Clinical Phenotypes of Parkinson’s Disease Associate with Distinct Gut Microbiota and Metabolome Enterotypes
Abstract
:1. Introduction
2. Materials and Method
2.1. Patients and Clinical Assessment
2.2. Gut Microbiota Analysis
2.3. Gut Metabolome Analysis
2.4. Data and Statistical Analysis
3. Results
3.1. PD Patient Subtypes
3.2. Demographic and Clinical Characteristics
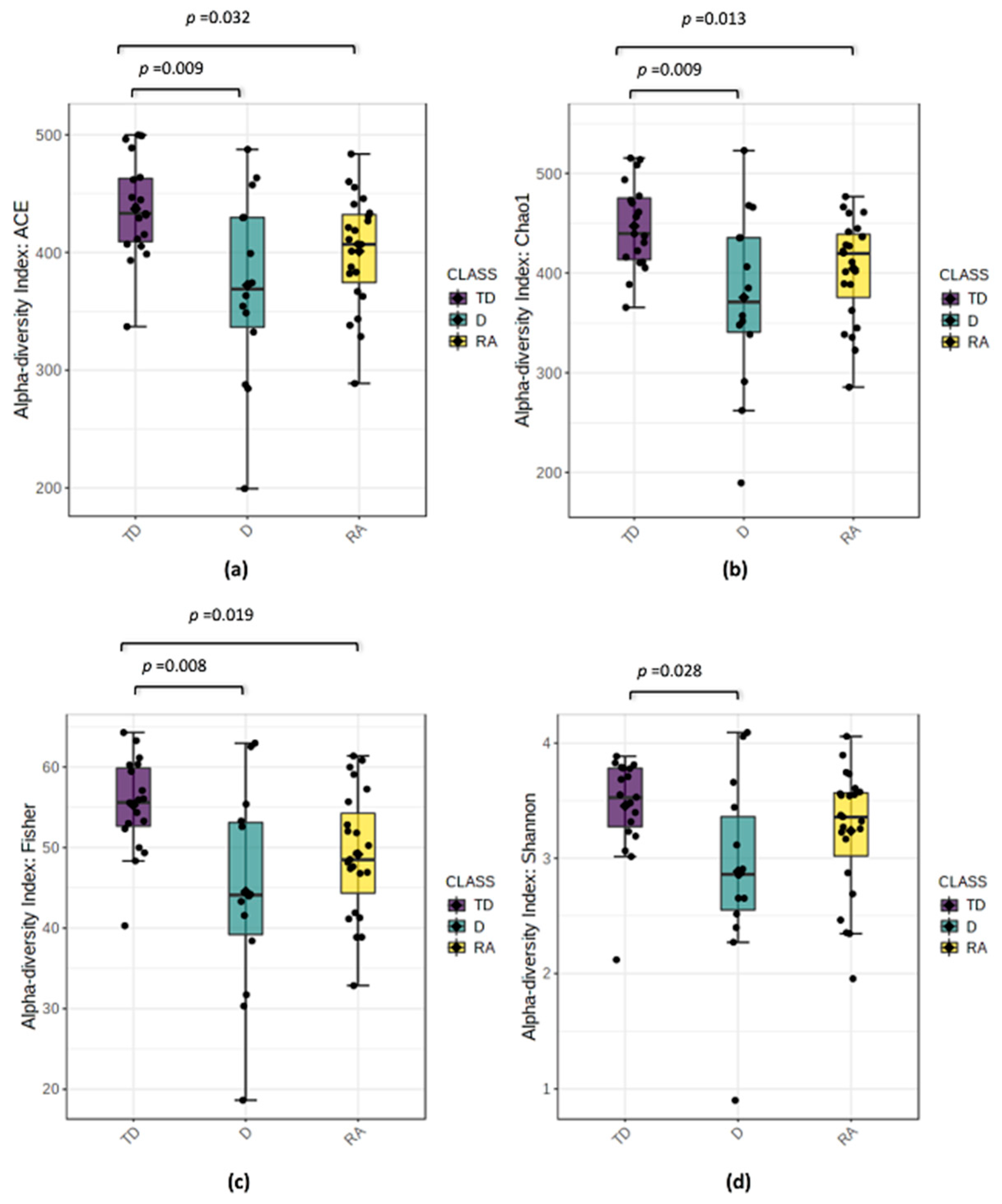
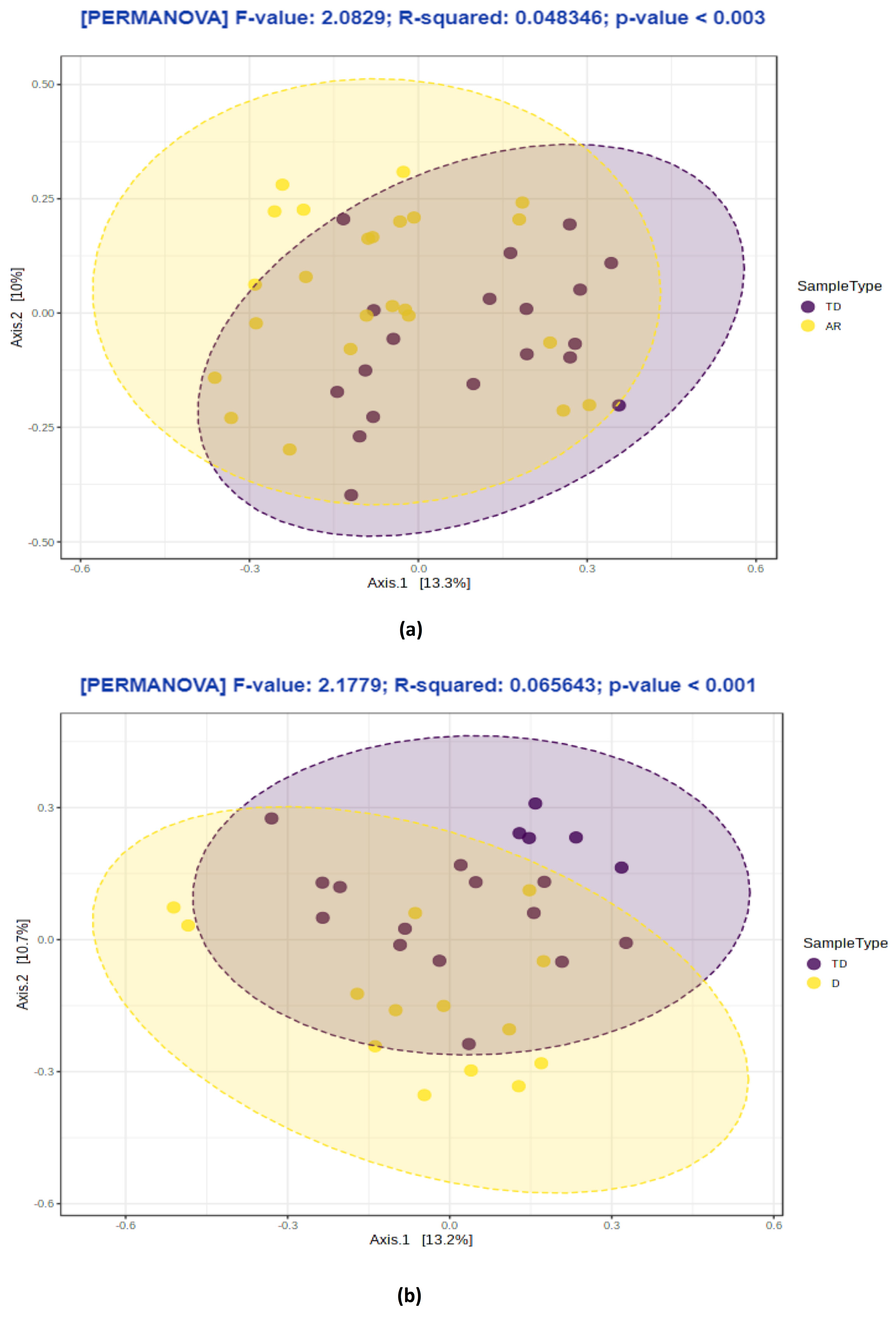
3.3. Changes in Bacterial Diversity Associate with Distinct PD Phenotypes
3.4. PD Phenotypes Differs in the Taxonomic Composition of Gut Microbiota
3.5. Distinct Metabolic Changes Associate with PD Phenotypes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Jankovic, J. Parkinson’s disease: Clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 2008, 79, 368–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, M.C.; Myers, P.S.; Weigand, A.J.; Foster, E.R.; Cairns, N.J.; Jackson, J.J.; Lessov-Schlaggar, C.N.; Perlmutter, J.S. Parkinson disease clinical subtypes: Key features & clinical milestones. Ann. Clin. Transl. Neurol. 2020, 7, 1272–1283. [Google Scholar] [PubMed]
- Mu, J.; Chaudhuri, K.R.; Bielza, C.; de Pedro-Cuesta, J.; Larrañaga, P.; Martinez-Martin, P. Parkinson’s disease subtypes identified from cluster analysis of motor and non-motor symptoms. Front. Aging Neurosci. 2017, 9, 301. [Google Scholar] [CrossRef]
- Nutt, J.G. Motor subtype in Parkinson’s disease: Different disorders or different stages of disease? Mov. Disord. 2016, 31, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Jankovic, J.; McDermott, M.; Carter, J.; Gauthier, S.; Goetz, C.; Golbe, L.; Huber, S.; Koller, W.; Olanow, C.; Shoulson, I.; et al. Variable expression of parkinson’s disease: A base-line analysis of the datatop cohort. Neurology 1990, 40, 1529. [Google Scholar] [CrossRef]
- Gelb, D.J.; Oliver, E.; Gilman, S. Diagnostic criteria for Parkinson disease. Arch. Neurol. 1999, 56, 33–39. [Google Scholar] [CrossRef]
- Lenz, F.A.; Kwan, H.C.; Martin, R.L.; Tasker, R.R.; Dostrovsky, J.O.; Lenz, Y.E. Single unit analysis of the human ventral thalamic nuclear group: Tremor-related activity in functionally identified cells. Brain 1994, 117, 531–543. [Google Scholar] [CrossRef]
- Deuschl, G.; Raethjen, J.; Baron, R.; Lindemann, M.; Wilms, H.; Krack, P. The pathophysiology of parkinsonian tremor: A review Introduction. J. Neurol. 2000, 247, V33–V48. [Google Scholar] [CrossRef]
- Zetusky, W.J.; Jankovic, J.; Pirozzolo, F.J. The heterogeneity of parkinson’s disease: Clinical and prognostic implications. Neurology 1985, 35, 522. [Google Scholar] [CrossRef]
- Eggers, C.; Kahraman, D.; Fink, G.R.; Schmidt, M.; Timmermann, L. Akinetic-rigid and tremor-dominant Parkinson’s disease patients show different patterns of FP-CIT Single photon emission computed tomography. Mov. Disord. 2011, 26, 416–423. [Google Scholar] [CrossRef]
