Analysis of the Dynamic Proteasome Structure by Cross-Linking Mass Spectrometry
Abstract
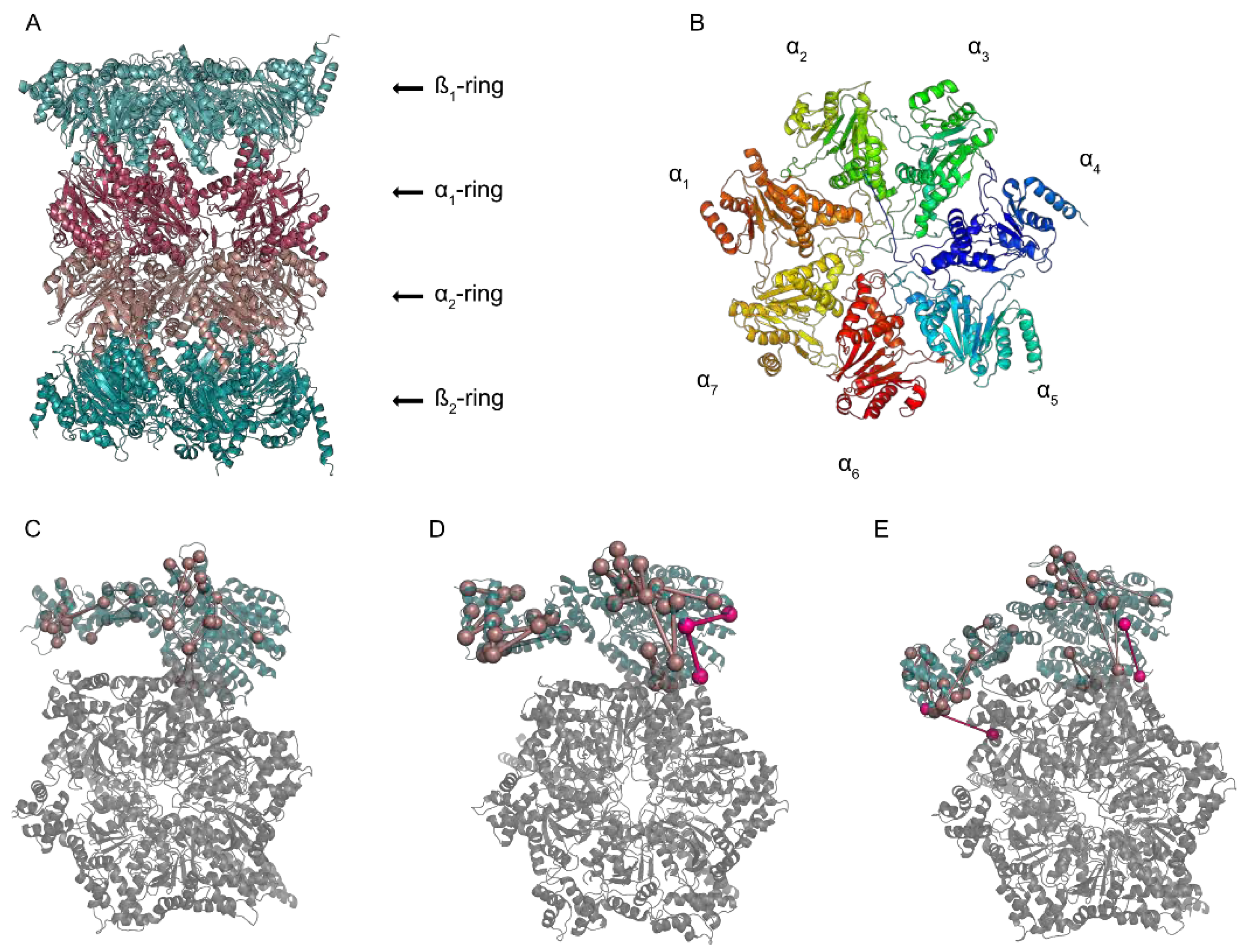
1. A Macromolecular Degradation Machine
2. The Pieces of the Puzzle
3. Solving the Structure of the Proteasome
4. The New Runner in the Race for Dynamic Structures—Cross-Linking Mass Spectrometry
5. Where Are We Heading?
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sahu, I.; Glickman, M.H. Structural Insights into Substrate Recognition and Processing by the 20S Proteasome. Biomolecules 2021, 11, 148. [Google Scholar] [CrossRef] [PubMed]
- Cascio, P. PA28γ: New Insights on an Ancient Proteasome Activator. Biomolecules 2021, 11, 228. [Google Scholar] [CrossRef]
- Aladdin, A.; Yao, Y.; Yang, C.; Kahlert, G.; Ghani, M.; Király, N.; Boratkó, A.; Uray, K.; Dittmar, G.; Tar, K. The Proteasome Activators Blm10/PA200 Enhance the Proteasomal Degradation of N-Terminal Huntingtin. Biomolecules 2020, 10, 1581. [Google Scholar] [CrossRef]
- Raule, M.; Cerruti, F.; Benaroudj, N.; Migotti, R.; Kikuchi, J.; Bachi, A.; Navon, A.; Dittmar, G.; Cascio, P. PA28αβ reduces size and increases hydrophilicity of 20S immunoproteasome peptide products. Chem. Biol. 2014, 21, 470–480. [Google Scholar] [CrossRef]
- Raule, M.; Cerruti, F.; Cascio, P. Enhanced rate of degradation of basic proteins by 26S immunoproteasomes. Biochim. Biophys. Acta 2014, 1843, 1942–1947. [Google Scholar] [CrossRef]
- Mendes, M.L.; Fischer, L.; Chen, Z.A.; Barbon, M.; O’Reilly, F.J.; Giese, S.H.; Bohlke-Schneider, M.; Belsom, A.; Dau, T.; Combe, C.W.; et al. An integrated workflow for crosslinking mass spectrometry. Mol. Syst. Biol. 2019, 15, e8994. [Google Scholar] [CrossRef]
- Löwe, J.; Stock, D.; Jap, B.; Zwickl, P.; Baumeister, W.; Huber, R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 1995, 268, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Ditzel, L.; Löwe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 1997, 386, 463–471. [Google Scholar] [CrossRef]
- Glickman, M.H.; Rubin, D.M.; Coux, O.; Wefes, I.; Pfeifer, G.; Cjeka, Z.; Baumeister, W.; Fried, V.A.; Finley, D. A Subcomplex of the Proteasome Regulatory Particle Required for Ubiquitin-Conjugate Degradation and Related to the COP9-Signalosome and eIF3. Cell 1998, 94, 615–623. [Google Scholar] [CrossRef]
- Mao, Y. Structure, Dynamics and Function of the 26S Proteasome. Subcell. Biochem. 2021, 96, 1–151. [Google Scholar]
- Deveraux, Q.; Ustrell, V.; Pickart, C.; Rechsteiner, M. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 1994, 269, 7059–7061. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, X.; Elsasser, S.; Stocks, B.B.; Tian, G.; Lee, B.-H.; Shi, Y.; Zhang, N.; de Poot, S.A.H.; Tuebing, F.; et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 2016, 351, aad9421. [Google Scholar] [CrossRef]
- Husnjak, K.; Elsasser, S.; Zhang, N.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef]
