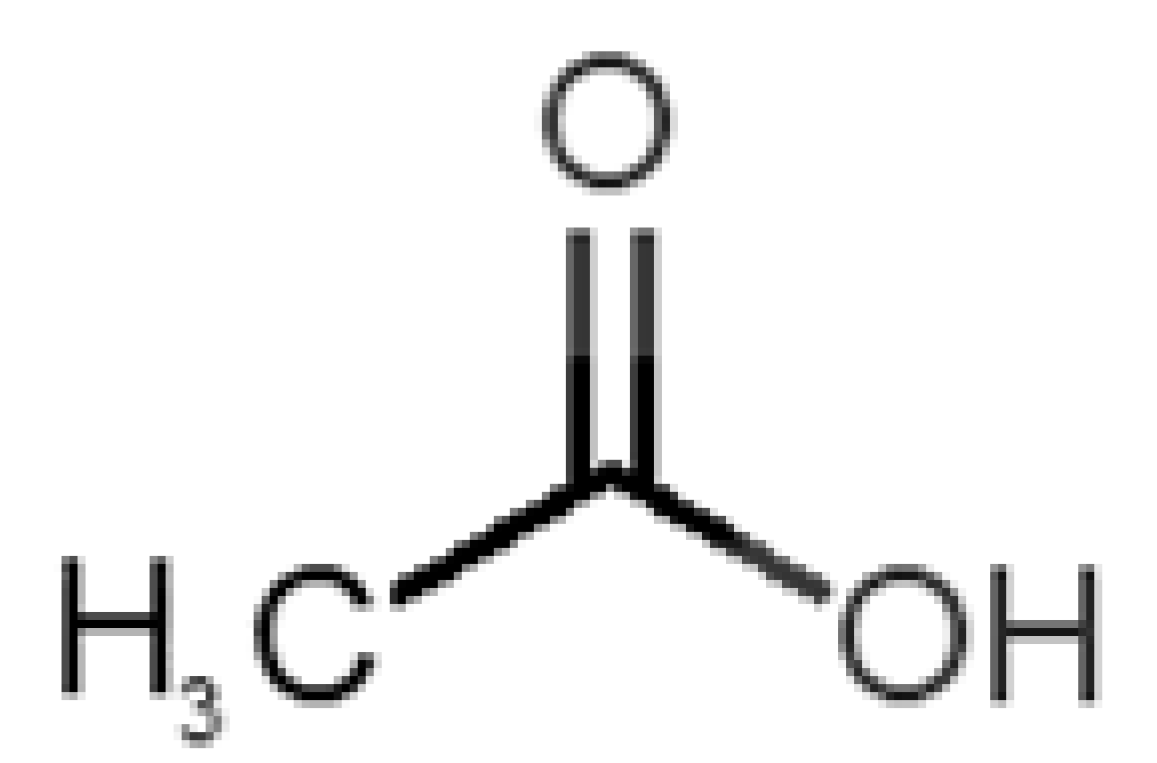
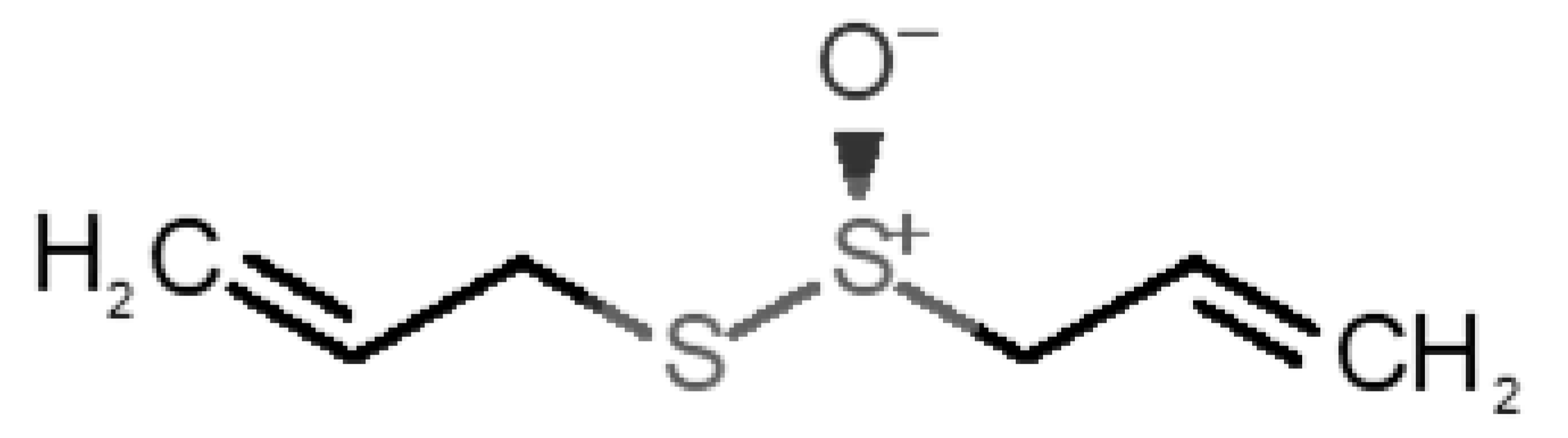
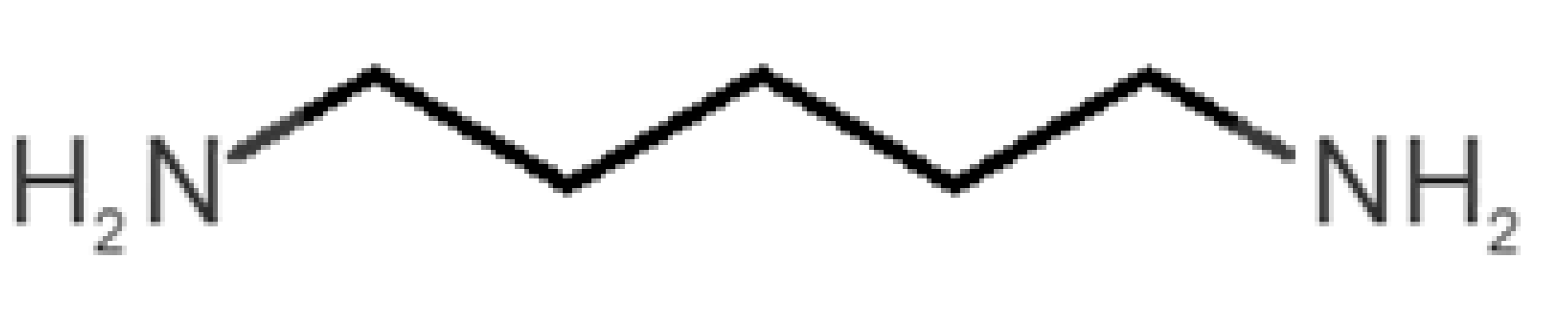
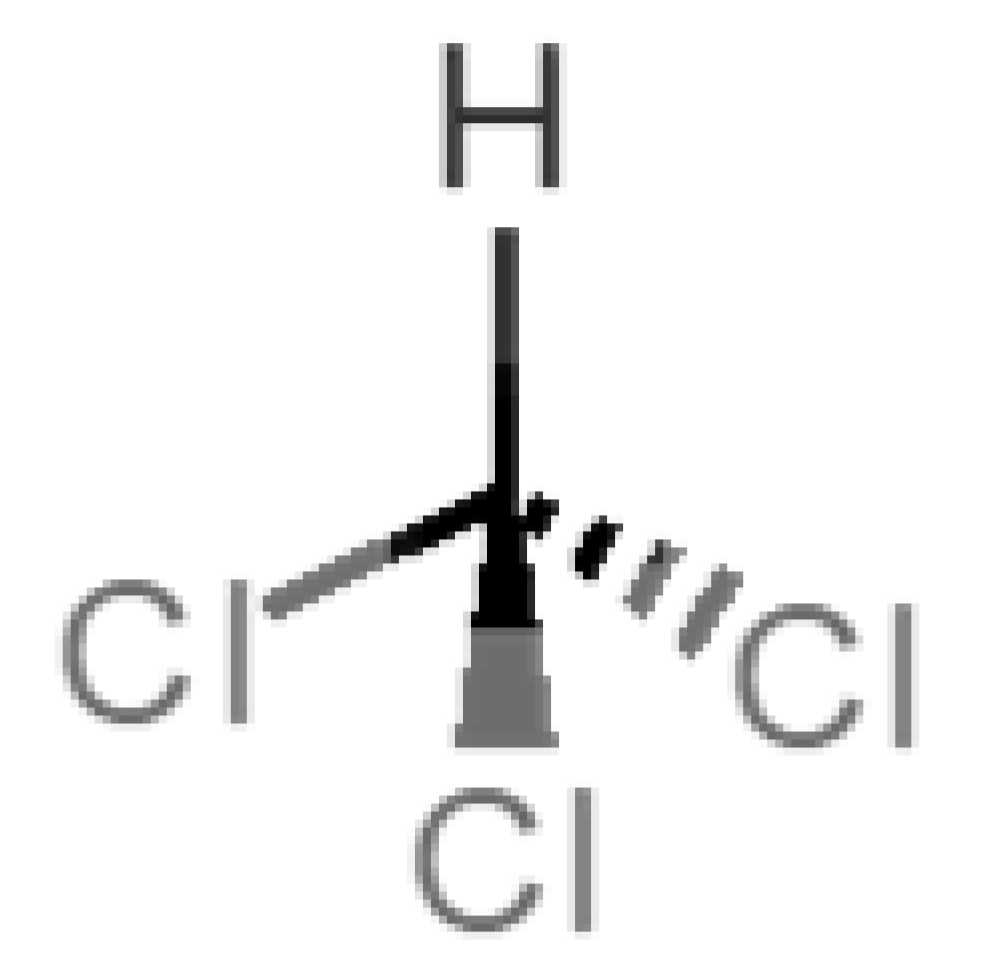
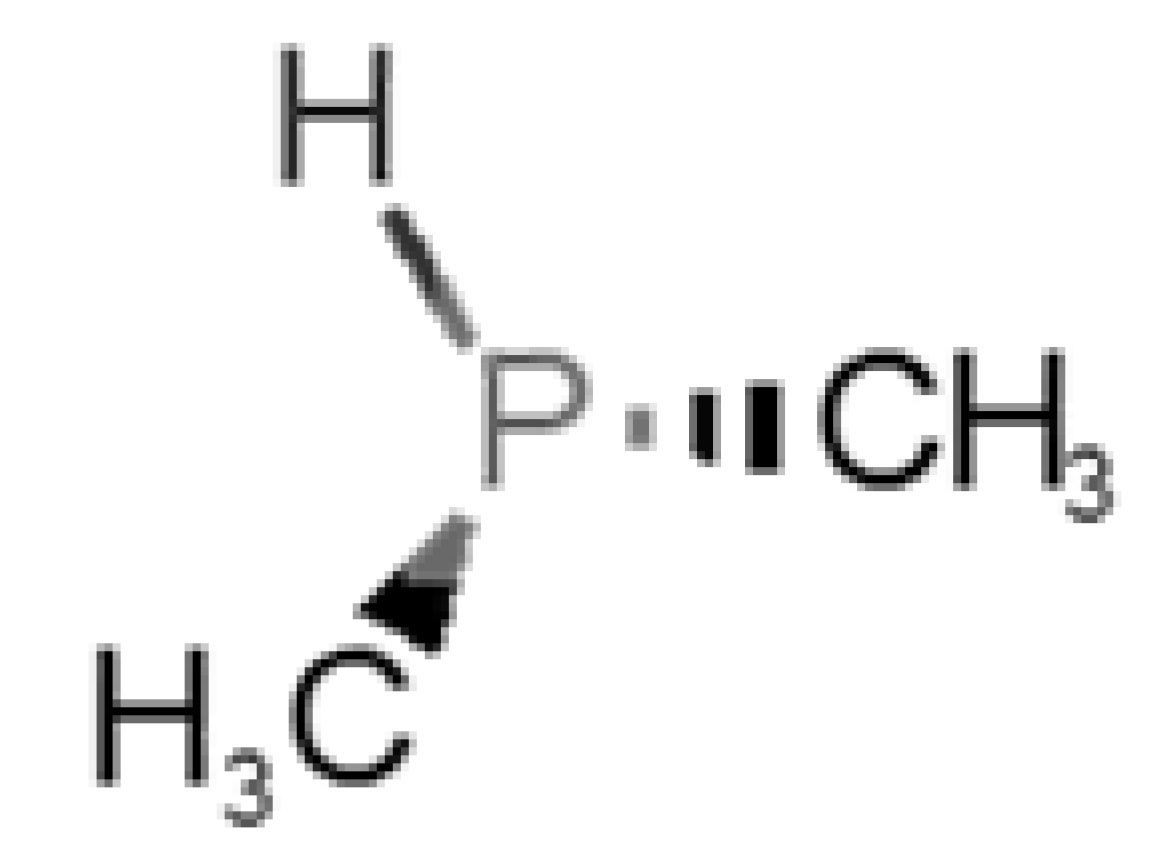
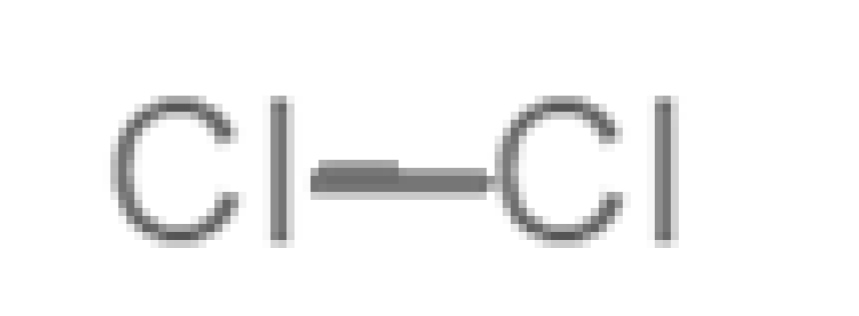Abstract
The binding of known odorant molecules to the human odorant-binding protein (hOBP) was evaluated in silico. Docking experiments elucidate the preferable binding site and binding affinity of odorant molecules to hOBP. The physicochemical properties molecular weight (MW), vapor pressure (Vp), hydrophobicity level (logP), number of double bonds (NºDB), degree of unsaturation (DoU) and the chemical classification, were selected for the study of odorant modulation. Here, these properties were analyzed concerning 30 pleasant and 30 unpleasant odorants, chosen to represent a wide variety of compounds and to determine their influence on the binding energy to hOBP. Our findings indicate that MW, logP and Vp are the most important odorant variables, directly correlated to odorant-binding energies (ΔGbinding) towards hOBP. Understanding how the odorants behave when complexed with the OBP in human olfaction opens new possibilities for the development of future biotechnological applications, including sensory devices, medical diagnosis, among others.
1. Introduction
Olfaction functions as a chemosensing system, allowing the detection and discrimination of millions of different volatile molecules, the odorants, which provide extremely important information about the surrounding environment. In humans, odorants recognition is mediated by a large repertoire of olfactory receptors (ORs), coded by 391 functional OR genes [1]. The ORs are located across the plasma membranes of the ciliated dendrites of olfactory sensory neurons, localized in the olfactory epithelium. Each sensory neuron expresses a single allele of a single OR gene to ensure a distinct pattern of neuronal activation for every odorant [2]. Mammalian ORs belong to one of two classes, according to the recognized odorant type: Class I, ORs mostly bind hydrophilic odorants, and Class II, ORs bind hydrophobic odorants. In order for the ligand–receptor binding to occur, the odorant must cross a hydrophilic barrier—the mucus, where the ciliated dendrites of olfactory neurons are immersed. The hydrophobic odorants need to be transported, which is thought to be the role of the small soluble proteins, the odorant-binding proteins (OBPs).
In vertebrates, OBPs are highly expressed in the nasal epithelia, where they bind and carry, through the aqueous mucus, hydrophobic and volatile odorant molecules. Humans express two “classical” OBP genes, OBP2A and OBP2B, but, in contrast to other mammals, their expression in the human olfactory epithelium is not enhanced [1]. In vertebrates, OBP presents the typical folding of lipocalins, a hydrophobic β-barrel core that encloses an internal ligand-binding site. The obvious and crucial physiological role of OBP in olfaction is to facilitate the transport of odorant molecules to the ORs. Based on experimental evidence, an active role also has been proposed for OBPs in addition to the passive transport of odorants [2,3]. It was