Natural Extracts to Augment Energy Expenditure as a Complementary Approach to Tackle Obesity and Associated Metabolic Alterations
Abstract
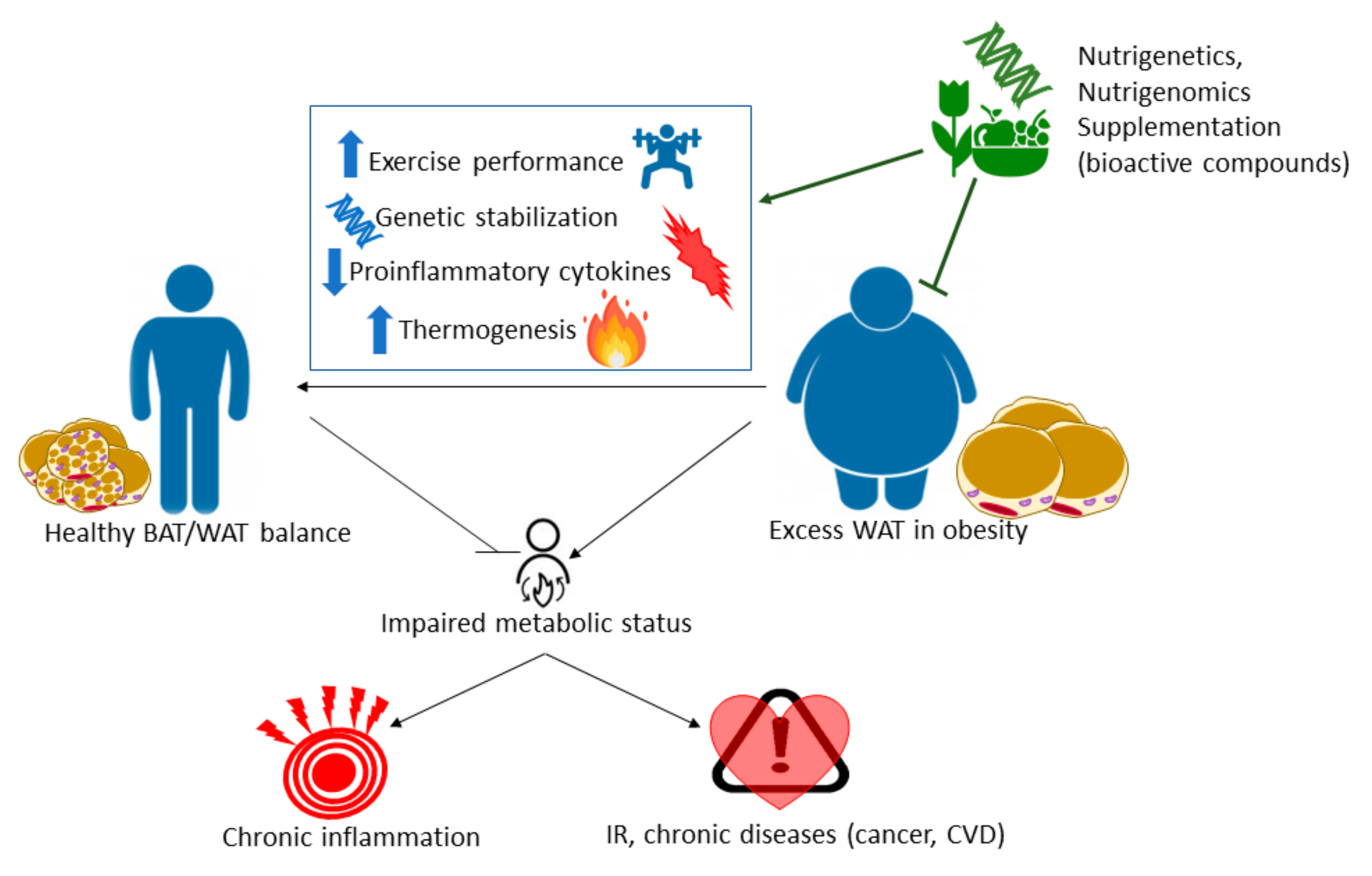
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Reagents
2.2. Natural Plant-Derived Extracts
2.3. Cell Culture
2.3.1. Cell Culture of Human HEK293
2.3.2. Human Adipocytes Cell Culture and In Vitro Differentiation
2.3.3. Human Myocytes Cell Culture and In Vitro Differentiation
2.4. MTT Assay
2.5. Gene Expression Analysis
2.6. Lipid Accumulation: Oil Red O Staining
2.7. Analysis of Mitochondrial Respiration
2.8. Data and Statistical Analysis
3. Results
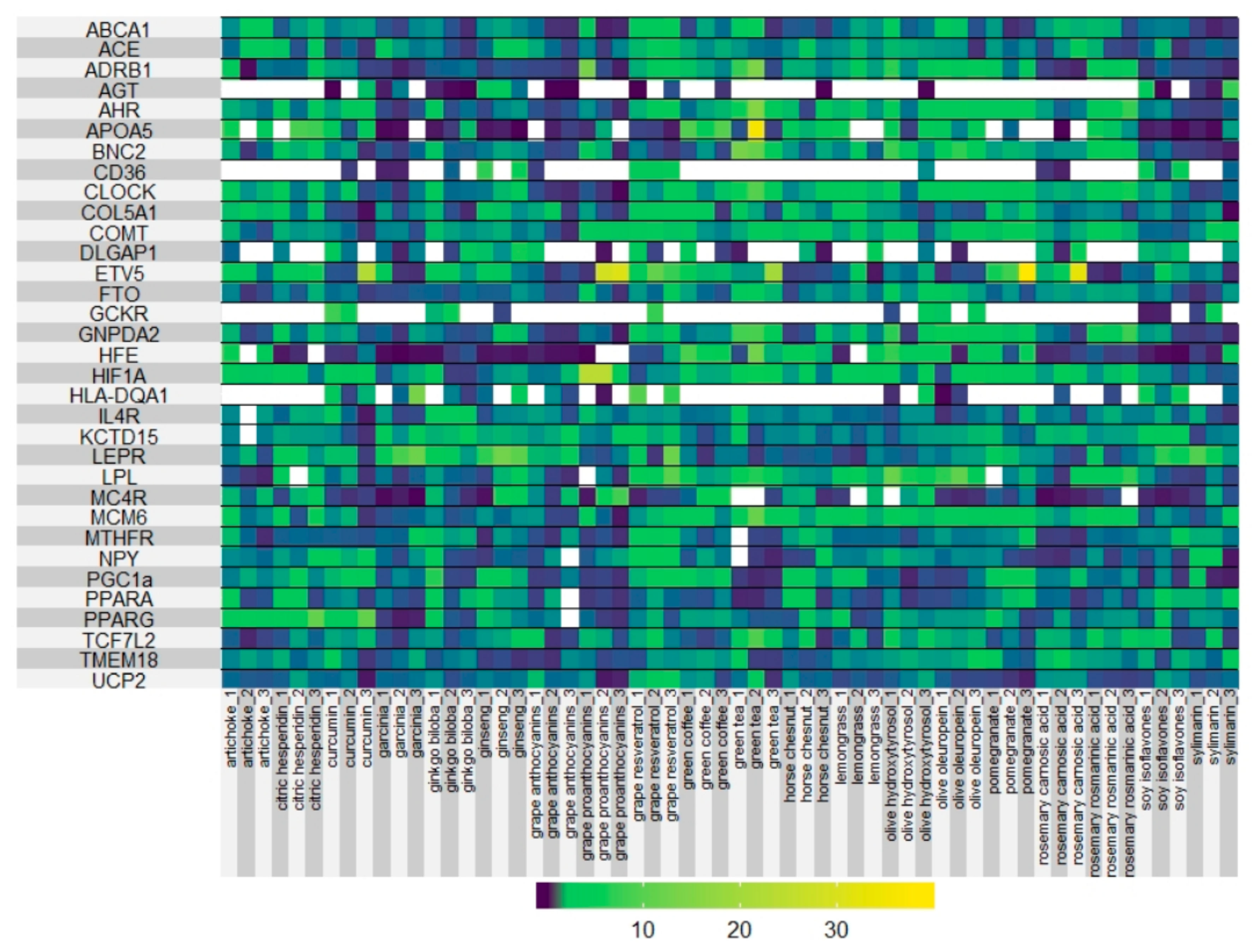
3.1. Screening of Plant-Derived Extracts in HEK293T Cell Line: MTT Assay and Gene Expression of Selected Metabolic Pathway
3.2. Screening of Plant-Derived Extracts in the Expression of Genes Implicated in the Activation of Thermogenesis and Mitochondrial Oxidative Phosphorylation
3.2.1. Effect of the Extracts on Cell Viability of Human Pre-Adipocytes and Human My-Blasts
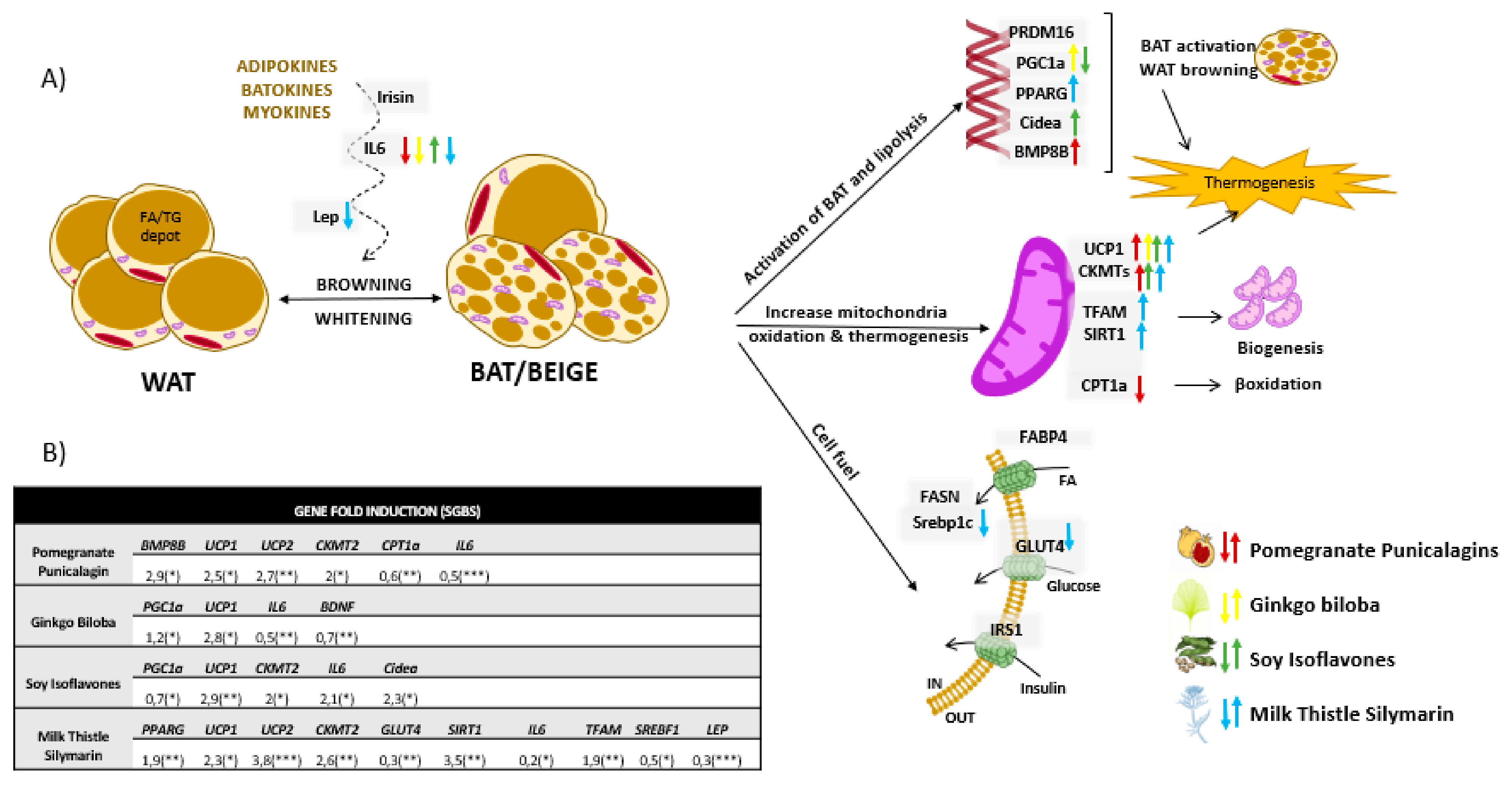
3.2.2. Effect of the Extracts in the Expression of Genes Related to the Activation of Thermogenesis and Mitochondrial Oxidative Phosphorylation
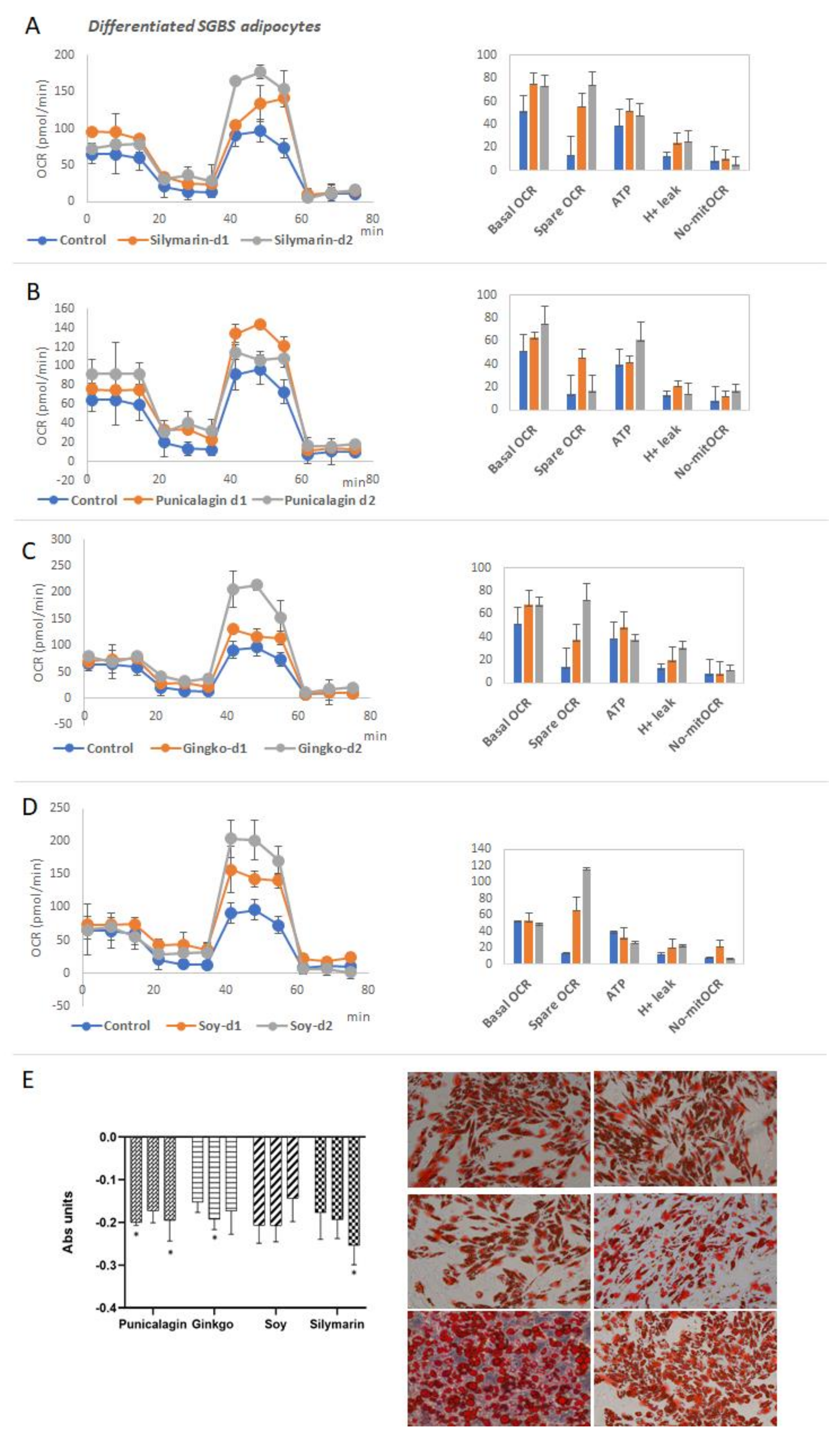
3.2.3. Functional Analysis of Mitochondrial Oxidative Phosphorylation and Lipid Accumulation in Differentiated Adipocytes
3.3. Functional Analysis of Mitochondrial Oxidative Phosphorylation in Differentiated Myocytes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Doo, M.; Kim, Y. Obesity: Interactions of Genome and Nutrients Intake. Prev. Nutr. Food Sci. 2015, 20, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ellulu, M.S.; Patimah, I.; Khaza’ai, H.; Rahmat, A.; Abed, Y. Obesity and Inflammation: The Linking Mechanism and the Complications. Arch. Med. Sci. AMS 2017, 13, 851–863. [Google Scholar] [CrossRef]
- Karastergiou, K.; Mohamed-Ali, V. The Autocrine and Paracrine Roles of Adipokines. Mol. Cell. Endocrinol. 2010, 318, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Iyengar, N.M.; Gucalp, A.; Dannenberg, A.J.; Hudis, C.A. Obesity and Cancer Mechanisms: Tumor Microenvironment and Inflammation. J. Clin. Oncol. 2016, 34, 4270–4276. [Google Scholar] [CrossRef] [PubMed]
- Michalakis, K.; Ilias, I. SARS-CoV-2 Infection and Obesity: Common Inflammatory and Metabolic Aspects. Diabetes Metab. Syndr. 2020, 14, 469–471. [Google Scholar] [CrossRef]
- Strieter, L.; Laddu, D.R.; Sainsbury, J.; Arena, R. The Importance of School-Based Healthy Living Initiatives: Introducing the Health and Wellness Academy Concept. Prog. Cardiovasc. Dis. 2019, 62, 68–73. [Google Scholar] [CrossRef]
- Hecht, E.M.; Layton, M.R.; Abrams, G.A.; Rabil, A.M.; Landy, D.C. Healthy Behavior Adherence: The National Health and Nutrition Examination Survey, 2005–2016. Am. J. Prev. Med. 2020, 59, 270–273. [Google Scholar] [CrossRef]
- Benito, P.J.; López-Plaza, B.; Bermejo, L.M.; Peinado, A.B.; Cupeiro, R.; Butragueño, J.; Rojo-Tirado, M.A.; González-Lamuño, D.; Gómez-Candela, C. Strength plus Endurance Training and Individualized Diet Reduce Fat Mass in Overweight Subjects: A Randomized Clinical Trial. Int. J. Environ. Res. Public Health 2020, 17, 2596. [Google Scholar] [CrossRef]
- Sarzynski, M.A.; Loos, R.J.F.; Lucia, A.; Pérusse, L.; Roth, S.M.; Wolfarth, B.; Rankinen, T.; Bouchard, C. Advances in Exercise, Fitness, and Performance Genomics in 2015. Med. Sci. Sports Exerc. 2016, 48, 1906–1916. [Google Scholar] [CrossRef]
- Hara, M.; Hachiya, T.; Sutoh, Y.; Matsuo, K.; Nishida, Y.; Shimanoe, C.; Tanaka, K.; Shimizu, A.; Ohnaka, K.; Kawaguchi, T.; et al. Genomewide Association Study of Leisure-Time Exercise Behavior in Japanese Adults. Med. Sci. Sports Exerc. 2018, 50, 2433–2441. [Google Scholar] [CrossRef]
- Kikuchi, N.; Miyamoto-Mikami, E.; Murakami, H.; Nakamura, T.; Min, S.-K.; Mizuno, M.; Naito, H.; Miyachi, M.; Nakazato, K.; Fuku, N. ACTN3 R577X Genotype and Athletic Performance in a Large Cohort of Japanese Athletes. Eur. J. Sport Sci. 2016, 16, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Braconi, D.; Bernardini, G.; Millucci, L.; Santucci, A. Foodomics for Human Health: Current Status and Perspectives. Expert Rev. Proteomics 2018, 15, 153–164. [Google Scholar] [CrossRef]
- Heianza, Y.; Qi, L. Gene-Diet Interaction and Precision Nutrition in Obesity. Int. J. Mol. Sci. 2017, 18, 787. [Google Scholar] [CrossRef] [PubMed]