- Verma, R.; Aravind, L.; Oania, R.; McDonald, W.H.; Yates, J.R., 3rd; Koonin, E.V.; Deshaies, R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 2002, 298, 611–615. [Google Scholar] [CrossRef]
- Yao, T.; Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 2002, 419, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Braun, B.C.; Glickman, M.; Kraft, R.; Dahlmann, B.; Kloetzel, P.M.; Finley, D.; Schmidt, M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol. 1999, 1, 221–226. [Google Scholar] [CrossRef]
- Bard, J.A.M.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef] [PubMed]
- Elsasser, S.; Chandler-Militello, D.; Müller, B.; Hanna, J.; Finley, D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 2004, 279, 26817–26822. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Chen, T.; Ziv, I.; Rosenzweig, R.; Matiuhin, Y.; Bronner, V.; Glickman, M.H.; Fushman, D. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol. Cell 2009, 36, 1018–1033. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-H.; Lu, Y.; Prado, M.A.; Shi, Y.; Tian, G.; Sun, S.; Elsasser, S.; Gygi, S.P.; King, R.W.; Finley, D. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature 2016, 532, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Vander Linden, R.T.; Hemmis, C.W.; Schmitt, B.; Ndoja, A.; Whitby, F.G.; Robinson, H.; Cohen, R.E.; Yao, T.; Hill, C.P. Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Mol. Cell 2015, 57, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, M.G.; Morais, M.C.; Leiman, P.G.; Zhang, W. Combining X-ray crystallography and electron microscopy. Structure 2005, 13, 355–362. [Google Scholar] [CrossRef]
- Whitby, F.G.; Masters, E.I.; Kramer, L.; Knowlton, J.R.; Yao, Y.; Wang, C.C.; Hill, C.P. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature 2000, 408, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Harshbarger, W.; Miller, C.; Diedrich, C.; Sacchettini, J. Crystal structure of the human 20S proteasome in complex with carfilzomib. Structure 2015, 23, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Schrader, J.; Henneberg, F.; Mata, R.A.; Tittmann, K.; Schneider, T.R.; Stark, H.; Bourenkov, G.; Chari, A. The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design. Science 2016, 353, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Van Drie, J.H.; Tong, L. Cryo-EM as a powerful tool for drug discovery. Bioorg. Med. Chem. Lett. 2020, 30, 127524. [Google Scholar] [CrossRef]
- Bohn, S.; Beck, F.; Sakata, E.; Walzthoeni, T.; Beck, M.; Aebersold, R.; Förster, F.; Baumeister, W.; Nickell, S. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc. Natl. Acad. Sci. USA 2010, 107, 20992–20997. [Google Scholar] [CrossRef] [PubMed]
- Nickell, S.; Beck, F.; Scheres, S.H.W.; Korinek, A.; Förster, F.; Lasker, K.; Mihalache, O.; Sun, N.; Nagy, I.; Sali, A.; et al. Insights into the molecular architecture of the 26S proteasome. Proc. Natl. Acad. Sci. USA 2009, 106, 11943–11947. [Google Scholar] [CrossRef]