- Nutt, J.G. Motor fluctuations and dyskinesia in Parkinson’s disease. Park. Relat. Disord. 2001, 8, 101–108. [Google Scholar] [CrossRef]
- Nutt, J.G. Levodopa-induced dyskinesia: Review, observations, and speculations. Neurology 1990, 40, 340. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.; Tang, B.S.; Song, C.Y.; Xu, Q.; Lou, M.X.; Liu, Z.H.; Yu, R.H.; Yan, X.X.; Guo, J.F. The relationship between the phenotype of Parkinson’s disease and levodopa-induced dyskinesia. Neurosci. Lett. 2013, 556, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Cersosimo, M.G.; Benarroch, E.E. Pathological correlates of gastrointestinal dysfunction in Parkinson’s disease. Neurobiol. Dis. 2012, 46, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Poewe, W. Non-motor symptoms in Parkinson’s disease. Eur. J. Neurol. 2008, 15, 14–20. [Google Scholar] [CrossRef]
- Korczyn, A.D. Autonomic nervous system disturbances in Parkinson’s disease. Adv. Neurol. 1990, 53, 463–468. [Google Scholar] [PubMed]
- Savica, R.; Carlin, J.M.; Grossardt, B.R.; Bower, J.H.; Ahlskog, J.E.; Maraganore, D.M.; Bharucha, A.E.; Rocca, W.A. Medical records documentation of constipation preceding Parkinson disease: A case-control study. Neurology 2009, 73, 1752–1758. [Google Scholar] [CrossRef] [Green Version]
- Noyce, A.J.; Bestwick, J.P.; Silveira-Moriyama, L.; Hawkes, C.H.; Giovannoni, G.; Lees, A.J.; Schrag, A. Meta-analysis of early nonmotor features and risk factors for Parkinson disease. Ann. Neurol. 2012, 72, 893–901. [Google Scholar] [CrossRef]
- Pfeiffer, R.F. Gastrointestinal dysfunction in Parkinson’s disease. Park. Relat. Disord. 2011, 17, 10–15. [Google Scholar] [CrossRef]
- Forsyth, C.B.; Shannon, K.M.; Kordower, J.H.; Voigt, R.M.; Shaikh, M.; Jaglin, J.A.; Estes, J.D.; Dodiya, H.B.; Keshavarzian, A. Increased intestinal permeability correlates with sigmoid mucosa alpha-synuclein staining and endotoxin exposure markers in early Parkinson’s disease. PLoS ONE 2011, 6, e28032. [Google Scholar] [CrossRef] [Green Version]
- Devos, D.; Lebouvier, T.; Lardeux, B.; Biraud, M.; Rouaud, T.; Pouclet, H.; Coron, E.; Bruley des Varannes, S.; Naveilhan, P.; Nguyen, J.M.; et al. Colonic inflammation in Parkinson’s disease. Neurobiol. Dis. 2013, 50, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Lebouvier, T.; Neunlist, M.; des Varannes, S.B.; Coron, E.; Drouard, A.; N’Guyen, J.M.; Chaumette, T.; Tasselli, M.; Paillusson, S.; Flamand, M.; et al. Colonic biopsies to assess the neuropathology of parkinson’s disease and its relationship with symptoms. PLoS ONE 2010, 5, e12728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunze, W.A.; Mao, Y.K.; Wang, B.; Huizinga, J.D.; Ma, X.; Forsythe, P.; Bienenstock, J. Lactobacillus reuteri enhances excitability of colonic AH neurons by inhibiting calcium-dependent potassium channel opening. J. Cell. Mol. Med. 2009, 13, 2261–2270. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yu, Y.B. Intestinal microbiota and chronic constipation. Springerplus 2016, 5, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef]