- Leońska-Duniec, A.; Ahmetov, I.; Zmijewski, P. Genetic Variants Influencing Effectiveness of Exercise Training Programmes in Obesity—An Overview of Human Studies. Biol. Sport 2016, 33, 207–214. [Google Scholar] [CrossRef] [PubMed]
- de Toro-Martín, J.; Arsenault, B.J.; Després, J.-P.; Vohl, M.-C. Precision Nutrition: A Review of Personalized Nutritional Approaches for the Prevention and Management of Metabolic Syndrome. Nutrients 2017, 9, 913. [Google Scholar] [CrossRef]
- Bush, C.L.; Blumberg, J.B.; El-Sohemy, A.; Minich, D.M.; Ordovás, J.M.; Reed, D.G.; Behm, V.A.Y. Toward the Definition of Personalized Nutrition: A Proposal by The American Nutrition Association. J. Am. Coll. Nutr. 2020, 39, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Salinas, I.; de la Iglesia, R.; Colmenarejo, G.; Molina, S.; Reglero, G.; Martinez, J.A.; Loria-Kohen, V.; Ramirez de Molina, A. GCKR Rs780094 Polymorphism as A Genetic Variant Involved in Physical Exercise. Genes 2019, 10, 570. [Google Scholar] [CrossRef]
- Marcos-Pasero, H.; Aguilar-Aguilar, E.; de la Iglesia, R.; Espinosa-Salinas, I.; Gómez-Patiño, M.; Colmenarejo, G.; de Molina, A.R.; Reglero, G.; Loria-Kohen, V. Association of Calcium and Dairy Product Consumption with Childhood Obesity and the Presence of a Brain Derived Neurotropic Factor-Antisense (BDNF-AS) Polymorphism. Clin. Nutr. 2019, 38, 2616–2622. [Google Scholar] [CrossRef]
- Fraga, C.G.; Croft, K.D.; Kennedy, D.O.; Tomás-Barberán, F.A. The Effects of Polyphenols and Other Bioactives on Human Health. Food Funct. 2019, 10, 514–528. [Google Scholar] [CrossRef]
- Ferguson, L.R. Nutrigenomics Approaches to Functional Foods. J. Am. Diet. Assoc. 2009, 109, 452–458. [Google Scholar] [CrossRef]
- Gupta, A.K.; Rather, M.A.; Kumar Jha, A.; Shashank, A.; Singhal, S.; Sharma, M.; Pathak, U.; Sharma, D.; Mastinu, A. Artocarpus Lakoocha Roxb. and Artocarpus Heterophyllus Lam. Flowers: New Sources of Bioactive Compounds. Plants 2020, 9, 1329. [Google Scholar] [CrossRef] [PubMed]
- Mastinu, A.; Bonini, S.; Rungratanawanich, W.; Aria, F.; Marziano, M.; Maccarinelli, G.; Abate, G.; Premoli, M.; Memo, M.; Uberti, D. Gamma-Oryzanol Prevents LPS-Induced Brain Inflammation and Cognitive Impairment in Adult Mice. Nutrients 2019, 11, 728. [Google Scholar] [CrossRef] [PubMed]
- Shabani, M.; Sadeghi, A.; Hosseini, H.; Teimouri, M.; Babaei Khorzoughi, R.; Pasalar, P.; Meshkani, R. Resveratrol Alleviates Obesity-Induced Skeletal Muscle Inflammation via Decreasing M1 Macrophage Polarization and Increasing the Regulatory T Cell Population. Sci. Rep. 2020, 10, 3791. [Google Scholar] [CrossRef]
- Song, D.; Cheng, L.; Zhang, X.; Wu, Z.; Zheng, X. The Modulatory Effect and the Mechanism of Flavonoids on Obesity. J. Food Biochem. 2019, 43. [Google Scholar] [CrossRef]
- Konstantinidi, M.; Koutelidakis, A.E. Functional Foods and Bioactive Compounds: A Review of Its Possible Role on Weight Management and Obesity’s Metabolic Consequences. Medicines 2019, 6, 94. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.Y.; Brychta, R.J.; Abdul Sater, Z.; Cassimatis, T.M.; Cero, C.; Fletcher, L.A.; Israni, N.S.; Johnson, J.W.; Lea, H.J.; Linderman, J.D.; et al. Opportunities and Challenges in the Therapeutic Activation of Human Energy Expenditure and Thermogenesis to Manage Obesity. J. Biol. Chem. 2020, 295, 1926–1942. [Google Scholar] [CrossRef] [PubMed]
- van Marken Lichtenbelt, W.D.; Vanhommerig, J.W.; Smulders, N.M.; Drossaerts, J.M.A.F.L.; Kemerink, G.J.; Bouvy, N.D.; Schrauwen, P.; Teule, G.J.J. Cold-Activated Brown Adipose Tissue in Healthy Men. N. Engl. J. Med. 2009, 360, 1500–1508. [Google Scholar] [CrossRef] [PubMed]
- Trayhurn, P. Origins and Early Development of the Concept That Brown Adipose Tissue Thermogenesis Is Linked to Energy Balance and Obesity. Biochimie 2017, 134, 62–70. [Google Scholar] [CrossRef]
- Betz, M.J.; Enerbäck, S. Targeting Thermogenesis in Brown Fat and Muscle to Treat Obesity and Metabolic Disease. Nat. Rev. Endocrinol. 2018, 14, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, E.M.; Steinberg, G.R. Emerging Role of AMPK in Brown and Beige Adipose Tissue (BAT): Implications for Obesity, Insulin Resistance, and Type 2 Diabetes. Curr. Diab. Rep. 2018, 18, 80. [Google Scholar] [CrossRef]
- Bal, N.C.; Periasamy, M. Uncoupling of Sarcoendoplasmic Reticulum Calcium ATPase Pump Activity by Sarcolipin as the Basis for Muscle Non-Shivering Thermogenesis. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190135. [Google Scholar] [CrossRef] [PubMed]