demonstrated in an in vitro assay that the rat OBP-F1 restores a OR activity at high odorant doses, changing the response curve from bell-shaped to sigmoidal, which is the characteristic curve of the OR in vivo response [3]. This active role was proposed to be mediated by a physical interaction between the OBP and the receptor, an interaction that is affected by the presence of the OR ligand. Furthermore, in another work, the authors have demonstrated that ORs constitutively form homodimers [4]. OR dimers displayed different conformational changes upon stimulation with various odorant doses, corresponding to different levels of activity. At low ligand concentrations, OR dimer would bind only one odorant molecule staying in an active form. While, at high ligand doses, the OR dimer would bind a second odorant molecule, leading to an inactive conformation. The authors assumed that in the presence of OBP, the second ligand would not be able to bind due to the OBP binding to OR dimer. In this way, the active role of OBP could be an allosteric control of OR dimer activity, at high ligand doses, besides the intrinsic and passive ability of OBP to buffer the levels of odorants.
OBP has been used in the biotechnology field for many purposes [5,6,7], as in the development of sensors [8] or smart textiles [9]. From a biochemical point of view, we have studied porcine OBP (pOBP) for its functionalization with small peptides, which promoted the liposomal transduction of a small molecule, as well as for the ability of a truncated form to have a thermal response [10,11]. These studies have shown the versatile profile of this protein under different conditions and environments.
Odorant molecules are volatile and structurally diverse compounds, which are perceived by the ORs, carrying information about the surrounding environment. The odorants can be detected directly from the inhaled air through the nose or by the throat, after chewing food. Yet, often, these molecules cannot cross the nasal mucus without support [12,13], which is the transport role of OBP.
The organoleptic classification of an odor substance should be understood loosely, i.e., the olfactive memory of each person, as well as the habituation to a specific aroma, may cause one to find an aroma more pleasant rather than other. That is why, herein, the odor compounds were categorized as generally pleasant or unpleasant, in contrast to terms like good/bad aromas. The selection of odors was thought to represent a wide variety of compounds in terms of the addressed physicochemical properties. In addition, we have prioritized the evaluation of aromas commonly used in industry or, in the case of unpleasant, body odors or decaying food smells.
Hence, far, the human OBP itself has been the subject of few studies [14], as the experimental characterization of its structure was only achieved in 2015. Recently, some works related an hOBP lysine to aldehyde odorants-binding [15,16]. These studies were conducted experimentally and by molecular dynamics (MD) simulations using an hOBP designed by homology modeling. Apart from that, the research in hOBP is still very incipient in contrast to OBPs from other vertebrates.