- Forsythe, P.; Kunze, W.A. Voices from within: Gut microbes and the CNS. Cell. Mol. Life Sci. 2013, 70, 55–69. [Google Scholar] [CrossRef]
- Paillusson, S.; Clairembault, T.; Biraud, M.; Neunlist, M.; Derkinderen, P. Activity-dependent secretion of alpha-synuclein by enteric neurons. J. Neurochem. 2013, 125, 512–517. [Google Scholar] [CrossRef]
- Sampson, T.R.; Debelius, J.W.; Thron, T.; Janssen, S.; Shastri, G.G.; Ilhan, Z.E.; Challis, C.; Schretter, C.E.; Rocha, S.; Gradinaru, V.; et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 2016, 167, 1469–1480.e12. [Google Scholar] [CrossRef] [Green Version]
- Del Tredici, K.; Braak, H. Sporadic Parkinson’s disease: Development and distribution of α-synuclein pathology. Neuropathol. Appl. Neurobiol. 2016, 42, 33–50. [Google Scholar] [CrossRef]
- Bedarf, J.R.; Hildebrand, F.; Coelho, L.P.; Sunagawa, S.; Bahram, M.; Goeser, F.; Bork, P.; Wüllner, U. Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med. 2017, 9, 1–13. [Google Scholar]
- Hasegawa, S.; Goto, S.; Tsuji, H.; Okuno, T.; Asahara, T.; Nomoto, K.; Shibata, A.; Fujisawa, Y.; Minato, T.; Okamoto, A.; et al. Intestinal dysbiosis and lowered serum lipopolysaccharide-binding protein in Parkinson’s disease. PLoS ONE 2015, 10, e0142164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill-Burns, E.M.; Debelius, J.W.; Morton, J.T.; Wissemann, W.T.; Lewis, M.R.; Wallen, Z.D.; Peddada, S.D.; Factor, S.A.; Molho, E.; Zabetian, C.P.; et al. Parkinson’s disease and Parkinson’s disease medications have distinct signatures of the gut microbiome. Mov. Disord. 2017, 32, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Hopfner, F.; Künstner, A.; Müller, S.H.; Künzel, S.; Zeuner, K.E.; Margraf, N.G.; Deuschl, G.; Baines, J.F.; Kuhlenbäumer, G. Gut microbiota in Parkinson disease in a northern German cohort. Brain Res. 2017, 1667, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Keshavarzian, A.; Green, S.J.; Engen, P.A.; Voigt, R.M.; Naqib, A.; Forsyth, C.B.; Mutlu, E.; Shannon, K.M. Colonic bacterial composition in Parkinson’s disease. Mov. Disord. 2015, 30, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wu, X.; Hu, X.; Wang, T.; Liang, S.; Duan, Y.; Jin, F.; Qin, B. Structural changes of gut microbiota in Parkinson’s disease and its correlation with clinical features. Sci. China Life Sci. 2017, 60, 1223–1233. [Google Scholar] [CrossRef]
- Scheperjans, F.; Aho, V.; Pereira, P.A.B.; Koskinen, K.; Paulin, L.; Pekkonen, E.; Haapaniemi, E.; Kaakkola, S.; Eerola-Rautio, J.; Pohja, M.; et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 2015, 30, 350–358. [Google Scholar] [CrossRef]