- Palmer, B.F.; Clegg, D.J. Non-Shivering Thermogenesis as a Mechanism to Facilitate Sustainable Weight Loss: Non-Shivering Thermogenesis and Weight Loss. Obes. Rev. 2017, 18, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Fischer-Posovszky, P.; Newell, F.S.; Wabitsch, M.; Tornqvist, H.E. Human SGBS Cells—A Unique Tool for Studies of Human Fat Cell Biology. Obes. Facts 2008, 1, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Klusóczki, Á.; Veréb, Z.; Vámos, A.; Fischer-Posovszky, P.; Wabitsch, M.; Bacso, Z.; Fésüs, L.; Kristóf, E. Differentiating SGBS Adipocytes Respond to PPARγ Stimulation, Irisin and BMP7 by Functional Browning and Beige Characteristics. Sci. Rep. 2019, 9, s41598-s019. [Google Scholar] [CrossRef]
- Owens, J.; Moreira, K.; Bain, G. Characterization of Primary Human Skeletal Muscle Cells from Multiple Commercial Sources. In Vitro Cell. Dev. Biol. Anim. 2013, 49, 695–705. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, Y.-F.; Han, Y.; Zhang, R.; Qin, L.; Wang, M.-X.; Ma, L. CETP/LPL/LIPC Gene Polymorphisms and Susceptibility to Age-Related Macular Degeneration. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Vaughan, D.; Huber-Abel, F.A.; Graber, F.; Hoppeler, H.; Flück, M. The Angiotensin Converting Enzyme Insertion/Deletion Polymorphism Alters the Response of Muscle Energy Supply Lines to Exercise. Eur. J. Appl. Physiol. 2013, 113, 1719–1729. [Google Scholar] [CrossRef]
- Vaughan, D.; Brogioli, M.; Maier, T.; White, A.; Waldron, S.; Rittweger, J.; Toigo, M.; Wettstein, J.; Laczko, E.; Flück, M. The Angiotensin Converting Enzyme Insertion/Deletion Polymorphism Modifies Exercise-Induced Muscle Metabolism. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Yu, K.; Li, L.; Zhang, L.; Guo, L.; Wang, C. Association between MC4R Rs17782313 Genotype and Obesity: A Meta-Analysis. Gene 2020, 733, 144372. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Qi, Q.; Zheng, Y.; Huang, T.; Lathrop, M.; Zelenika, D.; Bray, G.A.; Sacks, F.M.; Liang, L.; Qi, L. Neuropeptide Y Genotype, Central Obesity, and Abdominal Fat Distribution: The POUNDS LOST Trial1,2. Am. J. Clin. Nutr. 2015, 102, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Poher, A.-L.; Altirriba, J.; Veyrat-Durebex, C.; Rohner-Jeanrenaud, F. Brown Adipose Tissue Activity as a Target for the Treatment of Obesity/Insulin Resistance. Front. Physiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Okla, M.; Kim, J.; Koehler, K.; Chung, S. Dietary Factors Promoting Brown and Beige Fat Development and Thermogenesis. Adv. Nutr. Int. Rev. J. 2017, 8, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Kalinovich, A.V.; de Jong, J.M.A.; Cannon, B.; Nedergaard, J. UCP1 in Adipose Tissues: Two Steps to Full Browning. Biochimie 2017, 134, 127–137. [Google Scholar] [CrossRef]
- Shabalina, I.G.; Petrovic, N.; de Jong, J.M.A.; Kalinovich, A.V.; Cannon, B.; Nedergaard, J. UCP1 in Brite/Beige Adipose Tissue Mitochondria Is Functionally Thermogenic. Cell Rep. 2013, 5, 1196–1203. [Google Scholar] [CrossRef]
- Bonet, M.L.; Mercader, J.; Palou, A. A Nutritional Perspective on UCP1-Dependent Thermogenesis. Biochimie 2017, 134, 99–117. [Google Scholar] [CrossRef]
- Jorge, A.; Jorge, G.; Paraíso, A.; Franco, R.; Vieira, L.; Hilzenderger, A.; Guimarães, A.; Andrade, J.; De-Paula, A.; Santos, S. Brown and White Adipose Tissue Expression of IL6, UCP1 and SIRT1 Are Associated with Alterations in Clinical, Metabolic and Anthropometric Parameters in Obese Humans. Exp. Clin. Endocrinol. Diabetes 2017, 125, 163–170. [Google Scholar] [CrossRef]
- Hilse, K.E.; Kalinovich, A.V.; Rupprecht, A.; Smorodchenko, A.; Zeitz, U.; Staniek, K.; Erben, R.G.; Pohl, E.E. The Expression of UCP3 Directly Correlates to UCP1 Abundance in Brown Adipose Tissue. Biochim. Biophys. Acta 2016, 1857, 72–78. [Google Scholar] [CrossRef]
- Lomax, T.M.; Ashraf, S.; Yilmaz, G.; Harmancey, R. Loss of Uncoupling Protein 3 Attenuates Western Diet–Induced Obesity, Systemic Inflammation, and Insulin Resistance in Rats. Obesity 2020, 28, 1687–1697. [Google Scholar] [CrossRef]
- Riley, C.L.; Dao, C.; Kenaston, M.A.; Muto, L.; Kohno, S.; Nowinski, S.M.; Solmonson, A.D.; Pfeiffer, M.; Sack, M.N.; Lu, Z.; et al. The Complementary and Divergent Roles of Uncoupling Proteins 1 and 3 in Thermoregulation. J. Physiol. 2016, 594, 7455–7464. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Cao, C.; Tao, C.; Ye, R.; Dong, M.; Zheng, Q.; Wang, C.; Jiang, X.; Qin, G.; Yan, C.; et al. Cold Adaptation in Pigs Depends on UCP3 in Beige Adipocytes. J. Mol. Cell Biol. 2017, 9, 364–375. [Google Scholar] [CrossRef]