The present study is focused on the in silico analysis of human OBP (hOBP), aiming for a deeper understanding of the binding and transport of odorants by this protein while establishing a correlation between the type and strength of binding and the physicochemical properties of the odorants. Thus, a comprehensive library of 60 odorant compounds, 30 commonly perceived as pleasant and 30 as unpleasant, were analyzed regarding the binding affinity to the hOBP. These molecules were selected according to the established application in the industry (cosmetics and textiles) as well as to the structural characteristics, including size/volume, saturate/unsaturated bonds and functional groups. The functional groups included amines, alcohols, aldehydes, ketones, thiols, acids, phosphines, organosulfur compounds, esters, lactones and volatile gases, among others. The first binding studies in OBP were performed using heterocyclic and terpenoid derivatives and medium-size aliphatic alcohols and aldehydes [17,18]. The same rationale was used here; small to medium size odorant molecules were addressed. Docking virtual screening was applied to disclose the hOPB binding site, the interaction pattern and the affinity towards the 60 odorant molecules. The selected methodology elucidated about hOBP properties, including the binding modulation to pleasant and unpleasant odorants.
2. Methods
2.1. Human Odorant Binding Protein Structure
The UniProt [19] was used to obtain the full canonical amino acid sequence for hOBP, corresponding to the entry Q9NY56. Then, the hOBP full structure was predicted using the iterative threading assembly refinement (I-TASSER) server [20], a method to predict protein structure and function that uses a multiple threading approach based on templates from the Protein Data Bank (PDB) [21]. The threading templates used to generate the full hOBP structure were the proteins with the following codes: 5X7Y, 3CBC, 1EW3, 1GM6, 4RUN and 1EXS, all lipocalin-type proteins as the OBP. The I-TASSER server-generated five-hit models, and from this list, we chose the one with the best C-score (confidence).
The differences between the modeled conformation and the available hOBP X-ray structure (PDB ID: 4RUN) consist in the lack of the N-terminal in chain A, which is the chain carrying the ligand, but also the crystal structure has two different amino acids placed at the barrel core. As the binding site for hOBP is not established prior to the docking experiments or in literature, these two positions could be interacting sites with odorant molecules, hence the importance of modeling the canonical sequence. Nevertheless, the hOBP model was compared to the X-ray results, both at the conformational level and regarding docking affinities (Figures S2 and S4 and Table S3).
2.2. Molecular Dynamics Simulations and Choice of hOBP Conformation
Molecular dynamics (MD) simulations were performed on the hOBP complete sequence-structure model to equilibrate its conformation, although highly similar to human 4RUN X-ray conformation. The protein was inserted in a cubic box with an SPC water model, comprising a volume of 385.53 nm3. One stage of energy minimization was performed using a maximum of 50,000 steps and the steepest descent method. Initialization steps using canonical NVT (constant number of particles, volume and temperature) and isothermal-isobaric NPT (constant number of particles, pressure and temperature) ensembles were performed applying position restraints (with force constant of 1000 kJ·mol−1·nm−2) to all heavy atoms in both procedures. After initialization, 50 ns of production simulation took place, without restraints. All simulations were performed using the GROMACS 5.1.4 version [22,23] within the GROMOS 54a7 force field (FF) [24,25]. The Lennard-Jones interactions were truncated at 1.4 nm, and the particle-mesh Ewald (PME) [26] method for electrostatic interactions with a cutoff of 1.4 nm was used. The algorithm LINCS [27] was used to constrain the chemical bonds of the protein as well as the algorithm SETTLE [28] in the case of water.
After the simulation, RMSD and cluster analysis were used to determine the system equilibration at the most representative conformation for hOBP under solution. The single-linkage method, with a cutoff of 0.1 nm, was implemented for the last 25 ns of unrestrained simulation time, from which the RMSD is equilibrated (Figure S3). This technique clusters structures that are below the RMSD cutoff. For hOBP, only two clusters were computed, one containing 26 conformations and the second comprising 2473 structures. Within the most populated cluster, the structure which minimizes the RMSD variance among the others sampled (time 48,300 ps) was chosen to be used for docking experiments, i.e., the most representative structure of the whole simulation.
2.3. Odorant Molecules Setup
Quantum chemical calculations, at the DFT level, were used to prepare the odorant molecules for docking, i.e., to obtain an optimized structure for each odorant. Calculations were done with the hybrid density functional B3LYP [29] together with the 6-31 + G (d,p) basis set. All molecules were computed with the Gaussian 09 [30] suite of programs in a vacuum and without vibrational corrections. After obtaining the most stable/probable conformation, OpenBabel [31] was used to transform Gaussian outputs to the PDBQT format, suitable to be used in AutoDock Vina [32].
The physicochemical properties were collected from three preferential databases: PubChem [33], ChemSpider [34] and The Good Scents Company [35]. Whenever possible, values obtained experimentally were selected. Further, in the case of stereoisomers, preference was given to the most abundant structure in nature. The properties listed were molecular weight (MW), logP, vapor pressure (Vp; logVp was used), number of double bonds (nº DB) and the degree of unsaturation (DoU). This last property is calculated according to Equation (1), where C is the number of carbons, N the number of nitrogens, X is the number of halogens and H the number of hydrogens. For a saturated molecule (only single bonds and no ring), DoU will be 0.
2.4. Principal Component Analysis
Principal component analysis (PCA) from SPSS software [36] was used to correlate the data listed or obtained for the 60 odorant molecules against the binding energy predicted by using docking experiments. This methodology allows to reduce the dimensionality of data and to perceive straightforward the linear correlation among the variables. It is a way of in a single graph, to follow the relationship between the binding energy and all the properties of all odorant molecules, instead of using several linear correlation graphs (scatter plots) between only two variables (see SM), that will not reveal some pattern as in PCA.
2.5. Virtual Screening of Odorant Molecules
The affinity (ΔGbinding) of our odorant library was estimated by virtual screening all the 60 molecules against the hOBP structure, using AutoDock Vina [32]. The aim was to determine if when molecules share a particular structural or physicochemical characteristic, the binding mode and energy will be similar. In addition, docking experiments allowed us to search for different interaction patterns between pleasant and unpleasant odorant molecules.
In the hOBP middle structure, the grid box was settle comprising the barrel region with 22 × 32 × 24 grid points in a grid spacing of 1 Å. An exhaustiveness of 20 and num_modes = 20 was used for each docking run. Figure S1 shows the Vina grid box settled for virtual screening, where the barrel loops and bends (extremities) were also contemplated.
3. Results and Discussion
The Protein Data Bank (PDB) brings together some OBP X-ray structures from vertebrates, but only one structure is available for hOBP (ID: 4RUN) [37]. In this structure, chain A (containing the ligand) lacks the N-terminal portion, which could be important for protein function, as it is placed near the bottom of the barrel, but most important two residues placed at the barrel core differ from the hOBP canonical sequence, the Ser99 and the Asn112 (see alignment in Figure S2, Supplementary Materials), and the canonical residues in place (Cys and Lys, respectively) may be interacting residues with odor molecules.
For the docking experiments, we opted to use one MD structure (not flexible docking), which was the middle structure obtained through cluster analysis. This conformation represents the most frequent arrangement of hOBP in solution, therefore indirectly reflecting a dynamic perspective of the protein. Supplementary Materials (SM) shows the RMSD curves (Figure S3) for modeled hOBP, which is very stable, considering the number of flexible turns and bends connecting the β-sheet barrel. Additionally, Figure S4 (SM) presents the superposition of our middle structure to hOBP X-ray, revealing great structural similarity and conservation.