- Unger, M.M.; Spiegel, J.; Dillmann, K.U.; Grundmann, D.; Philippeit, H.; Bürmann, J.; Faßbender, K.; Schwiertz, A.; Schäfer, K.H. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Park. Relat. Disord. 2016, 32, 66–72. [Google Scholar] [CrossRef]
- Vascellari, S.; Palmas, V.; Melis, M.; Pisanu, S.; Cusano, R.; Uva, P.; Perra, D.; Madau, V.; Sarchioto, M.; Oppo, V.; et al. Gut Microbiota and Metabolome Alterations Associated with Parkinson’s Disease. mSystems 2020, 5. [Google Scholar] [CrossRef]
- Van Kessel, S.P.; Frye, A.K.; El-Gendy, A.O.; Castejon, M.; Keshavarzian, A.; van Dijk, G.; El Aidy, S. Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson’s disease. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Goldin, B.R.; Peppercorn, M.A.; Goldman, P. Contributions of host and intestinal microflora in the metabolism of L dopa by the rat. J. Pharmacol. Exp. Ther. 1973, 186, 160–166. [Google Scholar]
- Rekdal, V.M.; Bess, E.N.; Bisanz, J.E.; Turnbaugh, P.J.; Balskus, E.P. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science 2019, 364, eaau6323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, A.; Zheng, W.; He, Y.; Tang, W.; Wei, X.; He, R.; Huang, W.; Su, Y.; Huang, Y.; Zhou, H.; et al. Gut microbiota in patients with Parkinson’s disease in southern China. Park. Relat. Disord. 2018, 53, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Heintz-Buschart, A.; Pandey, U.; Wicke, T.; Sixel-Döring, F.; Janzen, A.; Sittig-Wiegand, E.; Trenkwalder, C.; Oertel, W.H.; Mollenhauer, B.; Wilmes, P. The nasal and gut microbiome in Parkinson’s disease and idiopathic rapid eye movement sleep behavior disorder. Mov. Disord. 2018, 33, 88–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabbrini, G.; Brotchie, J.M.; Grandas, F.; Nomoto, M.; Goetz, C.G. Levodopa-induced dyskinesias. Mov. Disord. 2007, 22, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Sofi, F.; Dinu, M.; Pagliai, G.; Marcucci, R.; Casini, A. Validation of a literature-based adherence score to Mediterranean diet: The MEDI-LITE score. Int. J. Food Sci. Nutr. 2017, 68, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Vascellari, S.; Melis, M.; Cossu, G.; Melis, M.; Serra, A.; Palmas, V.; Perra, D.; Oppo, V.; Fiorini, M.; Cusano, R.; et al. Genetic variants of TAS2R38 bitter taste receptor associate with distinct gut microbiota traits in Parkinson’s disease: A pilot study. Int. J. Biol. Macromol. 2020, 165, 665–674. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Zeng, M.Y.; Inohara, N.; Nuñez, G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017, 10, 18–26. [Google Scholar] [CrossRef] [Green Version]
- Niehaus, I.; Lange, J.H. Endotoxin: Is it an environmental factor in the cause of Parkinson’s disease? Occup. Environ. Med. 2003, 60, 378. [Google Scholar] [CrossRef] [Green Version]