- Caron, A.; Labbé, S.M.; Carter, S.; Roy, M.-C.; Lecomte, R.; Ricquier, D.; Picard, F.; Richard, D. Loss of UCP2 Impairs Cold-Induced Non-Shivering Thermogenesis by Promoting a Shift toward Glucose Utilization in Brown Adipose Tissue. Biochimie 2017, 134, 118–126. [Google Scholar] [CrossRef]
- Whittle, A.J.; Carobbio, S.; Martins, L.; Slawik, M.; Hondares, E.; Vázquez, M.J.; Morgan, D.; Csikasz, R.I.; Gallego, R.; Rodriguez-Cuenca, S.; et al. BMP8B Increases Brown Adipose Tissue Thermogenesis through Both Central and Peripheral Actions. Cell 2012, 149, 871–885. [Google Scholar] [CrossRef]
- Jiang, J.; Li, P.; Ling, H.; Xu, Z.; Yi, B.; Zhu, S. MiR-499/PRDM16 Axis Modulates the Adipogenic Differentiation of Mouse Skeletal Muscle Satellite Cells. Hum. Cell 2018, 31, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Wafer, R.; Tandon, P.; Minchin, J.E.N. The Role of Peroxisome Proliferator-Activated Receptor Gamma (PPARG) in Adipogenesis: Applying Knowledge from the Fish Aquaculture Industry to Biomedical Research. Front. Endocrinol. 2017, 8. [Google Scholar] [CrossRef]
- Dammone, G.; Karaz, S.; Lukjanenko, L.; Winkler, C.; Sizzano, F.; Jacot, G.; Migliavacca, E.; Palini, A.; Desvergne, B.; Gilardi, F.; et al. PPARγ Controls Ectopic Adipogenesis and Cross-Talks with Myogenesis During Skeletal Muscle Regeneration. Int. J. Mol. Sci. 2018, 19, 2044. [Google Scholar] [CrossRef]
- Chang, J.S.; Ghosh, S.; Newman, S.; Salbaum, J.M. A Map of the PGC-1α and NT-PGC-1α-Regulated Transcriptional Network in Brown Adipose Tissue. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Schnyder, S.; Handschin, C. Skeletal Muscle as an Endocrine Organ: PGC-1α, Myokines and Exercise. Bone 2015, 80, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.L. Sirt1 and the Mitochondria. Mol. Cells 2016, 39, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Khanh, V.C.; Zulkifli, A.F.; Tokunaga, C.; Yamashita, T.; Hiramatsu, Y.; Ohneda, O. Aging Impairs Beige Adipocyte Differentiation of Mesenchymal Stem Cells via the Reduced Expression of Sirtuin 1. Biochem. Biophys. Res. Commun. 2018, 500, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Artsi, H.; Gurt, I.; El-Haj, M.; Müller, R.; Kuhn, G.A.; Ben Shalom, G.; Cohen-Kfir, E.; Abramowitz, E.; Kandel, L.; Safran, O.; et al. Sirt1 Promotes a Thermogenic Gene Program in Bone Marrow Adipocytes: From Mice to (Wo)Men. Front. Endocrinol. 2019, 10. [Google Scholar] [CrossRef]
- Koh, J.-H.; Johnson, M.L.; Dasari, S.; LeBrasseur, N.K.; Vuckovic, I.; Henderson, G.C.; Cooper, S.A.; Manjunatha, S.; Ruegsegger, G.N.; Shulman, G.I.; et al. TFAM Enhances Fat Oxidation and Attenuates High-Fat Diet–Induced Insulin Resistance in Skeletal Muscle. Diabetes 2019, 68, 1552–1564. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.T.; Kolesar, J.E.; Kaufman, B.A. Mitochondrial Transcription Factor A Regulates Mitochondrial Transcription Initiation, DNA Packaging, and Genome Copy Number. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2012, 1819, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A Nutrient and Energy Sensor That Maintains Energy Homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Cantó, C.; Gerhart-Hines, Z.; Feige, J.N.; Lagouge, M.; Noriega, L.; Milne, J.C.; Elliott, P.J.; Puigserver, P.; Auwerx, J. AMPK Regulates Energy Expenditure by Modulating NAD+ Metabolism and SIRT1 Activity. Nature 2009, 458, 1056–1060. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, L.; Li, B.; Jiang, H.; Duan, Y.; Xie, Z.; Shuai, L.; Li, J.; Li, J. AMP-Activated Protein Kinase (AMPK) Regulates Energy Metabolism through Modulating Thermogenesis in Adipose Tissue. Front. Physiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Kazak, L.; Chouchani, E.T.; Jedrychowski, M.P.; Erickson, B.K.; Shinoda, K.; Cohen, P.; Vetrivelan, R.; Lu, G.Z.; Laznik-Bogoslavski, D.; Hasenfuss, S.C.; et al. A Creatine-Driven Substrate Cycle Enhances Energy Expenditure and Thermogenesis in Beige Fat. Cell 2015, 163, 643–655. [Google Scholar] [CrossRef]
- Kazak, L.; Rahbani, J.F.; Samborska, B.; Lu, G.Z.; Jedrychowski, M.P.; Lajoie, M.; Zhang, S.; Ramsay, L.C.; Dou, F.Y.; Tenen, D.; et al. Ablation of Adipocyte Creatine Transport Impairs Thermogenesis and Causes Diet-Induced Obesity. Nat. Metab. 2019, 1, 360–370. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, D.; Yuan, J.; Song, L.; Zhang, C.; Lin, Q.; Li, M.; Sheng, Z.; Ma, Z.; Lv, F.; et al. Hyperbaric Oxygen Ameliorates Insulin Sensitivity by Increasing GLUT4 Expression in Skeletal Muscle and Stimulating UCP1 in Brown Adipose Tissue in T2DM Mice. Front. Endocrinol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yu, H.; Shi, X.; Warren, C.R.; Lotta, L.A.; Friesen, M.; Meissner, T.B.; Langenberg, C.; Wabitsch, M.; Wareham, N.; et al. Functional Screening of Candidate Causal Genes for Insulin Resistance in Human Preadipocytes and Adipocytes. Circ. Res. 2020, 126, 330–346. [Google Scholar] [CrossRef]