- Unverdorben, P.; Beck, F.; Śledź, P.; Schweitzer, A.; Pfeifer, G.; Plitzko, J.M.; Baumeister, W.; Förster, F. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc. Natl. Acad. Sci. USA 2014, 111, 5544–5549. [Google Scholar] [CrossRef]
- Chen, S.; Wu, J.; Lu, Y.; Ma, Y.-B.; Lee, B.-H.; Yu, Z.; Ouyang, Q.; Finley, D.J.; Kirschner, M.W.; Mao, Y. Structural basis for dynamic regulation of the human 26S proteasome. Proc. Natl. Acad. Sci. USA 2016, 113, 12991–12996. [Google Scholar] [CrossRef]
- Eisele, M.R.; Reed, R.G.; Rudack, T.; Schweitzer, A.; Beck, F.; Nagy, I.; Pfeifer, G.; Plitzko, J.M.; Baumeister, W.; Tomko, R.J., Jr.; et al. Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating. Cell Rep. 2018, 24, 1301–1315.e5. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, S.; Wu, Z.; Li, X.; Wang, W.L.; Zhu, Y.; Stoilova-McPhie, S.; Lu, Y.; Finley, D.; Mao, Y. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 2019, 565, 49–55. [Google Scholar] [CrossRef]
- Farmer, T.B.; Caprioli, R.M. Assessing the multimeric states of proteins: Studies using laser desorption mass spectrometry. Biol. Mass Spectrom. 1991, 20, 796–800. [Google Scholar] [CrossRef] [PubMed]
- Haniu, M.; Narhi, L.O.; Arakawa, T.; Elliott, S.; Rohde, M.F. Recombinant human erythropoietin (rHuEPO): Cross-linking with disuccinimidyl esters and identification of the interfacing domains in EPO. Protein Sci. 1993, 2, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- Leitner, A.; Faini, M.; Stengel, F.; Aebersold, R. Crosslinking and Mass Spectrometry: An Integrated Technology to Understand the Structure and Function of Molecular Machines. Trends Biochem. Sci. 2016, 41, 20–32. [Google Scholar] [CrossRef]
- O’Reilly, F.J.; Rappsilber, J. Cross-linking mass spectrometry: Methods and applications in structural, molecular and systems biology. Nat. Struct. Mol. Biol. 2018, 25, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Sinz, A. Cross-Linking/Mass Spectrometry for Studying Protein Structures and Protein-Protein Interactions: Where Are We Now and Where Should We Go from Here? Angew. Chem. Int. Ed. Engl. 2018, 57, 6390–6396. [Google Scholar] [CrossRef]
- Belsom, A.; Rappsilber, J. Anatomy of a crosslinker. Curr. Opin. Chem. Biol. 2021, 60, 39–46. [Google Scholar] [CrossRef]
- Hofmann, T.; Fischer, A.W.; Meiler, J.; Kalkhof, S. Protein structure prediction guided by crosslinking restraints—A systematic evaluation of the impact of the crosslinking spacer length. Methods 2015, 89, 79–90. [Google Scholar] [CrossRef]
- Belsom, A.; Schneider, M.; Fischer, L.; Brock, O.; Rappsilber, J. Serum Albumin Domain Structures in Human Blood Serum by Mass Spectrometry and Computational Biology. Mol. Cell. Proteom. 2016, 15, 1105–1116. [Google Scholar] [CrossRef]
- Trnka, M.J.; Baker, P.R.; Robinson, P.J.J.; Burlingame, A.L.; Chalkley, R.J. Matching cross-linked peptide spectra: Only as good as the worse identification. Mol. Cell. Proteom. 2014, 13, 420–434. [Google Scholar] [CrossRef] [PubMed]