- Kelly, L.P.; Carvey, P.M.; Keshavarzian, A.; Shannon, K.M.; Shaikh, M.; Bakay, R.A.E.; Kordower, J.H. Progression of intestinal permeability changes and alpha-synuclein expression in a mouse model of Parkinson’s disease. Mov. Disord. 2014, 29, 999–1009. [Google Scholar] [CrossRef]
- Gao, H.M.; Zhang, F.; Zhou, H.; Kam, W.; Wilson, B.; Hong, J.S. Neuroinflammation and α-synuclein dysfunction potentiate each other, driving chronic progression of neurodegeneration in a mouse model of Parkinson’s disease. Environ. Health Perspect. 2011, 119, 807–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soret, R.; Chevalier, J.; De Coppet, P.; Poupeau, G.; Derkinderen, P.; Segain, J.P.; Neunlist, M. Short-Chain Fatty Acids Regulate the Enteric Neurons and Control Gastrointestinal Motility in Rats. Gastroenterology 2010, 138, 1772–1782.e4. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marras, C.; Lang, A. Parkinson’s disease subtypes: Lost in translation? J. Neurol. Neurosurg. Psychiatry 2013, 84, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Moretti, R.; Peinkhofer, C. B vitamins and fatty acids: What do they share with small vessel disease-related dementia? Int. J. Mol. Sci. 2019, 20, 5797. [Google Scholar] [CrossRef] [Green Version]
- Fricker, R.A.; Green, E.L.; Jenkins, S.I.; Griffin, S.M. The Influence of Nicotinamide on Health and Disease in the Central Nervous System. Int. J. Tryptophan Res. 2018, 11. [Google Scholar] [CrossRef] [Green Version]
- Yoshii, K.; Hosomi, K.; Sawane, K.; Kunisawa, J. Metabolism of dietary and microbial vitamin b family in the regulation of host immunity. Front. Nutr. 2019, 6, 48. [Google Scholar] [CrossRef] [Green Version]
- Pugin, B.; Barcik, W.; Westermann, P.; Heider, A.; Wawrzyniak, M.; Hellings, P.; Akdis, C.A.; O’Mahony, L. A wide diversity of bacteria from the human gut produces and degrades biogenic amines. Microb. Ecol. Health Dis. 2017, 28, 1353881. [Google Scholar] [CrossRef]
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef]
- Makletsova, M.G.; Syatkin, S.P.; Poleshchuk, V.V.; Urazgildeeva, G.R.; Chigaleychik, L.A.; Sungrapova, C.Y.; Illarioshkin, S.N. Polyamines in Parkinson’s Disease: Their Role in Oxidative Stress Induction and Protein Aggregation. J. Neurol. Res. 2019, 9, 1–7. [Google Scholar] [CrossRef]
- Pavlin, M.; Repič, M.; Vianello, R.; Mavri, J. The Chemistry of Neurodegeneration: Kinetic Data and Their Implications. Mol. Neurobiol. 2016, 53, 3400–3415. [Google Scholar] [CrossRef] [PubMed]
- Seiler, N. Oxidation of polyamines and brain injury. Neurochem. Res. 2000, 25, 471–490. [Google Scholar] [CrossRef] [PubMed]