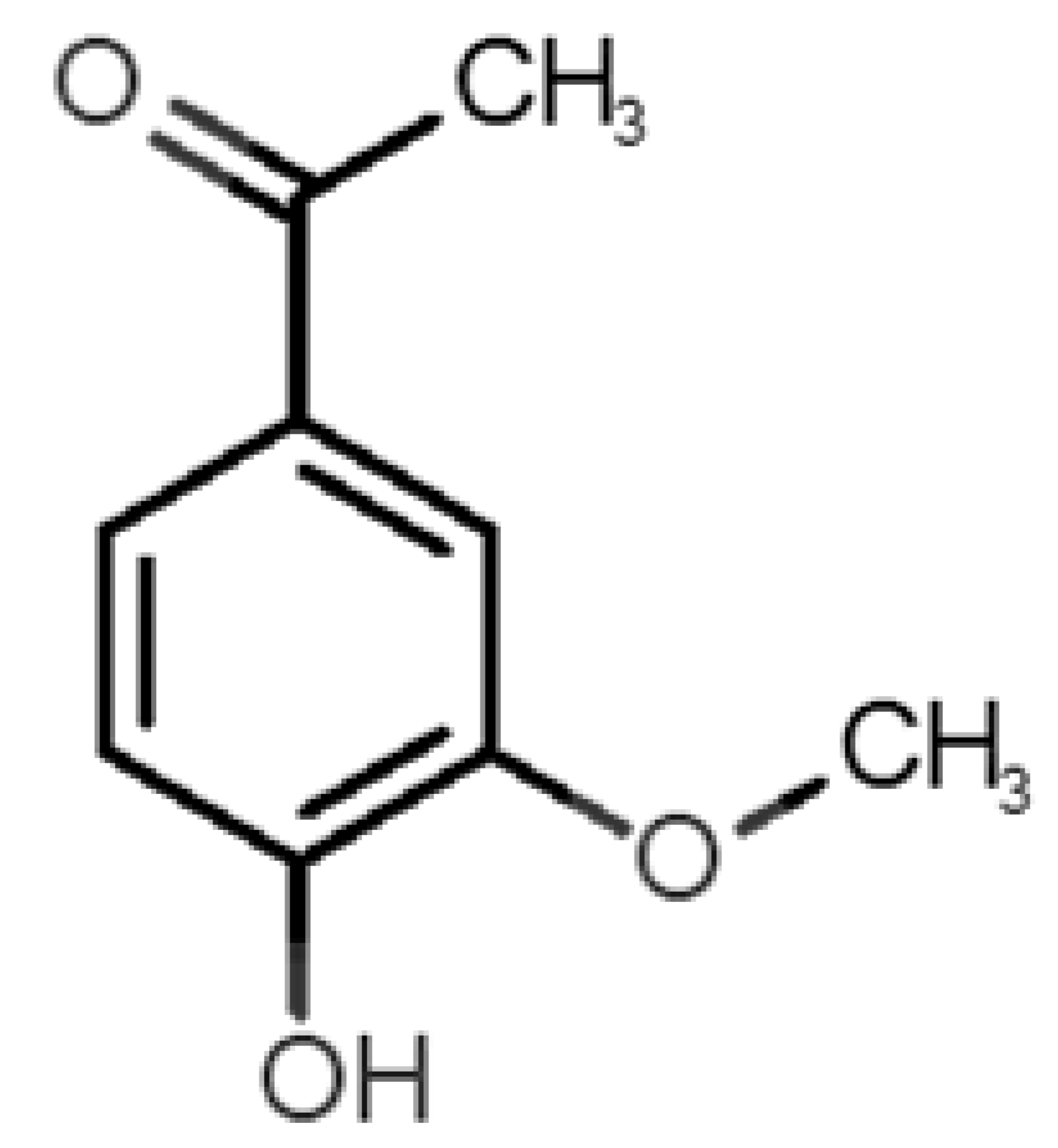
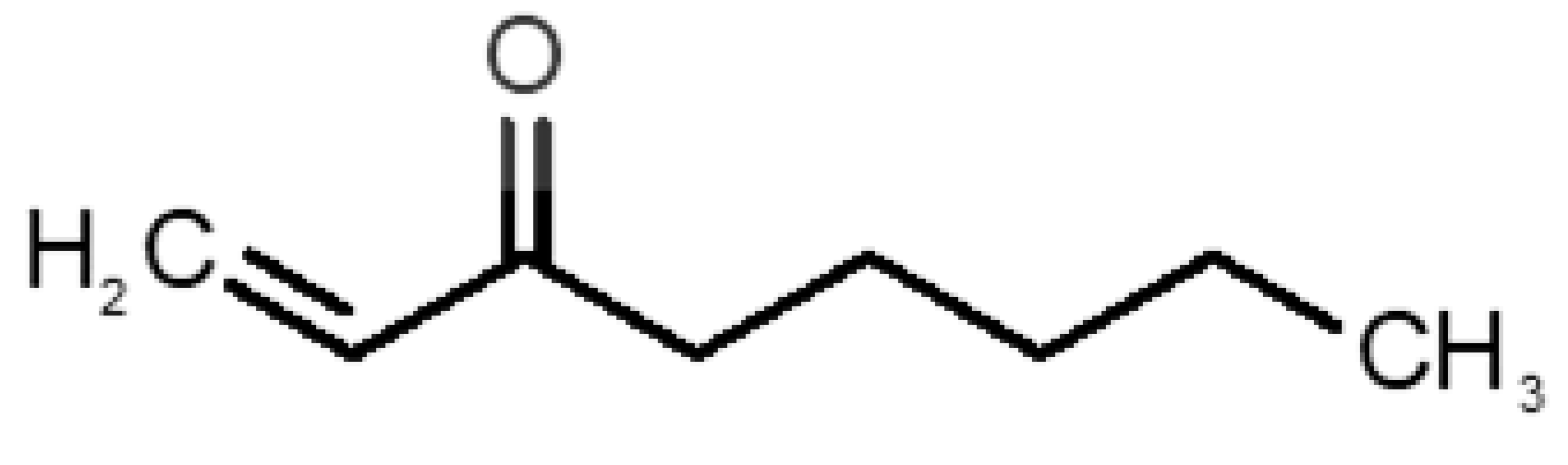
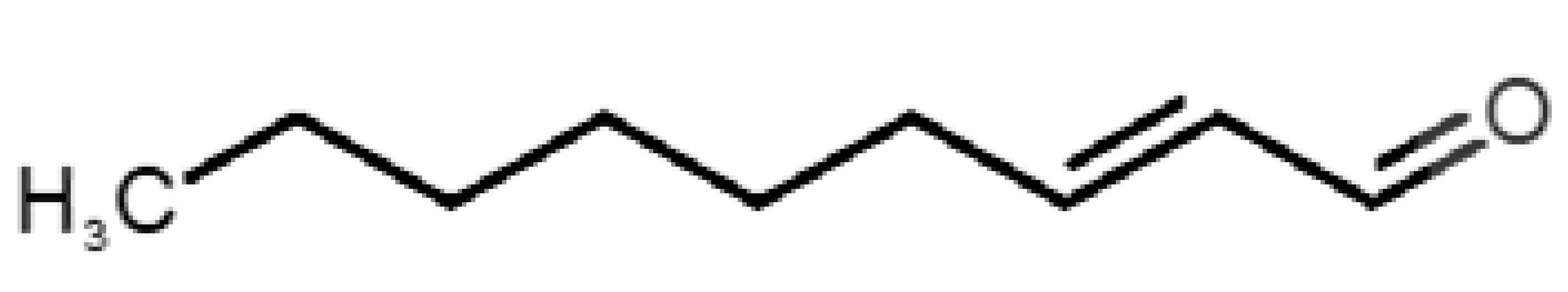
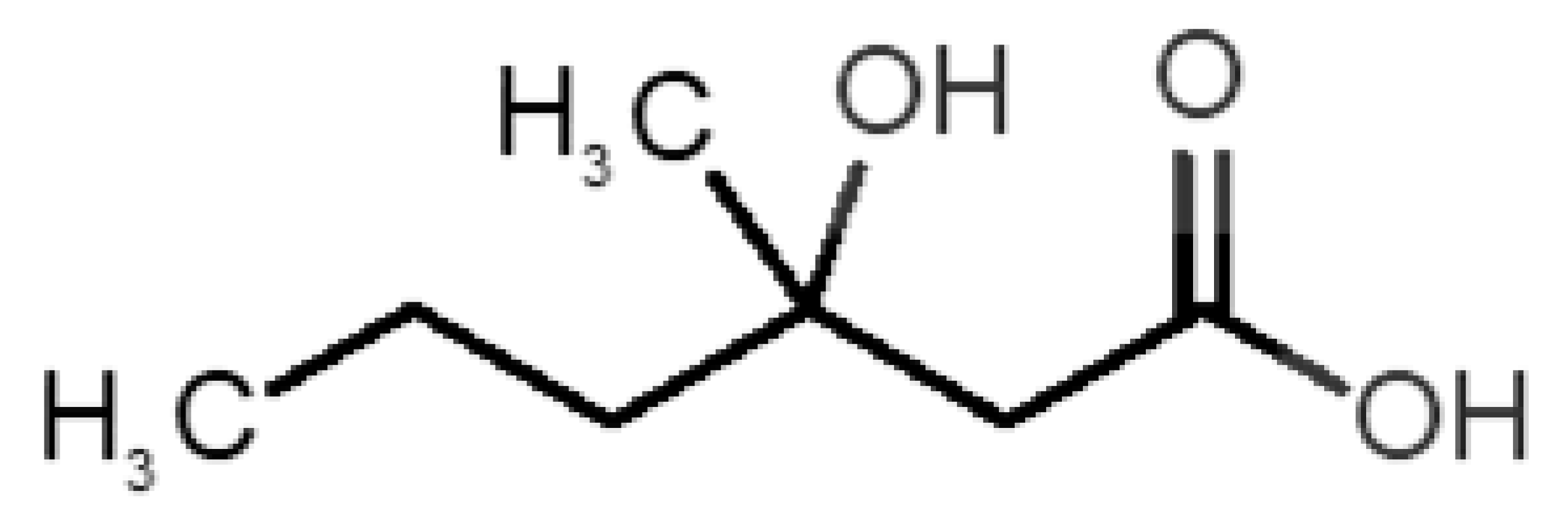
Table 1 and Table 2, along with Figure 1 and Figure 2, present a full description of the pleasant and unpleasant odorants under study. We set a diverse range of properties to infer if odorant molecules will cluster accordingly to physical or structural properties, or, for instance, if there is a dominant property, such as hydrophobicity, volatility, etc. Considering downstream biotechnology applications of hOBP, we also find it very important to understand how the general odorant profile (pleasant versus unpleasant) correlates with the hOBP-binding properties, namely location and affinity.

Table 1.
Description of pleasant odorant molecules under study, according to the physicochemical and structural characteristics.

Table 2.
Description of unpleasant odorant molecules under study, according to physicochemical and structural characteristics.
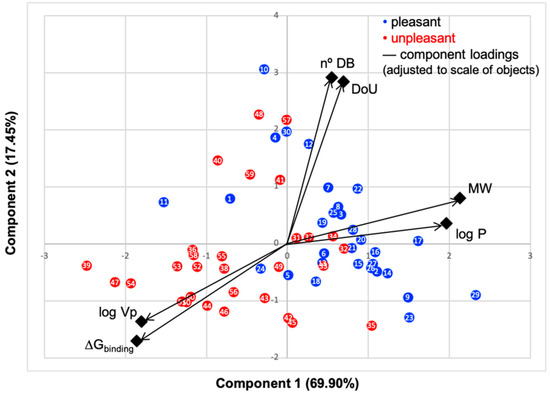
Figure 1.