- Man, X.; Hu, N.; Tan, S.; Tang, H.; Guo, Y.; Tang, C.; Liu, Y.; Tang, J.; Zhou, C.; Wang, F.; et al. Insulin Receptor Substrate-1 Inhibits High-fat Diet-induced Obesity by Browning of White Adipose Tissue through MiR-503. FASEB J. 2020, 34, 12308–12323. [Google Scholar] [CrossRef]
- Moseti, D.; Regassa, A.; Kim, W.-K. Molecular Regulation of Adipogenesis and Potential Anti-Adipogenic Bioactive Molecules. Int. J. Mol. Sci. 2016, 17, 124. [Google Scholar] [CrossRef] [PubMed]
- Crewe, C.; Zhu, Y.; Paschoal, V.A.; Joffin, N.; Ghaben, A.L.; Gordillo, R.; Oh, D.Y.; Liang, G.; Horton, J.D.; Scherer, P.E. SREBP-Regulated Adipocyte Lipogenesis Is Dependent on Substrate Availability and Redox Modulation of MTORC1. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Kobayashi, M.; Uta, S.; Otsubo, M.; Deguchi, Y.; Tagawa, R.; Mizunoe, Y.; Nakagawa, Y.; Shimano, H.; Higami, Y. Srebp-1c/Fgf21/Pgc-1α Axis Regulated by Leptin Signaling in Adipocytes—Possible Mechanism of Caloric Restriction-Associated Metabolic Remodeling of White Adipose Tissue. Nutrients 2020, 12, 2054. [Google Scholar] [CrossRef]
- Song, Z.; Xiaoli, A.M.; Yang, F. Regulation and Metabolic Significance of De Novo Lipogenesis in Adipose Tissues. Nutrients 2018, 10, 1383. [Google Scholar] [CrossRef] [PubMed]
- Guilherme, A.; Pedersen, D.J.; Henchey, E.; Henriques, F.S.; Danai, L.V.; Shen, Y.; Yenilmez, B.; Jung, D.; Kim, J.K.; Lodhi, I.J.; et al. Adipocyte Lipid Synthesis Coupled to Neuronal Control of Thermogenic Programming. Mol. Metab. 2017, 6, 781–796. [Google Scholar] [CrossRef]
- Prentice, K.J.; Saksi, J.; Hotamisligil, G.S. Adipokine FABP4 Integrates Energy Stores and Counterregulatory Metabolic Responses. J. Lipid Res. 2019, 60, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Dou, H.-X.; Wang, T.; Su, H.-X.; Gao, D.-D.; Xu, Y.-C.; Li, Y.-X.; Wang, H.-Y. Exogenous FABP4 Interferes with Differentiation, Promotes Lipolysis and Inflammation in Adipocytes. Endocrine 2020, 67, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Calderon-Dominguez, M.; Sebastián, D.; Fucho, R.; Weber, M.; Mir, J.F.; García-Casarrubios, E.; Obregón, M.J.; Zorzano, A.; Valverde, Á.M.; Serra, D.; et al. Carnitine Palmitoyltransferase 1 Increases Lipolysis, UCP1 Protein Expression and Mitochondrial Activity in Brown Adipocytes. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Warfel, J.D.; Vandanmagsar, B.; Dubuisson, O.S.; Hodgeson, S.M.; Elks, C.M.; Ravussin, E.; Mynatt, R.L. Examination of Carnitine Palmitoyltransferase 1 Abundance in White Adipose Tissue: Implications in Obesity Research. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2017, 312, R816–R820. [Google Scholar] [CrossRef]
- Kirk, B.; Feehan, J.; Lombardi, G.; Duque, G. Muscle, Bone, and Fat Crosstalk: The Biological Role of Myokines, Osteokines, and Adipokines. Curr. Osteoporos. Rep. 2020, 18, 388–400. [Google Scholar] [CrossRef]
- Rodríguez, A.; Catalán, V.; Ramírez, B.; Unamuno, X.; Portincasa, P.; Gómez-Ambrosi, J.; Frühbeck, G.; Becerril, S. Impact of Adipokines and Myokines on Fat Browning. J. Physiol. Biochem. 2020, 76, 227–240. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.W.; Cannon, B.; Nedergaard, J. Leptin: Is It Thermogenic? Endocr. Rev. 2019, 41, 232–260. [Google Scholar] [CrossRef] [PubMed]
- Caron, A.; Lee, S.; Elmquist, J.K.; Gautron, L. Leptin and Brain–Adipose Crosstalks. Nat. Rev. Neurosci. 2018, 19, 153–165. [Google Scholar] [CrossRef]
- Pandit, R.; Beerens, S.; Adan, R.A.H. Role of Leptin in Energy Expenditure: The Hypothalamic Perspective. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2017, 312, R938–R947. [Google Scholar] [CrossRef]
- Gaspar, R.C.; Muñoz, V.R.; Kuga, G.K.; Nakandakari, S.C.B.R.; Minuzzi, L.G.; Botezelli, J.D.; da Silva, A.S.R.; Cintra, D.E.; de Moura, L.P.; Ropelle, E.R.; et al. Acute Physical Exercise Increases Leptin-induced Hypothalamic Extracellular Signal-regulated Kinase1/2 Phosphorylation and Thermogenesis of Obese Mice. J. Cell. Biochem. 2019, 120, 697–704. [Google Scholar] [CrossRef]
- Palhinha, L.; Liechocki, S.; Hottz, E.D.; Pereira, J.A.; da Silva Pereira, J.A.; de Almeida, C.J.; Moraes-Vieira, P.M.M.; Bozza, P.T. Leptin Induces Proadipogenic and Proinflammatory Signaling in Adipocytes. Front. Endocrinol. 2019, 10. [Google Scholar] [CrossRef]