- Takano, K.; Ogura, M.; Yoneda, Y.; Nakamura, Y. Oxidative metabolites are involved in polyamine-induced microglial cell death. Neuroscience 2005, 134, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Santoru, M.L.; Piras, C.; Murgia, A.; Palmas, V.; Camboni, T.; Liggi, S.; Ibba, I.; Lai, M.A.; Orrù, S.; Blois, S.; et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.; Sinick, J.; Esko, T.; Fischer, K.; Menni, C.; Zierer, J.; Matey-Hernandez, M.; Fortney, K.; Morgen, E.K. Circulating glucuronic acid predicts healthspan and longevity in humans and mice. Aging 2019, 11, 7694–7706. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, F.M.; Maison, N.; Holtrop, G.; Young, P.; Stevens, V.J.; Ince, J.; Johnstone, A.M.; Lobley, G.E.; Flint, H.J.; Louis, P. Phylogenetic distribution of genes encoding β-glucuronidase activity in human colonic bacteria and the impact of diet on faecal glycosidase activities. Environ. Microbiol. 2012, 14, 1876–1887. [Google Scholar] [CrossRef]
- Dashnyam, P.; Mudududdla, R.; Hsieh, T.J.; Lin, T.C.; Lin, H.Y.; Chen, P.Y.; Hsu, C.Y.; Lin, C.H. β-Glucuronidases of opportunistic bacteria are the major contributors to xenobiotic-induced toxicity in the gut. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Pellock, S.J.; Redinbo, M.R. Glucuronides in the gut: Sugar-driven symbioses between microbe and host. J. Biol. Chem. 2017, 292, 8569–8576. [Google Scholar] [CrossRef] [Green Version]
- Freire, C.; Koifman, S. Pesticide exposure and Parkinson’s disease: Epidemiological evidence of association. Neurotoxicology 2012, 33, 947–971. [Google Scholar] [CrossRef]
- Eisenhofer, G.; Aneman, Å.; Friberg, P.; Hooper, D.; Fåndriks, L.; Lonroth, H.; Hunyady, B.; Mezey, E. Substantial production of Dopamine in the human gastrointestinal tract. J. Clin. Endocrinol. Metab. 1997, 82, 3864–3871. [Google Scholar] [CrossRef]
- Reigstad, C.S.; Salmonson, C.E.; Rainey, J.F.; Szurszewski, J.H.; Linden, D.R.; Sonnenburg, J.L.; Farrugia, G.; Kashyap, P.C. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015, 29, 1395–1403. [Google Scholar] [CrossRef] [Green Version]
- Haskel, Y.; Hanani, M. Inhibition of gastrointestinal motility by MPTP via adrenergic and dopaminergic mechanisms. Dig. Dis. Sci. 1994, 39, 2364–2367. [Google Scholar] [CrossRef] [PubMed]
- Sikander, A.; Rana, S.V.; Prasad, K.K. Role of serotonin in gastrointestinal motility and irritable bowel syndrome. Clin. Chim. Acta 2009, 403, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Asano, Y.; Hiramoto, T.; Nishino, R.; Aiba, Y.; Kimura, T.; Yoshihara, K.; Koga, Y.; Sudo, N. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G1288–G1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritter, J.K. Roles of glucuronidation and UDP-glucuronosyltransferases in xenobiotic bioactivation reactions. Chem. Biol. Interact. 2000, 129, 171–193. [Google Scholar] [CrossRef]
- Braak, H.; Rüb, U.; Gai, W.P.; Del Tredici, K. Idiopathic Parkinson’s disease: Possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. J. Neural Transm. 2003, 110, 517–536. [Google Scholar] [CrossRef]