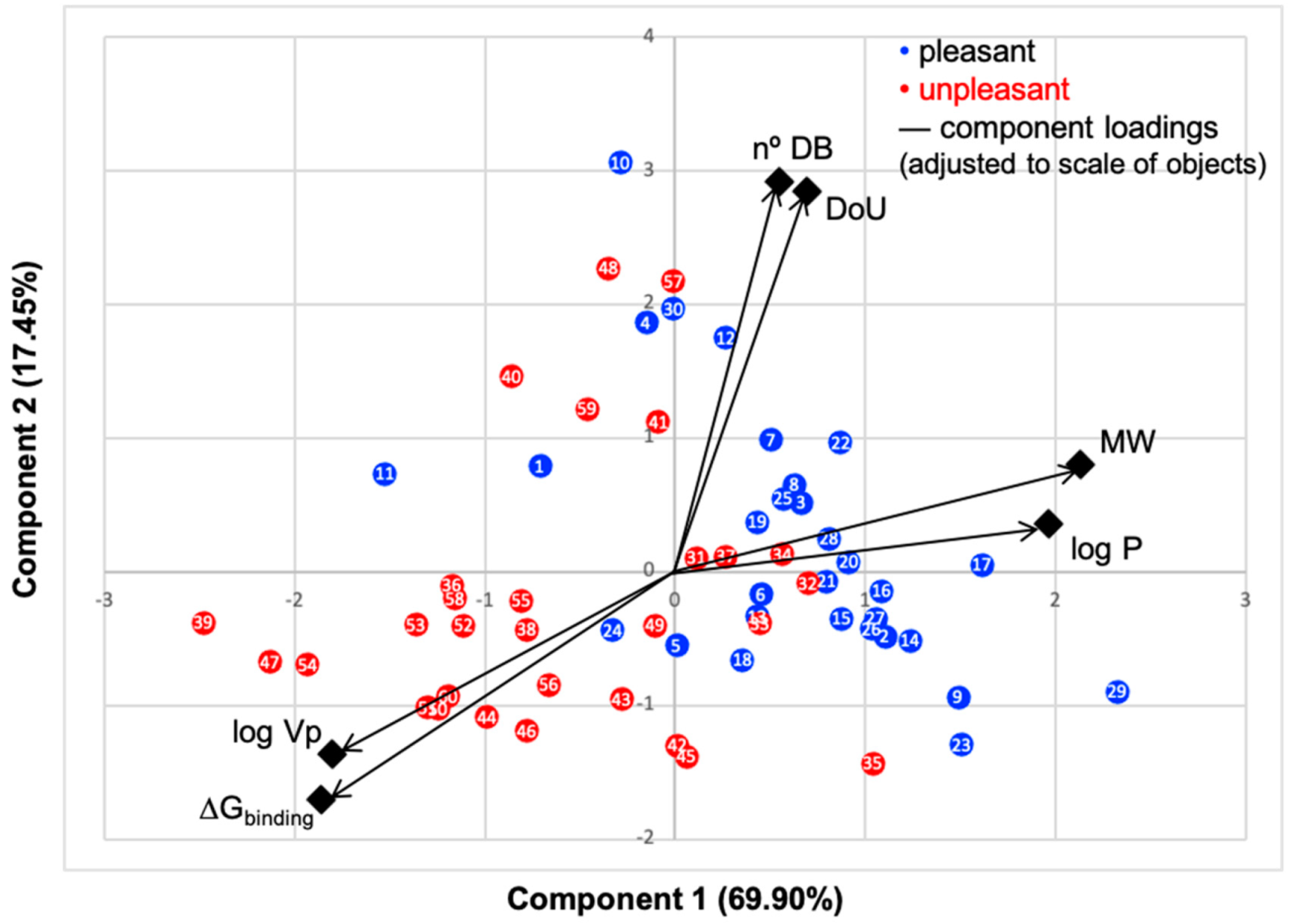
Principal component analysis (PCA) biplot of the first two components, objects factor scores and loadings, of data presented in Table 1 and Table 2. Rotated component matrix and scores were calculated using IBM SPSS Statistics software. Black color represents the loadings of the six variables scaled to objects’ values. Blue dots represent the molecules commonly perceived as pleasant odors, and red dots the unpleasant ones.
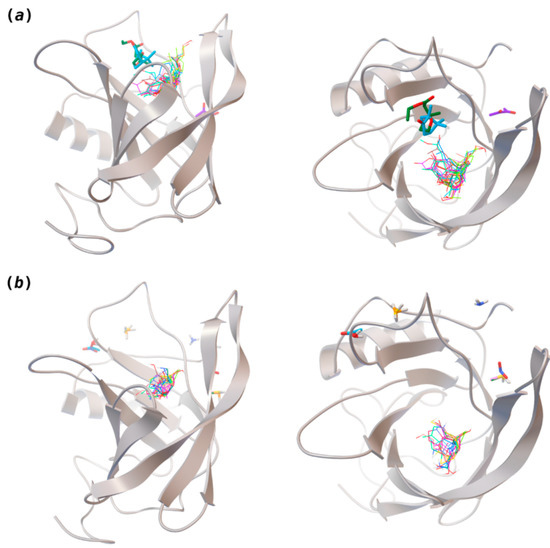
Figure 2.
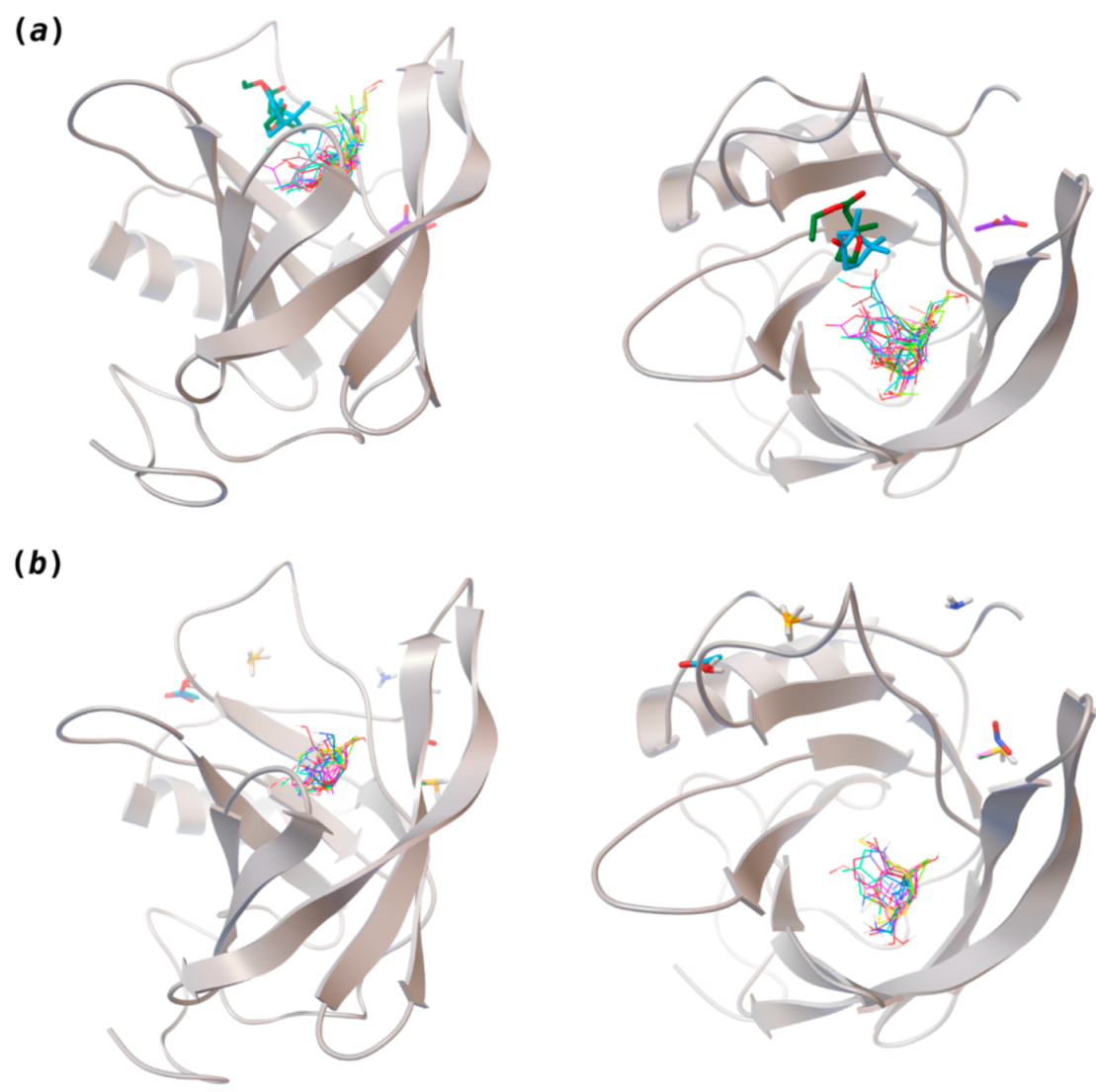
Interaction-binding modes of the selected (a) pleasant and (b) unpleasant odorant molecules, estimated with AutoDock Vina. Odorants in the most populated binding site are represented in diverse color lines, while odorants in other locations are shown in colored sticks. The human odorant-binding protein (hOBP) target is shown in the gray design.
The library of compounds herein was chosen to guarantee a diversified range of values in the addressed physicochemical properties. In addition, in the case of pleasant aromas, as the first studies on OBP-binding explored medium-size molecules from diverse chemical families [17,18,38], we adopted the same rationale, but taking into consideration, current odors generally used in the industry. Regarding unpleasant odors, their size range was widened, in part, due to our attempt to gather the principal body and decaying food odors or daily pungent aromas, more interesting according to our future purposes.