- Wang, P.; Loh, K.H.; Wu, M.; Morgan, D.A.; Schneeberger, M.; Yu, X.; Chi, J.; Kosse, C.; Kim, D.; Rahmouni, K.; et al. A Leptin–BDNF Pathway Regulating Sympathetic Innervation of Adipose Tissue. Nature 2020, 583, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Colitti, M.; Montanari, T. Brain-Derived Neurotrophic Factor Modulates Mitochondrial Dynamics and Thermogenic Phenotype on 3T3-L1 Adipocytes. Tissue Cell 2020, 66, 101388. [Google Scholar] [CrossRef]
- Boström, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Boström, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1α-Dependent Myokine That Drives Browning of White Fat and Thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Wu, Z.; Zhang, B.; Wang, C.; Mao, F.; Liu, X.; Hu, K.; Sun, X.; Jin, W.; Kuang, S. Fndc5 Loss-of-function Attenuates Exercise-induced Browning of White Adipose Tissue in Mice. FASEB J. 2019, 33, 5876–5886. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sotelo, D.; Roca-Rivada, A.; Baamonde, I.; Baltar, J.; Castro, A.I.; Domínguez, E.; Collado, M.; Casanueva, F.F.; Pardo, M. Lack of Adipocyte-Fndc5/Irisin Expression and Secretion Reduces Thermogenesis and Enhances Adipogenesis. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Knudsen, J.G.; Murholm, M.; Carey, A.L.; Biensø, R.S.; Basse, A.L.; Allen, T.L.; Hidalgo, J.; Kingwell, B.A.; Febbraio, M.A.; Hansen, J.B.; et al. Role of IL-6 in Exercise Training- and Cold-Induced UCP1 Expression in Subcutaneous White Adipose Tissue. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Mishra, D.; Richard, J.E.; Maric, I.; Porteiro, B.; Häring, M.; Kooijman, S.; Musovic, S.; Eerola, K.; López-Ferreras, L.; Peris, E.; et al. Parabrachial Interleukin-6 Reduces Body Weight and Food Intake and Increases Thermogenesis to Regulate Energy Metabolism. Cell Rep. 2019, 26, 3011–3026. [Google Scholar] [CrossRef]
- Milton-Laskibar, I.; Aguirre, L.; Etxeberria, U.; Milagro, F.; Martínez, J.; Portillo, M. Do the Effects of Resveratrol on Thermogenic and Oxidative Capacities in IBAT and Skeletal Muscle Depend on Feeding Conditions? Nutrients 2018, 10, 1446. [Google Scholar] [CrossRef]
- Andrade, J.M.O.; Barcala-Jorge, A.S.; Batista-Jorge, G.C.; Paraíso, A.F.; Freitas, K.M.; de Farias Lelis, D.; Guimarães, A.L.S.; de Paula, A.M.B.; Santos, S.H.S. Effect of Resveratrol on Expression of Genes Involved Thermogenesis in Mice and Humans. Biomed. Pharmacother. 2019, 112, 108634. [Google Scholar] [CrossRef]
- Meldrum, D.R.; Morris, M.A.; Gambone, J.C. Obesity Pandemic: Causes, Consequences, and Solutions—but Do We Have the Will? Fertil. Steril. 2017, 107, 833–839. [Google Scholar] [CrossRef]
- Unamuno, X.; Gómez-Ambrosi, J.; Rodríguez, A.; Becerril, S.; Frühbeck, G.; Catalán, V. Adipokine Dysregulation and Adipose Tissue Inflammation in Human Obesity. Eur. J. Clin. Investig. 2018, 48, e12997. [Google Scholar] [CrossRef]
- Villarroya, F.; Cereijo, R.; Gavaldà-Navarro, A.; Villarroya, J.; Giralt, M. Inflammation of Brown/Beige Adipose Tissues in Obesity and Metabolic Disease. J. Intern. Med. 2018, 284, 492–504. [Google Scholar] [CrossRef]
- Torres, S.; Fabersani, E.; Marquez, A.; Gauffin-Cano, P. Adipose Tissue Inflammation and Metabolic Syndrome. The Proactive Role of Probiotics. Eur. J. Nutr. 2019, 58, 27–43. [Google Scholar] [CrossRef]
- Stefania, D.S.; Clodoveo, M.L.; Cariello, M.; D’Amato, G.; Franchini, C.; Faienza, M.F.; Corbo, F. Polyphenols and Obesity Prevention: Critical Insights on Molecular Regulation, Bioavailability and Dose in Preclinical and Clinical Settings. Crit. Rev. Food Sci. Nutr. 2020, 1–23. [Google Scholar] [CrossRef]
- Lee, Y.-M.; Yoon, Y.; Yoon, H.; Park, H.-M.; Song, S.; Yeum, K.-J. Dietary Anthocyanins against Obesity and Inflammation. Nutrients 2017, 9, 1089. [Google Scholar] [CrossRef]
- Peña-Romero, A.C.; Navas-Carrillo, D.; Marín, F.; Orenes-Piñero, E. The Future of Nutrition: Nutrigenomics and Nutrigenetics in Obesity and Cardiovascular Diseases. Crit. Rev. Food Sci. Nutr. 2018, 58, 3030–3041. [Google Scholar] [CrossRef]