- Borghammer, P. How does parkinson’s disease begin? Perspectives on neuroanatomical pathways, prions, and histology. Mov. Disord. 2018, 33, 48–57. [Google Scholar] [CrossRef]
- Yang, D.; Zhao, D.; Ali Shah, S.Z.; Wu, W.; Lai, M.; Zhang, X.; Li, J.; Guan, Z.; Zhao, H.; Li, W.; et al. The Role of the Gut Microbiota in the Pathogenesis of Parkinson’s Disease. Front. Neurol. 2019, 10, 1155. [Google Scholar] [CrossRef] [Green Version]
- Uchihara, T.; Giasson, B.I. Propagation of alpha-synuclein pathology: Hypotheses, discoveries, and yet unresolved questions from experimental and human brain studies. Acta Neuropathol. 2016, 131, 49–73. [Google Scholar] [CrossRef] [Green Version]



| Phenotypes | TD | AR | D | p Value |
|---|---|---|---|---|
| 0.434 (TD-AR) | ||||
| n (%) | 19 (33.93%) | 23 (41.07%) | 14 (25.00%) | 0.301 (TD-D) |
| 0.070 (AR-D) |
| Variable | PD Patients (TD; n = 19) | PD Patients (AR; n = 23) | PD Patients (D; n = 14) |
|---|---|---|---|
| Age, mean ± SD | 72.00 ± 8.00 | 71.00 ± 11.07 | 68.00 ± 9.21 |
| BMI, mean ± SD | 27.00 ± 3.93 | 26.19 ± 3.05 | 25.61 ± 5.26 |
| Sex, n (%) | |||
| Male | 13 (68.42%) | 17 (73.91%) | 9 (64.29%) |
| Female | 6 (31.58%) | 6 (26.09%) | 5 (35.71%) |
| Constipation, n (%) | |||
| Yes | 9 (52.94%) | 9 (40.91%) | 9 (64.29%) |
| No | 8 (47.06%) | 13 (59.09%) | 5 (35.71%) |
| Missing | 2 | 1 | 0 |
| Coffee consumption, n (%) | |||
| Yes | 11 (64.71%) | 15 (71.43%) | 7 (50.00%) |
| No | 6 (35.29%) | 6 (28.57%) | 7 (50.00%) |
| Missing | 2 | 2 | 0 |
| Smoking status, n (%) | |||
| Yes | 1 (5.88%) | 0 (0%) | 3 (21.43%) |
| No | 16 (94.12%) | 19 (100%) | 11 (78.57%) |
| Missing | 2 | 4 | 0 |
| Duration of disease, n (%) | |||
| 0–3 years | 7 (41.18%) | 4 (21.05%) | 0 (0%) |
| 4–13 years | 10 (58.82%) | 10 (52.63%) | 7 (50.00%) |
| >13 years | 0 (0%) | 5 (26.32%) | 7 (50.00%) |
| Missing | 2 | 4 | 0 |
| Treatment, n (%) | |||
| Short LD | 8 (41.11%) | 5 (21.74%) | 1 (7.14%) |
| Long-term LD | 9 (47.37%) | 11 (47.83%) | 6 (42.85%) |
| L-CIG | 2 (10.53%) | 7 (30.43%) | 7 (50.00%) |
| Phenotype Comparison | Phylum | Family | Genus | ↓/↑ | MD | Adjusted p Value | |
|---|---|---|---|---|---|---|---|
| AR vs. TD | Actinobacteria | Brevibacteriaceae | ↓ | −0.417 | 0.000 | * 0.005 | |
| Brevibacteriun | ↓ | −0.417 | 0.000 | * 0.005 | |||
| Firmicutes | ↓ | −0.153 | 0.005 | − | |||
| Clostridiaceae | ↓ | −0.301 | 0.008 | − | |||
| Tindallia | ↓ | −0.744 | 0.003 | − | |||
| Gemellaceae | ↓ | −0.286 | 0.004 | − | |||
| Gemella | ↓ | −0.286 | 0.004 | − | |||
| Lachnospiraceae | ↓ | −0.366 | 0.004 | * 0.005 | |||
| Blautia | ↓ | −0.344 | 0.003 | * 0.014 | |||
| Coprococcus | ↓ | −0.931 | 0.002 | * 0.014 | |||
| Lachnospira | ↓ | −0.450 | 0.005 | * 0.036 | |||
| Ruminococcaceae | Faecalibacterium | ↓ | −0.403 | 0.009 | − | ||
| Peptostreptococcaceae | Sedimentibacter | ↑ | 0.424 | 0.000 | * 0.004 | ||