- Sinz, A. Divide and conquer: Cleavable cross-linkers to study protein conformation and protein-protein interactions. Anal. Bioanal. Chem. 2017, 409, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Chavez, J.D.; Weisbrod, C.R.; Zheng, C.; Eng, J.K.; Bruce, J.E. Protein interactions, post-translational modifications and topologies in human cells. Mol. Cell. Proteom. 2013, 12, 1451–1467. [Google Scholar] [CrossRef]
- Kaake, R.M.; Wang, X.; Burke, A.; Yu, C.; Kandur, W.; Yang, Y.; Novtisky, E.J.; Second, T.; Duan, J.; Kao, A.; et al. A new in vivo cross-linking mass spectrometry platform to define protein-protein interactions in living cells. Mol. Cell. Proteom. 2014, 13, 3533–3543. [Google Scholar] [CrossRef]
- Steigenberger, B.; Pieters, R.J.; Heck, A.J.R.; Scheltema, R.A. PhoX: An IMAC-Enrichable Cross-Linking Reagent. ACS Cent. Sci. 2019, 5, 1514–1522. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Zhu, W.; Wu, Y.; Chen, J.; Yu, J.; Jiang, B.; Chen, H.; Chen, W. A novel mass spectrometry-cleavable, phosphate-based enrichable and multi-targeting protein cross-linker. Chem. Sci. 2019, 10, 6443–6447. [Google Scholar] [CrossRef] [PubMed]
- Leitner, A.; Reischl, R.; Walzthoeni, T.; Herzog, F.; Bohn, S.; Förster, F.; Aebersold, R. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteom. 2012, 11, M111.014126. [Google Scholar] [CrossRef]
- Yang, B.; Wu, Y.-J.; Zhu, M.; Fan, S.-B.; Lin, J.; Zhang, K.; Li, S.; Chi, H.; Li, Y.-X.; Chen, H.-F.; et al. Identification of cross-linked peptides from complex samples. Nat. Methods 2012, 9, 904–906. [Google Scholar] [CrossRef]
- Hoopmann, M.R.; Zelter, A.; Johnson, R.S.; Riffle, M.; MacCoss, M.J.; Davis, T.N.; Moritz, R.L. Kojak: Efficient analysis of chemically cross-linked protein complexes. J. Proteome Res. 2015, 14, 2190–2198. [Google Scholar] [CrossRef]
- Liu, F.; Rijkers, D.T.S.; Post, H.; Heck, A.J.R. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods 2015, 12, 1179–1184. [Google Scholar] [CrossRef]
- Götze, M.; Pettelkau, J.; Schaks, S.; Bosse, K.; Ihling, C.H.; Krauth, F.; Fritzsche, R.; Kühn, U.; Sinz, A. StavroX--a software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectrom. 2012, 23, 76–87. [Google Scholar] [CrossRef]
- Yu, F.; Li, N.; Yu, W. Exhaustively Identifying Cross-Linked Peptides with a Linear Computational Complexity. J. Proteome Res. 2017, 16, 3942–3952. [Google Scholar] [CrossRef] [PubMed]
- Tran, B.Q.; Goodlett, D.R.; Goo, Y.A. Advances in protein complex analysis by chemical cross-linking coupled with mass spectrometry (CXMS) and bioinformatics. Biochim. Biophys. Acta 2016, 1864, 123–129. [Google Scholar] [CrossRef]