- Barrea, L.; Annunziata, G.; Bordoni, L.; Muscogiuri, G.; Colao, A.; Savastano, S. Nutrigenetics—Personalized Nutrition in Obesity and Cardiovascular Diseases. Int. J. Obes. Suppl. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Rizzi, F.; Conti, C.; Dogliotti, E.; Terranegra, A.; Salvi, E.; Braga, D.; Ricca, F.; Lupoli, S.; Mingione, A.; Pivari, F.; et al. Interaction between Polyphenols Intake and PON1 Gene Variants on Markers of Cardiovascular Disease: A Nutrigenetic Observational Study. J. Transl. Med. 2016, 14, 186. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Mateos, A.; Istas, G.; Boschek, L.; Feliciano, R.P.; Mills, C.E.; Boby, C.; Gomez-Alonso, S.; Milenkovic, D.; Heiss, C. Circulating Anthocyanin Metabolites Mediate Vascular Benefits of Blueberries: Insights From Randomized Controlled Trials, Metabolomics, and Nutrigenomics. J. Gerontol. Ser. A 2019, 74, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qi, J.; Li, L. Phytochemicals as Potential Candidates to Combat Obesity via Adipose Non-Shivering Thermogenesis. Pharmacol. Res. 2019, 147, 104393. [Google Scholar] [CrossRef]
- Horvath, C.; Wolfrum, C. Feeding Brown Fat: Dietary Phytochemicals Targeting Non-Shivering Thermogenesis to Control Body Weight. Proc. Nutr. Soc. 2020, 79, 338–356. [Google Scholar] [CrossRef]
- Pedersen, B.K.; Febbraio, M.A. Muscles, Exercise and Obesity: Skeletal Muscle as a Secretory Organ. Nat. Rev. Endocrinol. 2012, 8, 457–465. [Google Scholar] [CrossRef]
- Barbagallo, I.; Vanella, L.; Cambria, M.T.; Tibullo, D.; Godos, J.; Guarnaccia, L.; Zappalà, A.; Galvano, F.; Li Volti, G. Silibinin Regulates Lipid Metabolism and Differentiation in Functional Human Adipocytes. Front. Pharmacol. 2016, 6. [Google Scholar] [CrossRef]
- Suh, H.J.; Cho, S.Y.; Kim, E.Y.; Choi, H.-S. Blockade of Lipid Accumulation by Silibinin in Adipocytes and Zebrafish. Chem. Biol. Interact. 2015, 227, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, S.; Wang, Y.; Zhu, T. Silymarin Improved Diet-Induced Liver Damage and Insulin Resistance by Decreasing Inflammation in Mice. Pharm. Biol. 2016, 54, 2995–3000. [Google Scholar] [CrossRef]
- Alsaggar, M.; Bdour, S.; Ababneh, Q.; El-Elimat, T.; Qinna, N.; Alzoubi, K.H. Silibinin Attenuates Adipose Tissue Inflammation and Reverses Obesity and Its Complications in Diet-Induced Obesity Model in Mice. BMC Pharmacol. Toxicol. 2020, 21, 8. [Google Scholar] [CrossRef]
- Liu, G.; Grifman, M.; Macdonald, J.; Moller, P.; Wong-Staal, F.; Li, Q.-X. Isoginkgetin Enhances Adiponectin Secretion from Differentiated Adiposarcoma Cells via a Novel Pathway Involving AMP-Activated Protein Kinase. J. Endocrinol. 2007, 194, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Bu, S.; Yuan, C.Y.; Xue, Q.; Chen, Y.; Cao, F. Bilobalide Suppresses Adipogenesis in 3T3-L1 Adipocytes via the AMPK Signaling Pathway. Molecules 2019, 24, 3503. [Google Scholar] [CrossRef]
- Xia, B.; Shi, X.C.; Xie, B.C.; Zhu, M.Q.; Chen, Y.; Chu, X.Y.; Cai, G.H.; Liu, M.; Yang, S.Z.; Mitchell, G.A.; et al. Urolithin A Exerts Antiobesity Effects through Enhancing Adipose Tissue Thermogenesis in Mice. PLoS Biol. 2020, 18, e3000688. [Google Scholar] [CrossRef]
- Cao, K.; Xu, J.; Pu, W.; Dong, Z.; Sun, L.; Zang, W.; Gao, F.; Zhang, Y.; Feng, Z.; Liu, J. Punicalagin, an Active Component in Pomegranate, Ameliorates Cardiac Mitochondrial Impairment in Obese Rats via AMPK Activation. Sci. Rep. 2015, 5, 14014. [Google Scholar] [CrossRef]
- Wang, C.; Liu, W.; Nie, Y.; Qaher, M.; Horton, H.E.; Yue, F.; Asakura, A.; Kuang, S. Loss of MyoD Promotes Fate Transdifferentiation of Myoblasts Into Brown Adipocytes. EBioMedicine 2017, 16, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Palacios-González, B.; Zarain-Herzberg, A.; Flores-Galicia, I.; Noriega, L.G.; Alemán-Escondrillas, G.; Zariñan, T.; Ulloa-Aguirre, A.; Torres, N.; Tovar, A.R. Genistein Stimulates Fatty Acid Oxidation in a Leptin Receptor-Independent Manner through the JAK2-Mediated Phosphorylation and Activation of AMPK in Skeletal Muscle. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2014, 1841, 132–140. [Google Scholar] [CrossRef]
- Aziz, S.A.; Wakeling, L.A.; Miwa, S.; Alberdi, G.; Hesketh, J.E.; Ford, D. Metabolic Programming of a Beige Adipocyte Phenotype by Genistein. Mol. Nutr. Food Res. 2017, 61, 1600574. [Google Scholar] [CrossRef]
- Cederroth, C.R.; Vinciguerra, M.; Gjinovci, A.; Kuhne, F.; Klein, M.; Cederroth, M.; Caille, D.; Suter, M.; Neumann, D.; James, R.W.; et al. Dietary Phytoestrogens Activate AMP-Activated Protein Kinase With Improvement in Lipid and Glucose Metabolism. Diabetes 2008, 57, 1176–1185. [Google Scholar] [CrossRef]
- Yoshino, M.; Naka, A.; Sakamoto, Y.; Shibasaki, A.; Toh, M.; Tsukamoto, S.; Kondo, K.; Iida, K. Dietary Isoflavone Daidzein Promotes Tfam Expression That Increases Mitochondrial Biogenesis in C2C12 Muscle Cells. J. Nutr. Biochem. 2015, 26, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Calderon-Dominguez, M.; Alcalá, M.; Sebastián, D.; Zorzano, A.; Viana, M.; Serra, D.; Herrero, L. Brown Adipose Tissue Bioenergetics: A New Methodological Approach. Adv. Sci. 2017, 4, 1600274. [Google Scholar] [CrossRef] [PubMed]