Looking at Table 1 and Table 2, we can infer some general differences in the properties under study between the chosen group of pleasant and unpleasant odorants; these differences between the two groups of molecules are significant according to the low p values and high size effects of the performed nonparametric analysis are observed (Table S2). The MW values are, on average, higher for pleasant odors. Similarly, the logP reveals more hydrophobic molecules in this category. In addition, the nº DB and the DoU values are superior for pleasant molecules, while the Vp values are overall higher for the unpleasant odorants. Regarding the calculated Gibbs free energy of binding to hOBP, the pleasant odorants presented on average more negative values than the unpleasant ones. Complementarily, binding energies were computed by using the 4RUN X-ray structure, sampling very similar results in comparison with our hOBP model (Table S3). This result is also important in order to evaluate the robustness of modeling techniques and MD simulations in predict and equilibrate protein structures, which will result in docking results in line with the screening in crystal structures.
Therefore, the combination of the differences among the properties of the chosen library of odorants leads to the hypothesis that the pleasant profile of odorant molecules may imply a high dependency on a carrier protein in order them to reach the ORs. Yet only the docking screening and the PCA analysis can unveil how the binding is structural-dependent for both classes.
PC1 represents 69.60% and PC2 17.45%; thus, over 87% of data are described by the first two components. The N°DB and DoU have some of their correlation impaired by the pleasant molecules, that is, while these properties impact the binding energy of the unpleasant molecules, for pleasant odorants, the linear correlation is very low, with an R-squared around 20% (Figures S8 and S9, SM), which results in a lower correlation in PCA plot. Although many odorants present double bonds and unsaturated moieties, these characteristics prove to be less constant through the series in their impact on the binding mode and energy. The variables logVp and ΔGbinding positively correlate, as they present a small angle between them, and both negatively correlate with MW and logP, due to the large angles approaching 180°. We, thus, clearly verify that the variables that most influence the binding of molecules to hOBP are logVp (volatility), logP (hydrophobicity) and MW.
Looking at the scatter plot, a distinct pattern can be perceived for the chosen pleasant and unpleasant odorants (blue and red dots), as they form two groups with minimal overlapping, indicating that in general, these aroma types behave differently. Unpleasant molecule properties correlate more with the binding energy than the pleasant ones, also seen in SM linear regressions (Supplementary Materials Figures S5–S9). This is very interesting, as it suggests that, in the selected library, the unpleasant profile preferentially modulates the binding. A less negative ΔGbinding, indicates, in general, that the aromas are less strongly linked to hOBP. The fact that odorants from the unpleasant group present less affinity towards hOBP and that they are smaller and more hydrophilic led us to infer that, in general, they are more capable of crossing the mucus barrier by diffusion as free molecules rather than being transported by hOBP. Kinetically, this could mean a faster binding of these unpleasant odors to their specific ORs. Perhaps these unpleasant odors, connected with “hazard” or less favorable signs from the surroundings, can therefore be perceived faster than pleasant aromas. Yet, olfaction is a very complex process [39]; other factors will influence the binding, as odorant concentration, mucus viscosity and temperature, for example.
The chemical classification (see Table 1 and Table 2; 2D formula) was not tracked through PCA as a variable; nevertheless, the PCA plot can show clusters of odorants based on their similarity. For pleasant molecules, it is possible to see some aggregation consonant with the chemical function. The odorants 5, 13, 18 and 24, which are esters, appear near each other. Similarly, the terpenes 3, 7, 8, 16, 19, 20, 21, 25 and 28 are closer. Interestingly, the molecules 10 and 29, the most negatives ΔGbinding are farther from the other objects. Molecule 11 is an outlier in this series in terms of MW, logP and Vp, having values more similar to the ones in the unpleasant library, explaining why it is closer to the red dots.
Highlights in the unpleasant series point to molecules 39, 47 and 54, shifted from others, which are some of the molecules outside the barrel. In fact, these odorants have the highest Vp of the series, making them outliers from the series. The molecules 31, 32, 33, 34 and 37, all with MW above 120.00 g/mol, are the molecules that are mixed with the pleasant ones. Importantly, the molecules 48 and 57, which clusters together, have the higher DoU values and are the ones with most negative ΔGbinding, this is probably due to this characteristic jointly with the MW value. The compound classification, however, is not a strong differentiating factor for the binding location, as the majority of compounds bind to the same hOBP place (Figure 2).
The binding of pleasant odorants occurs mostly at the same location (Figure 2a), at the top of the barrel. The few exceptions are observed for camphor (6), and fructone (13), which bind a little shifted from the rest of the series, and for diacetyl (11), which binds out of the barrel core. Diacetyl is the lightest pleasant molecule and also the most hydrophilic and volatile, as indicated by the values of MW, logP and Vp, which may explain the differentiated binding to hOBP. Regarding camphor, fructone and diacetyl, their most negative binding energies are associated with a binding site different from the other pleasant odorants; however, their following docking positions resembled the preferable binding location as for the other pleasant molecules.
Looking at the unpleasant odors (Figure 2b), the location of the binding sites is much more variable among molecules. The two acids, acetic acid (36) and nitric acid (52), bind at a similar position and are external to the barrel, with a very similar ΔGbinding. Ammonia (39) is also outside the barrel, but in a different location than the acids, as well as the pair phosphine (54) and hydrogen sulfide (47), and the trio methanethiol (50), methyl phosphine (51) and nitrogen dioxide (53). For the latter, the binding energies vary from −0.6 up to −2.8 kcal, which may suggest that these molecules may not even bind to OBP at all. Remarkably, the Vp of these molecules is the biggest on the unpleasant list, being the most volatile odorants. In addition, they are small and hydrophilic molecules with lower logP, which may facilitate the direct access to olfactory receptors by diffusion in the mucus, without the participation of OBP as a carrier.
The majority of the unpleasant molecules bind to the same site as the pleasant ones, making it possible to establish a binding site for hOBP. Yet, it is important to highlight that the binding energy varies considerably between these groups as a consequence of the intrinsic properties of the odorants. In fact, in 2002, Briand and coworkers [40] suggested that hOBP can discriminate among odors. They stated that OBP binds fatty acids rather than aldehydes and larger aldehydes, preferably to other chemical functions, concluding that hOBP binds more efficiently longer chains. Throughout our odors’ list, longer and bulky molecules as hedione (17), mefrosol (22) and sandalore (29), with 13, 12 and 14 carbons, respectively, present strong affinity towards hOBP. Remarkably, in the unpleasant group, the aldehyde 2-nonenal (32), which has one of the most negative ΔGbinding, has the longest hydrocarbon chain from the series.
In 2015, Di Pietrantonio and colleagues [41] developed a bio-electronic nose based on OBPs from pig and bovine, which was able to distinguish between octenol (mushroom, human breath and sweat) and carvone (mint), revealing a different sensitivity to pleasant and unpleasant molecules, in line with our findings.
If we compare the binding of 1-aminoanthracene (1-AMA) to porcine OBP (pOBP) [11] with the bindings calculated here for hOBP, in the former, 1-AMA binds almost on the edge of the barrel, while in hOBP, the binding is slightly more internal. Although it can be mentioned that hOBP receives the typical binding of hydrophobic molecules to lipocalins barrels and, structurally, OBPs from vertebrates are very similar, the amino acid sequence differs considerably, which may affect the hydrophobic interactions between the pair ligand–receptor. Hence it is of most importance the understanding of the binding site and energy in hOBP, as it is a less studied target to date. The amino acids frequently participating in the binding are Val49, Phe 66, Phe68, Ile79, Ile99 and Lys97. Both Phe residues are able to interact via π-stacking in a few cases.