| Proteobacteria | Enterobacteriaceae | ↑ | 0.819 | 0.005 | * 0.010 | ||
| Escherichia | ↑ | 1.039 | 0.002 | − | |||
| Serratia | ↑ | 0.925 | 0.002 | * 0.026 | |||
| D vs. TD | Actinobacteria | Brevibacteriaceae | ↓ | −0.481 | 0.000 | * 0.004 | |
| Brevibacteriun | ↓ | −0.481 | 0.000 | * 0.004 | |||
| Firmicutes | ↓ | −0.215 | 0.002 | − | |||
| Clostridiaceae | ↓ | −0.454 | 0.002 | − | |||
| Tindallia | ↓ | −0.811 | 0.012 | − | |||
| Eubacteriaceae | ↓ | −0.199 | 0.002 | * 0.003 | |||
| Acetobacterium | ↓ | −0.422 | 0.002 | * 0.008 | |||
| Gemellaceae | ↓ | −0.394 | 0.000 | * 0.040 | |||
| Gemella | ↓ | −0.394 | 0.000 | * 0.040 | |||
| Lachnospiraceae | ↓ | −0.422 | 0.003 | * 0.008 | |||
| Blautia | ↓ | −0.407 | 0.004 | * 0.016 | |||
| Coprococcus | ↓ | −0.888 | 0.003 | * 0.036 | |||
| Lachnospira | ↓ | −0.740 | 0.000 | * 0.005 | |||
| Lactobacillaceae | ↑ | 0.095 | 0.002 | − | |||
| Lactobacillus | ↑ | 0.841 | 0.002 | − | |||
| Peptococcaceae | ↓ | −0.450 | 0.000 | * 0.037 | |||
| Peptostreptococcaceae | Sedimentibacter | ↓ | −0.893 | 0.000 | * 0.000 | ||
| Ruminococcaceae | ↓ | −0.487 | 0.002 | − | |||
| Faecalibacterium | ↓ | −0.493 | 0.004 | − | |||
| Proteobacteria | Enterobacteriaceae | ↑ | 1.061 | 0.003 | − | ||
| Escherichia | ↑ | 1.273 | 0.003 | * 0.019 | |||
| Serratia | ↑ | 1.062 | 0.004 | * 0.010 | |||
| D vs. AR | Firmicutes | Lactobacillaceae | ↑ | 0.814 | 0.008 | − | |
| Lactobacillus | ↑ | 0.749 | 0.007 | − | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vascellari, S.; Melis, M.; Palmas, V.; Pisanu, S.; Serra, A.; Perra, D.; Santoru, M.L.; Oppo, V.; Cusano, R.; Uva, P.; et al. Clinical Phenotypes of Parkinson’s Disease Associate with Distinct Gut Microbiota and Metabolome Enterotypes. Biomolecules 2021, 11, 144. https://doi.org/10.3390/biom11020144
Vascellari S, Melis M, Palmas V, Pisanu S, Serra A, Perra D, Santoru ML, Oppo V, Cusano R, Uva P, et al. Clinical Phenotypes of Parkinson’s Disease Associate with Distinct Gut Microbiota and Metabolome Enterotypes. Biomolecules. 2021; 11(2):144. https://doi.org/10.3390/biom11020144
Chicago/Turabian StyleVascellari, Sarah, Marta Melis, Vanessa Palmas, Silvia Pisanu, Alessandra Serra, Daniela Perra, Maria Laura Santoru, Valentina Oppo, Roberto Cusano, Paolo Uva, and et al. 2021. "Clinical Phenotypes of Parkinson’s Disease Associate with Distinct Gut Microbiota and Metabolome Enterotypes" Biomolecules 11, no. 2: 144. https://doi.org/10.3390/biom11020144
APA StyleVascellari, S., Melis, M., Palmas, V., Pisanu, S., Serra, A., Perra, D., Santoru, M. L., Oppo, V., Cusano, R., Uva, P., Atzori, L., Morelli, M., Cossu, G., & Manzin, A. (2021). Clinical Phenotypes of Parkinson’s Disease Associate with Distinct Gut Microbiota and Metabolome Enterotypes. Biomolecules, 11(2), 144. https://doi.org/10.3390/biom11020144