| Terpenes | Saponins | Aesculus hippocastanum L. (Horse chesnut) | Seed | Ethanol: water (70:30 V/V) | Aescin | 10, 30, 90 | >100 |
| Panax ginseng C.A.Mey (Ginseng) | Root | Ethanol: water (50:50 V/V) | Ginsenoside | 10, 30, 90 | >100 | ||
| Diterpenes | Rosmarinus officinalis L. (Rosemary) | Leaves | Ethanol | Carnosic acid | 5, 10, 15 | ≈18 | |
| Organic acids | Garcinia Cambogia L. (Garcinia) | Fruit | Water | Hydroxycitric acid | 10, 30, 90 | >100 | |
| Polyphenols | Phenolic acids | Cynara scolymus L. (Artichoke) | Leaves | Ethanol: water (60:40 V/V) | Hydroxycinnamic derivatives | 10, 30, 90 | >100 |
| Coffea Arabica L. (Green coffee) | Fruit | Ethanol: water (60:40 V/V) | Hydroxycinnamic derivatives | 10, 30, 90 | >100 | ||
| Punica granatum L. (Pomegranate) | Fruit | Ethanol: water (70:30 V/V) | Punicalagin | 10, 30, 90 | >100 | ||
| Olea europaea L. (Olive) | Fruit | Water | Hydroxytyrosol | 10, 30, 50 | ≈60 | ||
| Olea europaea L. (Olive) | Leaves | Ethanol: water (70:30 V/V) | Oleuropein | 10, 30, 90 | >100 | ||
| Rosmarinus officinalis L. (Rosemary) | Leaves | Ethanol: water (50:50 V/V) | Rosmarinic acid | 10, 30, 90 | >100 | ||
| Silbenes | Aloysia citrodora L. (Lemon verbena) | Leaves | Ethanol: water (60:40 V/V) | Verbascoside | 10, 30, 90 | >100 | |
| Vitis Vinifera L. (Grape) | Root | Ethanol: water (50:50 V/V) | Resveratrol | 5, 10, 20 | ≈20 | ||
| Diarylheptanoids | Curcuma longa L. (Tumeric) | Root | Ethanol | Curcuminoids | 2, 5, 10 | ≈8 | |
| Flavonoids | Ginkgo biloba L. (Ginkgo) | Leaves | Ethanol: water (70:30 V/V) | Flavonoid glycosides | 10, 30, 90 | >100 | |
| Glycine max. (L.) Merr (Soy) | Seed | Ethanol: water (50:50 V/V) | Isoflavone | 10, 30, 90 | >100 | ||
| Vitis Vinifera L. (Grape) | Fruit | Water | Anthocyanins | 10, 30, 90 | >100 | ||
| Camellia sinensis L. (Green tea) | Leaves | Ethanol: water (60:40 V/V) | Flavan-3-ols | 10, 30, 90 | >100 | ||
| Vitis Vinifera L. (Grape) | Seed | Water | Proanthocyanins | 10, 30, 70 | ≈75 | ||
| Citrus sp. (Orange) | Fruit | Ethanol: water (50:50 V/V) | Flavonoid glycosides (Hesperidin) | 10, 30, 90 | >100 | ||
| Silybum marianum L. Gaertn. (Milk thistle) | Seed | Ethanol: water (70:30 V/V) | Silymarin | 10, 30, 90 | >100 |
| Metabolic Process | Genes Implicated |
|---|---|
| Energetic metabolism, thermogenesis, and obesity | UCP2, HIF1A, PGC1a, FTO, MC4R, KCTD15, ETV5, NPY, |
| Xenobiotic metabolism | AHR |
| Lipid metabolism and adipogenesis | LEPR, LPL, PPARG, PPARA, PGC1a, AHR, LIPC, FTO, CD36 |
| Cholesterol and lipoprotein homeostasis | LPL, PPARA, ABCA1, APOA5 |
| Glucose homeostasis and diabetes | TCF7L2, PGC1a, TMEM18, GNPDA2, GCKR |
| Blood pressure | AGT, ACE |
| Mitochondrial biogenesis | PPARG, PGC1a |
| Metabolic response to exercise and muscle capacity | MTHFR, PPARG, HFE, HIF1A, PGC1a |
| Aerobic capacity and sport performance | ADRB1, ACE, HIF1A |
| Structural function | COL5A1, |
| Biorhythm | CLOCK |
| Appetite | NPY, LEPR |
| Immune response | IL4R, PPARA |
| Senescence and aging-related diseases | MTHFR, ABCA1, BNC2, COL51A, DLGAP1, CD36, |
| Cognitive function | DLGAP1, COMT, ETV5, NPY |
| Food-related intolerances (gluten, lactose, fructose) | HLA-DQA1, MCM6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reguero, M.; Gómez de Cedrón, M.; Reglero, G.; Quintela, J.C.; Ramírez de Molina, A. Natural Extracts to Augment Energy Expenditure as a Complementary Approach to Tackle Obesity and Associated Metabolic Alterations. Biomolecules 2021, 11, 412. https://doi.org/10.3390/biom11030412
Reguero M, Gómez de Cedrón M, Reglero G, Quintela JC, Ramírez de Molina A. Natural Extracts to Augment Energy Expenditure as a Complementary Approach to Tackle Obesity and Associated Metabolic Alterations. Biomolecules. 2021; 11(3):412. https://doi.org/10.3390/biom11030412
Chicago/Turabian StyleReguero, Marina, Marta Gómez de Cedrón, Guillermo Reglero, José Carlos Quintela, and Ana Ramírez de Molina. 2021. "Natural Extracts to Augment Energy Expenditure as a Complementary Approach to Tackle Obesity and Associated Metabolic Alterations" Biomolecules 11, no. 3: 412. https://doi.org/10.3390/biom11030412
APA StyleReguero, M., Gómez de Cedrón, M., Reglero, G., Quintela, J. C., & Ramírez de Molina, A. (2021). Natural Extracts to Augment Energy Expenditure as a Complementary Approach to Tackle Obesity and Associated Metabolic Alterations. Biomolecules, 11(3), 412. https://doi.org/10.3390/biom11030412