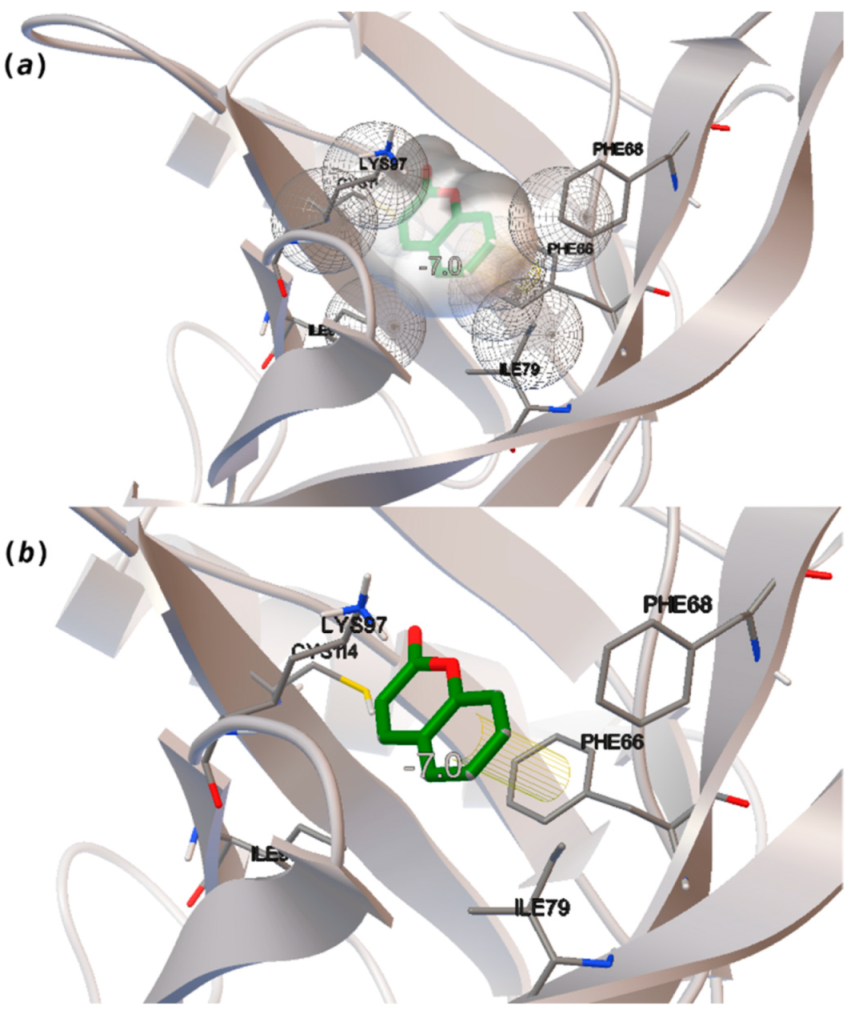
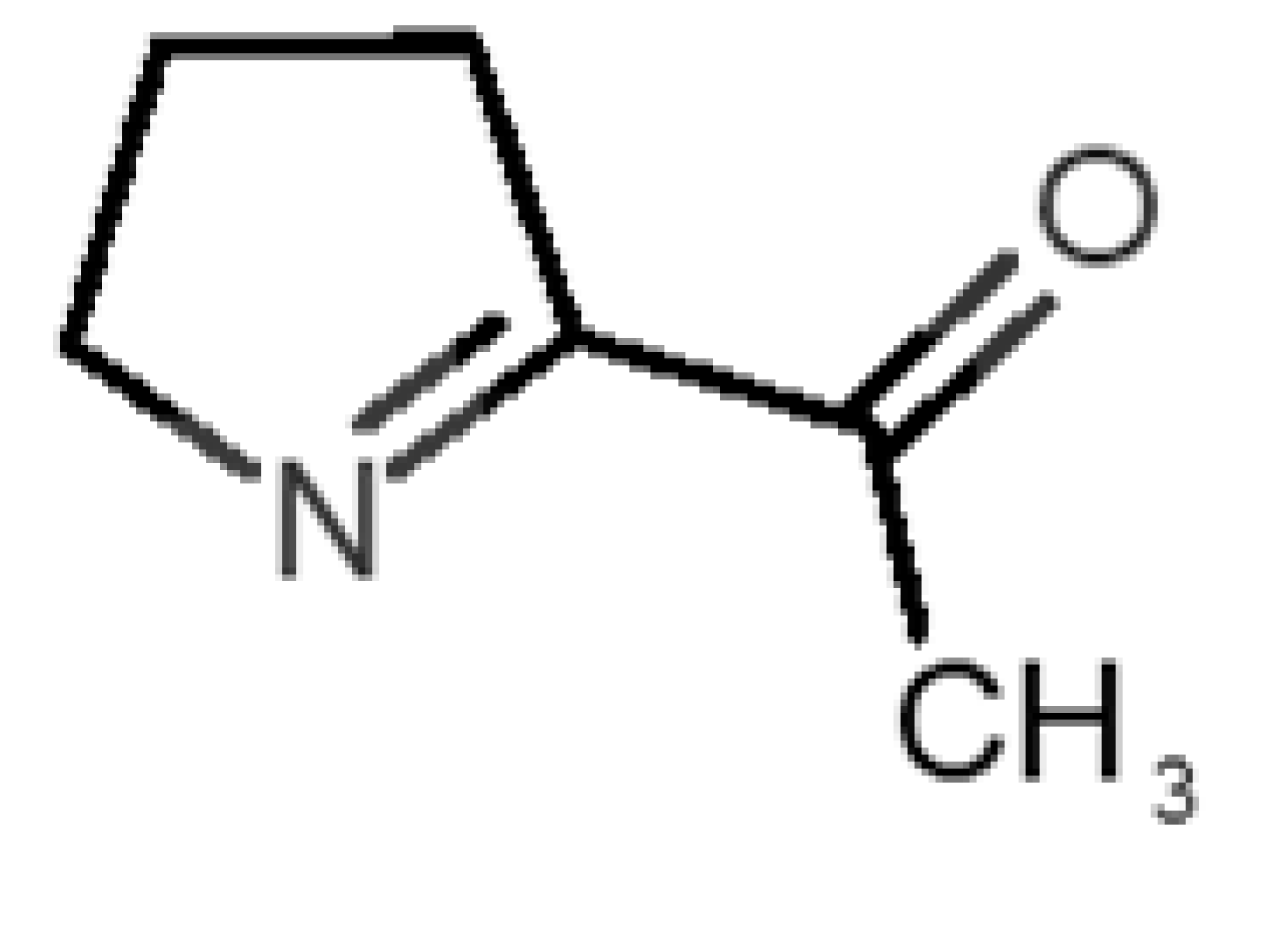
Coumarin (10) stands out from the other pleasant odorants with the higher ΔGbinding. The affinity to hOBP ranges from −3.6 kcal/mol to −7.0 kcal/mol within the selected group of pleasant odors, mostly resulting from a combination of the physicochemical properties MW, logP and Vp, where one or two of these variables have a greater role in binding. The MW and the logP are reflected in the number of hydrophobic atoms that interact with the hOBP, contributing in this way to the sum of forces in the docking algorithm, especially in van der Waals interactions. Yet, in the particular case of coumarin, a π-stacking interaction must have a higher weight leading to the highest binding affinity (Figure 3), even though coumarin does not have the highest MW or logP.
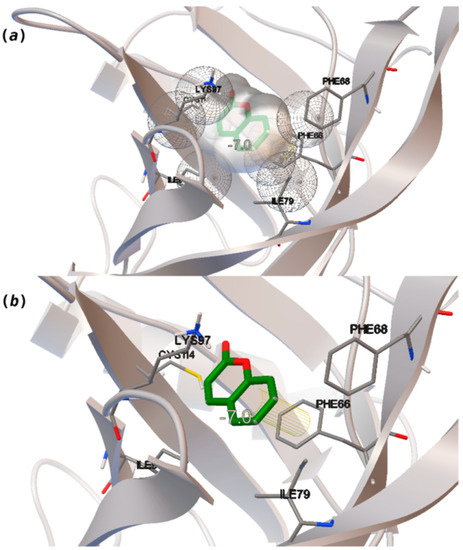
Figure 3.
Interaction binding modes of coumarin (10) in hOBP, estimated with AutoDock Vina. In (a), van der Waals/hydrophobic interactions are represented, and in (b), a pi-pi interaction between coumarin ring and Phe residue. Coumarin is shown in green sticks and amino acids participating in the binding, from hOBP, in gray lines.
4. Conclusions
OBP is well established as a key player in the sense of smell, being responsible for carrying and delivering the odorant molecules to the ORs [42]. In the present study, we address the binding of 60 odorant molecules to a very robust model of hOBP complete sequence, based on the 4RUN X-ray structure. Our findings demonstrate that the MW, the logP (hydrophobicity level), and the vapor pressure (Vp–volatility) are the physicochemical properties that impact more the ΔGbinding, being the chemical classification, the number of double bonds and the degree of unsaturation, less crucial variables for the binding event. Our findings suggested that for the selected unpleasant odorants, the above-cited properties correlate more with ΔGbinding than for the pleasant molecules. According to the here reported in silico data, OBP discriminates between molecules from the pleasant and unpleasant groups of chosen odorants, not only by the binding site but also and mainly through the “binding strength”. From a biotechnological perspective, the fact that OBP will bind preferentially “pleasant” odors than “unpleasant” ones or that pleasant odors may displace unpleasant ones from OBP is particularly important and useful for future OBP applications, such as sensors systems for the assessment of food contamination or for the evaluation of indoor air quality in buildings, as suggested by Di Pietrantonio [41].
Odor scientists, as well as fragrance professionals, have tried to establish comprehensive standards for the description, measurement, and prediction of odor quality characteristics. An olfactory classification system to define a perceptual space and facilitate objective communication about odors has not yet been found. Not all chemical molecules entering the nasal mucosa produce odor sensations, but those that do produce will vary in profile—pleasant or unpleasant—and intensity. Pleasantness is a significant aspect of odor perception: the neuronal processing of odors and emotions are partly overlapping in limbic structures, and the close connection is rooted in a point of human evolution when odors informed us on what to approach and what to avoid [43]. The odor quality and character also depend on odorant concentration, which was not contemplated in this work. The complexity is raised if the odor is a mixture of odorant molecules. In this case, the odor quality and character also depend on the organoleptic purity and not so much on the chemical purity. Looking just at the physicochemical properties here analyzed will hardly be sufficient to predict if a certain molecule can be perceived as pleasant or unpleasant. In spite of that, the findings of this work, in such a small group of odorant molecules compared to the odor space, are very relevant and may provide important clues for researchers devoted to developing predictive models of odor quality.
In this study, we found a clearly different pattern of hOBP-binding among the selected groups of pleasant and unpleasant odors, based on the MW, hydrophobicity and volatility of odorants. Given that, this work is important in emphasizing the role of OBP in human olfaction and in drawing attention to the possible role of this group of auxiliary proteins in the olfactory code.
This work is a first step in understanding the relationship between the odorant profile and its connection to hOBP, which can help in the technological application of aromas or in the development of sensors that mimic the function of this protein. Nevertheless, future steps in this research will go through the design of one or more OR structural models to assess the delivery of odorants to these important receptors and how OBP interacts with both classes of ORs.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-273X/11/2/145/s1. Odorant molecules CAS identification are listed in Table S1. Statistical analysis on the comparison of odorant properties (described in Table 1 and Table 2) in Table S2. The Vina grid box in Figure S1, the X-ray and Uniprot hOBP sequences alignment are presented in Figure S2 and the RMSD and cartoon structural superposition in Figures S3 and S4, respectively. Standard scatter plots/linear correlation regression are presented as SM, in Figures S5–S9, evaluating the relation of ΔGbinding to the other properties separately. The docking binding energies against the 4RUN X-ray structure are shown in Table S3.