- Hartmann-Petersen, R.; Tanaka, K.; Hendil, K.B. Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch. Biochem. Biophys. 2001, 386, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Tomko, R.J., Jr.; Funakoshi, M.; Schneider, K.; Wang, J.; Hochstrasser, M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: Implications for proteasome structure and assembly. Mol. Cell 2010, 38, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Förster, F.; Lasker, K.; Beck, F.; Nickell, S.; Sali, A.; Baumeister, W. An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem. Biophys. Res. Commun. 2009, 388, 228–233. [Google Scholar] [CrossRef]
- Lasker, K.; Förster, F.; Bohn, S.; Walzthoeni, T.; Villa, E.; Unverdorben, P.; Beck, F.; Aebersold, R.; Sali, A.; Baumeister, W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. USA 2012, 109, 1380–1387. [Google Scholar] [CrossRef] [PubMed]
- Tomko, R.J., Jr.; Taylor, D.W.; Chen, Z.A.; Wang, H.-W.; Rappsilber, J.; Hochstrasser, M. A Single α Helix Drives Extensive Remodeling of the Proteasome Lid and Completion of Regulatory Particle Assembly. Cell 2015, 163, 432–444. [Google Scholar] [CrossRef]
- Wang, X.; Cimermancic, P.; Yu, C.; Schweitzer, A.; Chopra, N.; Engel, J.L.; Greenberg, C.H.; Huszagh, A.S.; Beck, F.; Sakata, E.; et al. Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome. Mol. Cell. Proteom. 2017, 16, 840–854. [Google Scholar] [CrossRef]
- Yu, C.; Wang, X.; Huszagh, A.S.; Viner, R.; Novitsky, E.; Rychnovsky, S.D.; Huang, L. Probing H2O2-mediated Structural Dynamics of the Human 26S Proteasome Using Quantitative Cross-linking Mass Spectrometry (QXL-MS). Mol. Cell. Proteom. 2019, 18, 954–967. [Google Scholar] [CrossRef]
- Götze, M.; Iacobucci, C.; Ihling, C.H.; Sinz, A. A Simple Cross-Linking/Mass Spectrometry Workflow for Studying System-wide Protein Interactions. Anal. Chem. 2019, 91, 10236–10244. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, F.J.; Xue, L.; Graziadei, A.; Sinn, L.; Lenz, S.; Tegunov, D.; Blötz, C.; Singh, N.; Hagen, W.J.H.; Cramer, P.; et al. In-cell architecture of an actively transcribing-translating expressome. Science 2020, 369, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Schnirch, L.; Nadler-Holly, M.; Siao, S.-W.; Frese, C.K.; Viner, R.; Liu, F. Expanding the Depth and Sensitivity of Cross-Link Identification by Differential Ion Mobility Using High-Field Asymmetric Waveform Ion Mobility Spectrometry. Anal. Chem. 2020, 92, 10495–10503. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendes, M.L.; Dittmar, G. Analysis of the Dynamic Proteasome Structure by Cross-Linking Mass Spectrometry. Biomolecules 2021, 11, 505. https://doi.org/10.3390/biom11040505
Mendes ML, Dittmar G. Analysis of the Dynamic Proteasome Structure by Cross-Linking Mass Spectrometry. Biomolecules. 2021; 11(4):505. https://doi.org/10.3390/biom11040505
Chicago/Turabian StyleMendes, Marta L., and Gunnar Dittmar. 2021. "Analysis of the Dynamic Proteasome Structure by Cross-Linking Mass Spectrometry" Biomolecules 11, no. 4: 505. https://doi.org/10.3390/biom11040505
APA StyleMendes, M. L., & Dittmar, G. (2021). Analysis of the Dynamic Proteasome Structure by Cross-Linking Mass Spectrometry. Biomolecules, 11(4), 505. https://doi.org/10.3390/biom11040505