Author Contributions
The manuscript was written through the contributions of all authors. All authors have given approval to the final version of the manuscript. T.G.C. conducted the modeling experiments; all authors contributed to the overall design of the project, analyses and discussion. All authors have read and agreed to the published version of the manuscript.
Funding
The following funding is acknowledged: the authors thank the support received from the Portuguese Foundation for Science and Technology (FCT) through the strategic funding of UID/BIO/04469/2019 unit and BioTecNorte Operation (NORTE-01-0145-FEDER-000004) funded by the European Regional Development Fund under the scope of Norte2020—Programa Operacional Regional do Norte.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Access to computing resources funded by the Project “Search-ON2: Revitalization of HPC infrastructure of UMinho” (NORTE-07-0162-FEDER-000086), co-founded by the North Portugal Regional Operational Programme (ON.2–O Novo Norte), under the National Strategic Reference Framework (NSRF), through the European Regional Development Fund (ERDF), is gratefully acknowledged.
Conflicts of Interest
The authors declare no conflict of interest. The authors declare no competing financial interest.
Abbreviations
| hOBP | Human odorant-binding protein |
| PCA | Principal component analysis |
| OR | Olfactory receptor |
| MD | Molecular dynamics |
| MW | Molecular weight |
| Vp | Vapor pressure (volatility) |
| logP | Partition coefficient (hydrophobicity level) |
| N°DB | Number of double bonds |
| DoU | Degree of unsaturation |
References
- Olender, T.; Keydar, I.; Pinto, J.M.; Tatarskyy, P.; Alkelai, A.; Chien, M.-S.; Fishilevich, S.; Restrepo, D.; Matsunami, H.; Gilad, Y.; et al. The human olfactory transcriptome. BMC Genom. 2016, 17, 1–18. [Google Scholar] [CrossRef]
- Persuy, M.-A.; Sanz, G.; Tromelin, A.; Thomas-Danguin, T.; Gibrat, J.-F.; Pajot-Augy, E. Mammalian olfactory receptors: Molecular mechanisms of odorant detection, 3D-modeling, and structure–activity relationships. Prog. Mol. Biol. Transl. Sci. 2015, 130, 1–36. [Google Scholar] [CrossRef]
- Vidic, J.; Grosclaude, J.; Monnerie, R.; Persuy, M.-A.; Badonnel, K.; Baly, C.; Caillol, M.; Briand, L.; Salesse, R.; Pajot-Augy, E. On a chip demonstration of a functional role for odorant binding protein in the preservation of olfactory receptor activity at high odorant concentration. Lab Chip 2008, 8, 678–688. [Google Scholar] [CrossRef]
- Wade, F.; Espagne, A.; Persuy, M.-A.; Vidic, J.; Monnerie, R.; Merola, F.; Pajot-Augy, E.; Sanz, G. Relationship between homo-oligomerization of a mammalian olfactory receptor and its activation state demonstrated by bioluminescence resonance energy transfer. J. Biol. Chem. 2011, 286, 15252–15259. [Google Scholar] [CrossRef]
- Da Silva, C.M.P.M.; Matamá, M.T.; Azoia, N.G.; Mansilha, C.; Casal, M.; Cavaco-Paulo, A. Odorant binding proteins: A biotechnological tool for odour control. Appl. Microbiol. Biotechnol. 2013, 98, 3629–3638. [Google Scholar] [CrossRef]
- Gonçalves, F.; Ribeiro, A.; Silva, C.; Cavaco-Paulo, A. Biotechnological applications of mammalian odorant-binding proteins. Crit. Rev. Biotechnol 2021, in press. [Google Scholar] [CrossRef]
- Pelosi, P.; Zhu, J.; Knoll, W. Odorant-binding proteins as sensing elements for odour monitoring. Sensors 2018, 18, 3248. [Google Scholar] [CrossRef]
- Pelosi, P.; Mastrogiacomo, R.; Iovinella, I.; Tuccori, E.; Persaud, K. Structure and biotechnological applications of odorant-binding proteins. Appl. Microbiol. Biotechnol. 2014, 98, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, F.; Ribeiro, A.; Silva, C.; Cavaco-Paulo, A. Release of fragrances from cotton functionalized with carbohydrate-binding module proteins. ACS Appl. Mater. Interfaces 2019, 11, 28499–28506. [Google Scholar] [CrossRef]
- Gonçalves, F.; Castro, T.G.; Azoia, N.G.; Ribeiro, A.; Silva, C.; Cavaco-Paulo, A. Two engineered OBPs with opposite temperature-dependent affinities towards 1-aminoanthracene. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Gonçalves, F.; Castro, T.G.; Nogueira, E.S.D.C.; Pires, R.A.; Silva, C.; Ribeiro, A.; Cavaco-Paulo, A. OBP fused with cell-penetrating peptides promotes liposomal transduction. Colloids Surf. B Biointerfaces 2018, 161, 645–653. [Google Scholar] [CrossRef]
- Pelosi, P. Odorant-binding proteins. Crit. Rev. Biochem. Mol. Biol. 1994, 29, 199–228. [Google Scholar] [CrossRef]
- Pevsner, J.; Snyder, S.H. Odorant-binding protein: Odorant transport function in the vertebrate nasal epithelium. Chem. Sens. 1990, 15, 217–222. [Google Scholar] [CrossRef]
- Whitson, K.B.; Whitson, S.R. Human odorant binding Protein 2a has two affinity states and is capable of binding some uremic toxins. Biochem. Anal. Biochem. 2014, 3, 1. [Google Scholar] [CrossRef]
- Tcatchoff, L.; Nespoulous, C.; Pernollet, J.-C.; Briand, L. A single lysyl residue defines the binding specificity of a human odorant-binding protein for aldehydes. FEBS Lett. 2006, 580, 2102–2108. [Google Scholar] [CrossRef]
- Charlier, L.; Cabrol-Bass, D.; Golebiowski, J. How does human odorant binding protein bind odorants? The case of aldehydes studied by molecular dynamics. C. R. Chim. 2009, 12, 905–910. [Google Scholar] [CrossRef]
- Tegonia, M.; Pelosi, P.; Vincenta, F.; Spinellia, S.; Campanacci, V.; Grollic, S.; Ramonic, R.; Cambillaua, C. Mammalian odorant binding proteins. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzym. 2000, 1482, 229–240. [Google Scholar] [CrossRef]
- Pevsner, J.; Hou, V.; Snowman, A.M.; Snyder, S.H. Odorant-binding protein. characterization of ligand binding. J. Biol. Chem. 1990, 265, 6118–6125. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Softwarex 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Abraham, M.J.; van der Spoel, D.; Lindahl, E.; Hess, B. Development Team GROMACS User Manual Version 5.1.5; Groningen University: Groningen, The Netherlands, 2017. [Google Scholar]
- Huang, W.; Lin, Z.; Van Gunsteren, W.F. Validation of the GROMOS 54A7 force field with respect to β-peptide folding. J. Chem. Theory Comput. 2011, 7, 1237–1243. [Google Scholar] [CrossRef] [PubMed]
- Schmid, N.; Eichenberger, A.P.; Choutko, A.; Riniker, S.; Winger, M.; Mark, A.E.; Van Gunsteren, W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011, 40, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Darden, A.T.; York, D.M.; Pedersen, L. Particle mesh Ewald: AnN⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hess, B. P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2007, 4, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open babel: An open chemical toolbox. J. Chemin. 2011, 3, 33. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/ (accessed on 15 June 2020).
- Royal Society of Chemistry. ChemSpider. Available online: http://www.chemspider.com/ (accessed on 15 June 2020).
- The Good Scents Company. Available online: http://www.thegoodscentscompany.com/index.html (accessed on 15 June 2020).
- IBM SPSS Statistics for Macintosh; IBM Corp. Academic License: Armonk, NY, USA, 2019; Version 26.0, Released 2019.
- Schiefner, A.; Freier, R.; Eichinger, A.; Skerra, A. Crystal structure of the human odorant binding protein, OBPIIa. Proteins Struct. Funct. Bioinform. 2015, 83, 1180–1184. [Google Scholar] [CrossRef] [PubMed]
- Monte, M.D.; Centini, M.; Anselmi, C.; Pelosi, P. Binding of selected odorants to bovine and porcine odorant-binding proteins. Chem. Sens. 1993, 18, 713–721. [Google Scholar] [CrossRef]
- Grabe, V.; Sachse, S. Fundamental principles of the olfactory code. Biosystems 2018, 164, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Briand, L.; Eloit, C.; Nespoulous, C.; Bézirard, V.; Huet, J.-C.; Henry, C.; Blon, F.; Trotier, D.; Pernollet, J.-C. Evidence of an odorant-binding protein in the human olfactory mucus: Location, structural characterization, and odorant-binding properties†. Biochemical 2002, 41, 7241–7252. [Google Scholar] [CrossRef] [PubMed]
- Di Pietrantonio, F.; Benetti, M.; Cannatà, D.; Verona, E.; Palla-Papavlu, A.; Fernández-Pradas, J.; Serra, P.; Staiano, M.; Varriale, A.; D’Auria, S. A surface acoustic wave bio-electronic nose for detection of volatile odorant molecules. Biosens. Bioelectron. 2015, 67, 516–523. [Google Scholar] [CrossRef]
- Archunan, G. Odorant binding proteins: A key player in the sense of smell. Bioinformation 2018, 14, 36–37. [Google Scholar] [CrossRef]
- Kaeppler, K.; Mueller, F. Odor classification: A review of factors influencing perception-based odor arrangements. Chem. Sens. 2013, 38, 189–209. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).